Abstract
Observational studies revealed a relationship between changes in gastric mucosa and risk of esophageal squamous cell carcinoma (ESCC) which suggested a possible role for gastric microbiota in ESCC carcinogenesis. In this study we aimed to compare pattern of gastric corpus microbiota in ESCC with normal esophagus. Cases were included subjects with early ESCC (stage I–II) and esophageal squamous dysplasia (ESD) as the cancer precursor. Control groups included age and sex-matched subjects with mid-esophagus esophagitis (diseased-control) and histologically normal esophagus (healthy-control). DNA was extracted from snap-frozen gastric corpus tissues and 16S rRNA was sequenced on GS-FLX Titanium. After noise removal, an average of 3004 reads per sample was obtained from 93 subjects. We applied principal coordinate analysis to ordinate distances from beta diversity data. Pattern of gastric microbiota using Unifrac (p = 0.004) and weighted Unifrac distances (p = 0.018) statistically varied between cases and healthy controls. Sequences were aligned to SILVA database and Clostridiales and Erysipelotrichales orders were more abundant among cases after controling for multiple testing (p = 0.011). No such difference was observed between mid-esophagitis and healthy controls. This study is the first to show that composition of gastric corpus mucosal microbiota differs in early ESCC and ESD from healthy esophagus.
Similar content being viewed by others
Introduction
Cancer of the esophagus affects more than 450,000 people each year, of which 90% are squamous cell carcinomas (ESCC)1. Highest incidence rates have been reported from the “esophageal cancer belt”, an area that stretches from northern China to northern Iran2. The relationship between gastric environment and ESCC has been evaluated through observational studies3,4. As a link for this impact, atrophic gastritis has been shown to be associated with ESCC risk5, although no dose-response relation with severity of atrophy has been reported6. Human stomach was considered an inhospitable environment for bacteria until the recognition of H. pylori7 and most of research was focused on the relation between H. pylori and ESCC5,8,9,10,11,12,13,14 with showing little evidence of risk. Gastric acidity was believed to be a barrier against colonizing most of bacteria in the stomach, although 16S rRNA sequencing of gastric mucosa reveals a diverse bacterial community15. More than one hundred phylotypes have been detected in stomach of which 50% were from uncultivated bacteria15. Furthermore sequencing-based methods showed lower bacterial diversity associated with higher gastric pH16 even in the absence of atrophic gastritis. Even in H. pylori negative stomach high abundance of Streptococcus and Prevotella17 were observed, among them certain Streptococcus species are resistant to low pH18. Although risk of ESCC in relation with individual microorganisms has been tested, to date the association between gastric microbiota and ESCC has not been investigated.
Golestan province in Northern Iran is located in “esophageal cancer belt”19,20. We aimed to investigate associations between gastric mucosal microbiota and ESCC in this population. In this study in addition to early-stage ESCC cases and controls with healthy esophagus, we included mid-esophagitis as a diseased control group with inflammatory lesion in the esophagus and squamous dysplasia as the only known precursor of ESCC to compare the pattern of gastric microbiota.
Results
Table 1 summarizes characteristics of the subjects and the reads. There was no significant differences between cases and controls except for the illiteracy rate which diseased-controls were less illiterate than cases and healthy controls (p = 0.001). Tobacco and alcohol consumption are not major risk factors in study area and there was no difference in proportion of exposed cases and healthy controls in our study samples. Table 2 summarizes major histopathologic findings in gastric biopsy samples. There was no difference in proportion of nonatrophic gastritis and intestinal metaplasia between cases and healthy controls. No evidence of gastric corpus atrophy was observed among study subjects.
Figure 1 depicts sequence data processing steps. Briefly a total of 369,539 sequences with mean length of 419 nt from 93 tissue samples were evaluated (3004 average sequences per sample). A total of 25% of sequences was removed as noise or chimera. Two samples had less than 1000 reads (504 and 710 reads) and were excluded. Clusters of unique sequences at 3% of dissimilarity rate formed 2075 operational taxonomic units (OTUs). Of these OTUs, 1283 were assigned to bacterial taxa based on the SILVA database. The majority of unclassified OTUs (80%) appeared only once (n = 517) or twice (n = 123). The mean percentage of unknown OTUs per sample was 0.5%.
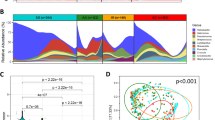
OTUs were assigned to 31 Phyla, 53 Classes, 90 Orders, 168 Families and 336 Genera. Five most abundant phyla were Firmicutes, Bacteroidetes Proteobacteria, Actinobacteria and Fusobacteria. Phyla composition was consistent across ESCC, ESD, healthy esophagus and mid-esophagitis groups. Table 3 summarizes the percentage of OTUs assigned to Order level among study cases and controls. Testing for differences in OTU abundance revealed higher abundance of Clostridiales (FDR = 0.011) and Erysipelotrichales Orders (FDR = 0.011) in the case group compared to the healthy esophagus group (Table 3). Helicobacteriacea composed nearly 43% of total reads amongst reads and all subjects except 3 cases and 2 controls had non-zero reads. Grouping abundance of Helicobacteriacea, no distinct cluster among cases was formed in relation to the quartile of reads. Alpha diversity measured by Chao1 was not significantly different among study groups though Helicobacteraceae abundance among cases (mean = 1059) was lower than healthy controls (mean = 1449) (p = 0.03) and diseased-controls (mean = 1715) (p = 0.02).
Unifrac and weighted Unifrac distances were ordinated by applying PCoA. Figure 2, depicted the percentage of variance coverage by Unifrac coordinates. Conditional logistic regression model was used to compare coordinates between cases and controls (Table 4). For Unifrac distance, a best model with three coordinates showed significant difference between cases and healthy esophagus group (p = 0.003) based on 37 matched pairs. Similar difference was found when OTU abundances were counted in the weighted Unifrac distances (p = 0.018). Removing ESCC cases and restricting the analysis to esophageal squamous dysplasia, did not change the results (based on 17 matched pairs). There was no individual taxonomic difference between the diseased-controls and healthy controls. We did not observe a significant difference in the coordinates based on Unifrac distances between diseased-control and healthy-control groups.
Discussion
We have observed significant differences in microbiota composition of gastric fundal mucosa in subjects with early ESCC and ESD compared to those with a normal esophagus.
So far no single or combination of environmental or genetic risk factors has been identified to explain the high incidence of ESCC in Asian Cancer belt. Although alcohol and tobacco consumption account for major proportion of the disease in low risk area, their contribution to the risk of ESCC in high incidence regions of Asian Cancer Belt – from where 90% of cases arise- is limited. One suggested possible explanation for this excess risk is intrinsic exposure to carcinogens. Gastric mucosal changes have been shown to be associated with ESCC risk and bacterial alteration in the stomach has been considered to be the possible link. This study is the first to evaluate the relevance of microbial link in this association.
Similar to studies on gastric mucosa microbiota15, the most common phyla in our samples were Proteobacteria, Firmicutes, Bacteroidetes, Actinobacteria and Fusobacteria. Our data suggests that presence and abundance of Clostridiales and Erysipelotrichales Orders were associated with early ESCC and ESD. Both microbial hints belong to Firmicutes phylum. In an animal study, presence of members of Clostridiales altered the pathogenicity of H. pylori by recruitment of CD4 T-cells to the gastric mucosa21 which suggests a possible impact of microbial composition on gastritis outcome. Higher abundance of Clostridiales might enhance more aggressive response to H. pylori and induces severe or pangastritis which may further develop to atrophic gastritis. The link between Clostridiales and the esophagus has been observed in response to proton pump inhibitor use, as abundance of its members increases in the lower esophagus. Such an effect has not been reported in gastric mucosa. Erysipelotrichales is highly dependent on the fat content in the diet of humanized mice model22, associated with periodontitis23 and gut inflammatory response in animal model24. No study has been published on its association with esophageal disorders, but its high abundance in periodontitis microbiota and the observed association of poor oral heath with ESCC risk25 may be one explanation for its role and our finding. Difference between diversity of oral microbiota in ESD patients compared to those with healthy esophagus has been shown in a cross-sectional study26.
The relation between H. pylori and ESCC risk is controversial. Most studies have not shown an excess risk including in our study population27 with more than 80% H. pylori seropositivity27. We observed lower abundance of Helicobacteriacea order among case group which might be secondary to subclinical atrophic changes in the stomach. Our finding on non-distinct microbial clusters in gastric mucosa in relation to different abundances of H. pylori among ESCC is similar to the report in healthy stomach15. Although given that the H. pylori infection is prevalent in catchment area (seropositivity >84% among older >55 years), any inference on relation between H. pylori and gastric microbiota in this study should be done with caution.
In this study, 16S rRNA with coverage of 464 nt of region V3–V4 was used to distinguish bacteria. Although it did not cover whole hyper variable V1–V9 regions, the most divergent regions were covered and it was possible to align with SILVA reference with 90.5% coverage. Furthermore, V4 and V5 are less specific for species level28 and we avoided assigning genus or species nomenclature to the OTUs. Although 16S is a gold standard for bacterial phylogeny, presence of its multiple copies in some bacteria with slight difference in sequence could lead to identifying multiple types of the same bacterium29. As our finding of different microbial composition did not rely only on presence or absence of taxa, possible discriminatory power of 16S for bacterial detection would not make limitation on inferring difference in this study.
Some of gastric microbiota are anaerobes and difficult to cultivate and some of them could not be assigned to reference databases. We tried to reduce this effect through OTU formation. In our data unassigned OTUs were unevenly distributed among samples, as five samples shared most of them. These rare species could be formed due to intrinsic errors of pyrosequencing30 though per-base error rate is comparable or lower for 454 sequencing compared to sanger sequencing31.
Modest sample size is one of the limitations of this study, though based on national cancer registry data less than 9% of ESCC cases are diagnosed at early stages32. In addition squamous dysplasia is a rare phenomenon even in high risk areas for ESCC, as its prevalence among endoscopy subjects is less than 5%33. Due to this we combined early ESCC and ESD as one case group. Though is probable that the low number of study subjects was in part compensated for by the higher depth of sequencing. Previous studies have considered a broad range of reads (from 100 to 10,000) to be sufficient depth to separate the microbiota of healthy and diseased organs34, though higher sequencing depth might amplify the difference in our study. However we did not observe difference in alpha diversity between healthy-controls and cases, we do not think that the depth of sequencing was enough for estimating alpha diversity of microbiota. It has been shown that at least 5000 clean reads are needed for valid estimation of diversity35.
Similar to every PCR-based methods, amplification bias is a concern as some less abundant taxa might be underestimated or ignored. Using endoscopy clinic controls may limit the generalizability of our results to healthy populations, although several biopsies from the esophagus were carefully examined before assigning subjects to healthy control group.
We used mid-esophagus esophagitis as a diseased-control to evaluate the specificity of our findings. Inflammation is a common pathway between neoplasia and esophagitis meanwhile esophagitis does not play a role in ESCC carcinogenesis. Detection of explicitly different microbiota pattern between cases and healthy controls and not with diseased-controls increases the validity of our findings. Number of esophagitis controls was less than healthy controls and 16 out of 17 were male, although no difference in microbiota was observed between male and female, reduced sample size might affect the precision of statistical tests.
In this study – similar to the most other studies- we used one snap frozen biopsy from mid corpus of the gastric greater curvature. In an early study of gastric biota15, it has been shown that phylogenic pattern of mucosal biota did not differ between the corpus and antrum, although corpus mucosal microbiota may not be representative of whole gastric microbiota.
The different patterns of gastric microbiota that we observed might be secondary to cancer. We used early-stage ESCC and patients with ESD (the asymptomatic precursor lesion of ESCC) as cases, to decrease this possibility. As a sensitivity test we excluded Early-stage ESCC and observed different microbiota pattern between healthy controls and ESD subjects (data was not shown), which lowered the possibility that our findings is secondary to cancer. Although it is probable that microbiota pattern changes secondary to dysplasia.
In summary we observed an altered gastric corpus microbiota in patients with early ESCC or ESD, compared to subjects with healthy esophagus, with higher abundance of bacteria in the Clostridiales and Erysipelotrichales Orders. Studies with higher sequencing depth and larger sample sizes are warranted.
Methods
Ethical approval
From all subjects, an informed consent was obtained. The case-control study was approved by the local ethical committee in the Digestive Disease Research Centre of Tehran University of Medical Sciences, Iran (IRB00001641), Institutional Review Board of National Cancer Institute, US and Stockholm Regional Ethics Vetting Board, Sweden (Dnr:2010/471-31/4). The methods were carried out in accordance with the approved guidelines.
Subjects
Details of study design have been reported earlier36. Briefly, subjects were recruited at Atrak clinic, the only specialized clinic for upper gastrointestinal cancer diagnosis and treatment in eastern Golestan, from December 2003 to June 2007. During study period, all physicians in catchment area were contacted and asked to refer patients with clinical suspicion for upper gastrointestinal malignancies to this clinic. For all subjects, upper gastrointestinal endoscopy was performed and at least 9 biopsies from the normal-looking esophagus and stomach were taken. To find squamous dysplasia, esophageal chromoendoscopy with 2% Lugol solution was done and biopsies were taken from unstained mucosa. Unstained lesions were further examined in Digestive Disease Research Center, Tehran University. Those with squamous dysplasia and no evidence of cancer were included as esophageal squamous dysplasia (ESD) subjects. Based on cancer registry data, during the study period it was estimated that about 70% of incident ESCC cases in catchment area were recruited. The majority of ESCC patients (>90%) were diagnosed with clinical Stage III–IV disease at the time of recruitment. To reduce the chance that differences in the gastric microbiota might be secondary to cancer, we included patients who were diagnosed with clinical Stage I–II ESCC plus all patients who were diagnosed with ESD during study in our case group. Two control groups were randomly selected from endoscopy clinic patients who had the same referral pattern as cases, including (1) healthy controls with endoscopically and histologically normal esophagus in all biopsies and (2) diseased controls with histologic esophagitis in mid-esophageal biopsies.
Sample size of original case-control study was calculated based on achieving power of 95% to detect odds ratio of 2 between matched cases and control and total of 300 cases and 600 controls were calculated. For present study, the difference between 2 taxonomic levels of more than 20% or odds ratio of 2.5 would require at least 30 pair of case-control to achieve 90% power. A priori for odds ratio was based on reports from other studies in gastrointestinal malignancies and microbiota37.
Among 579 endoscopy clinic controls and 300 ESCC cases, we found 19 early-stage ESCC and 18 ESD patients. One age/sex-matched control was randomly selected from endoscopy clinic controls with a healthy esophagus for each case. We also found 17 subjects with mid-esophageal esophagitis who were age and sex matched with the cases and included them as a diseased control group. For each subject, one snap-frozen biopsy from mid corpus of the stomach greater curvature was preserved in RNA-later preserved at −70 and send to Karolinska Institutet on dry ice for microbiota assessment.
DNA extraction and sequencing
DNA was extracted from snap frozen gastric tissue (approximately 5–6 mm in length) using DNeasy Blood & Tissue Kit (Qiagen. Inc., Valencia. CA). Tissues were treated with filtered-lysozyme in lysis buffer (Tris-EDTA-Triton) and overnight incubation at 56°C in buffer AL (Qiagen. Inc., Germany) and proteinase K and blended with glass beads (Tactum Lab. Sweden) with 0.1. 0.5 and 1 mm diameter for 1 minute in Bullet Blender (BBX24. Next Advance. Inc., NY). Mixture was incubated with RNase (Qiagen. Inc., Valencia. CA). Tubes containing only beads, lysozyme, lysis buffer and extraction kit substances were included as quality controls for contamination.
Small subunit of ribosomal RNA (16 S rRNA) gene was amplified from extracted DNA by using primers targeting region V3–V4. Forward primer (Bekt_341F: 5′-CCTACGGGNGGCWGCAG) and reverse primer (Bekt_805R: 5′-GACTACHVGGGTATCTAATCC) carried 454-adaptor sequences A and B38. For multiplexing, unique 7 bp barcode for each sample was included in reverse primer in a way that barcodes differ in 2 nucleotides. Polymerase chain reaction (PCR) included a mix containing 10 μl 5X PCR buffer HF. 0.65 μl Phusion high fidelity DNA polymerase (New England Biolab Inc.. MA), 1 μl PurePeak deoxyribonucleoside triphosphates (200 μM. ThermoScientific. Milwaukee) and 2.5 μl of each primer (MWG eurofins. Germany). Two μl of template DNA was added to this mix with total volume of 50 μl, GeneAmp PCR system 9700 cycler (Applied Biosystems. CA) was used with cycling parameters of initiation (95°C for 5 min), 30 cycles of denaturation (95°C for 40 s), annealing (58°C for 40 s) and elongation (72°C for 1 min) with a final extension at 72°C for 7 min. Each sample was put in triplet with one negative control for avoiding PCR cross contamination. Amplified products for each sample were pooled and verified by gel electrophoresis. PCR products were purified by Agencourt AMPure XP magnetic beads (Beckman Coulter, Inc.) with size selection through modifying PEG concentration to obtain 400–500 nt template. DNA concentration in each sample was measured using Qubit 2.0 fluorometer (Invitrogen Inc., UK). An amplicon pool was formed by pooling equimolar amounts of all DNA libraries for a minimum of 20 ng/dl. Pool amplicon was subsequently sent to SciLifeLab (Stockholm), where pooled DNA was amplified in PCR-mixture-in-oil emulsions and sequenced on whole PicoTiterPlate (PTP) on GS-FLX Titanium XLR-70 system (Roche Diagnostics Co. Sweden). Plate was physically divided into 2 lanes and duplicate samples were run on both lanes.
Sequence data processing
Standard flowgrams were split to individual samples. AmpliconNoise v1.25 with its default filtering setting (PCR noise precision = 30. pyro noise precision = 0.6) was used to remove noisy data39. Chimeric sequences were removed by applying Perseus39. All noise-free individual sequences were combined and clustered by applying complete linkage clustering algorithm in FCluster. Clusters were remapped to operational taxonomic units (OTUs) at the similarity level of 97%. The taxonomic position of the representative sequence of each OTU was identified using last common ancestor method implemented in SINA aligner40 which was run against SILVA SSU 111 reference database41 imported to ARB42.
Statistical analysis
Two sets of analysis were done:(a) Individual taxonomy comparison: Assigned OTUs were compared individually between study groups using conditional logistic regression with false discovery rate (FDR) control to correct for multiple comparisons43. To have power for analysis, a minimum of 8 reads per OTU were required. (b) Microbiota pattern analysis, including all assigned and unassigned OTUs: Unifrac and weighted Unifrac distances44 were fed into a principal coordinate analysis (PCoA). Conditional logistic regression and Akaike information criterion (AIC) were applied to select the best-fit model. To examine whether presence or abundance of H. pylori influences the microbiota composition, an average-linkage hierarchical clustering analysis was performed excluding H. pylori from microbiota and proportion of different quartiles of H. pylori was compared between clusters. R and phyloseq package were used for statistical analysis (www.r-project.org)43.
References
Jemal, A. et al. Global cancer statistics. CA Cancer J Clin 61, 69–90 (2011).
Boyle, P. & Levin, B. World cancer report 510–511. (International Agency for Research on Cancer, WHO Press, Geneva, 2008).
Uno, K. et al. Direct measurement of gastroesophageal reflux episodes in patients with squamous cell carcinoma by 24-h pH-impedance monitoring. Am J Gastroenterol 106, 1923–9 (2011).
Viani, F. et al. The effect of intra-gastric acidity and flora on the concentration of N-nitroso compounds in the stomach. Eur J Gastroenterol Hepatol 12, 165–73 (2000).
Ye, W. et al. Helicobacter pylori infection and gastric atrophy: risk of adenocarcinoma and squamous-cell carcinoma of the esophagus and adenocarcinoma of the gastric cardia. J Natl Cancer Inst 96, 388–96 (2004).
de Vries, A. C. et al. Increased risk of esophageal squamous cell carcinoma in patients with gastric atrophy: independent of the severity of atrophic changes. Int J Cancer 124, 2135–8 (2009).
Marshall, B. Helicobacter pylori: 20 years on. Clin Med 2, 147–152 (2002).
Kamangar, F. et al. Helicobacter pylori and oesophageal and gastric cancers in a prospective study in China. Br J Cancer 96, 172–6 (2007).
Simán, J. H., Forsgren, A., Berglund, G. & Florén, C. H. Association between Helicobacter pylori and gastric carcinoma in the city of Malmö, Sweden. A prospective study. Scand J Gastroenterol 32, 1215–21 (1997).
El-Omar, E. M. et al. Increased risk of noncardia gastric cancer associated with proinflammatory cytokine gene polymorphisms. Gastroenterology 124, 1193–201 (2003).
Wang, K. X. et al. Detection of serum anti-Helicobacter pylori immunoglobulin G in patients with different digestive malignant tumors. World J Gastroenterol 9, 2501–4 (2003).
Wu, D. C. et al. Helicobacter pylori infection: a protective factor for esophageal squamous cell carcinoma in a Taiwanese population. Am J Gastroenterol 100, 588–93 (2005).
Iijima, K. et al. Extensive gastric atrophy: an increased risk factor for superficial esophageal squamous cell carcinoma in Japan. Am J Gastroenterol 102, 1603–9 (2007).
Whiteman, D. C. et al. Association of Helicobacter pylori infection with reduced risk for esophageal cancer is independent of environmental and genetic modifiers. Gastroenterology 139, 73–83 (2010).
Bik, E. M. et al. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci U S A 103, 732–7 (2006).
von Rosenvinge, E. C. et al. Immune status, antibiotic medication and pH are associated with changes in the stomach fluid microbiota. ISME J 7, 1354–66 (2013).
Li, X. X. et al. Bacterial microbiota profiling in gastritis without Helicobacter pylori infection or non-steroidal anti-inflammatory drug use. PLoS One 4, e7985; 10.1371/journal.pone.0007985 (2009).
Quivey, R. G., Kuhnert, W. L. & Hahn, K. Genetics of acid adaptation in oral streptococci. Crit Rev Oral Biol Med 12, 301–14 (2001).
Mahboubi, E. et al. Oesophageal cancer studies in the Caspian Littoral of Iran: the Caspian cancer registry. Br J Cancer 28, 197–214 (1973).
Saidi, F. et al. Oesophageal cancer among the Turkomans of northeast Iran. Br J Cancer 83, 1249–54 (2000).
Rolig, A. S., Cech, C., Ahler, E., Carter, J. E. & Ottemann, K. M. The degree of Helicobacter pylori inflammation is manipulated by the pre-infection host microbiota. Infect Immun 81, 1382–9 (2013).
Turnbaugh, P. J., Backhed, F., Fulton, L. & Gordon, J. I. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3, 213–23 (2008).
Griffen, A. L. et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16 S pyrosequencing. ISME J 6, 1176–85 (2012).
Schwab, C. et al. Longitudinal study of murine microbiota activity and interactions with the host during acute inflammation and recovery. ISME J 8, 1101–14 (2014).
Abnet, C. C. et al. Tooth loss and lack of regular oral hygiene are associated with higher risk of esophageal squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev 17, 3062–8 (2008).
Yu, G. et al. Association between upper digestive tract microbiota and cancer-predisposing states in the esophagus and stomach. Cancer Epidemiol Biomarkers Prev 23, 735–41 (2014).
Nasrollahzadeh, D. et al. Gastric atrophy and oesophageal squamous cell carcinoma: possible interaction with dental health and oral hygiene habit. Br J Cancer 107, 888–94 (2012).
Chakravorty, S., Helb, D., Burday, M., Connell, N. & Alland, D. A detailed analysis of 16 S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J Microbiol Methods 69, 330–9 (2007).
Case, R. J. et al. Use of 16 S rRNA and rpoB genes as molecular markers for microbial ecology studies. Appl Environ Microbiol 73, 278–88 (2007).
Kunin, V., Engelbrektson, A., Ochman, H. & Hugenholtz, P. Wrinkles in the rare biosphere: pyrosequencing errors can lead to artificial inflation of diversity estimates. Environ Microbiol 12, 118–23 (2010).
Huse, S. M., Huber, J. A., Morrison, H. G., Sogin, M. L. & Welch, D. M. Accuracy and quality of massively parallel DNA pyrosequencing. Genome Biol 8, R143; 10.1186/gb-2007-8-7 (2007).
Mir, M. R. et al. Short- and long-term survival of esophageal cancer patients treated at the Cancer Institute of Iran. Dig Surg 30, 331–6 (2013).
Taylor, P. R., Abnet, C. C. & Dawsey, S. M. Squamous dysplasia--the precursor lesion for esophageal squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev 22, 540–52 (2013).
Kuczynski, J. et al. Microbial community resemblance methods differ in their ability to detect biologically relevant patterns. Nat Methods 7, 813–9 (2010).
Lundin, D. et al. Which sequencing depth is sufficient to describe patterns in bacterial α- and β-diversity? Environ Microbiol Rep 4, 367–72 (2012).
Nasrollahzadeh, D. et al. Opium, tobacco and alcohol use in relation to oesophageal squamous cell carcinoma in a high-risk area of Iran. Br J Cancer 98, 1857–63 (2008).
Ahn, J. et al. Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst 105, 1907–11 (2013).
Herlemann, D. P. et al. Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J 5, 1571–9 (2011).
Quince, C., Lanzen, A., Davenport, R. J. & Turnbaugh, P. J. Removing noise from pyrosequenced amplicons. BMC Bioinformatics 12, 38; 10.1186/1471-2105-12-38 (2011).
Pruesse, E., Peplies, J. & Glockner, F. O. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28, 1823–9 (2012).
Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41, 590–6 (2013).
Ludwig, W. et al. ARB: a software environment for sequence data. Nucleic Acids Res 32, 1363–71 (2004).
Yoav, B. & Yosef, H. Controlling the false discovery rate: apractical and powerful approach to multiple testing. J R Statisc Soc B 75, 289–300 (1995).
Lozupone, C. & Knight, R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71, 8228–35 (2005).
Acknowledgements
The authors would like to acknowledge support from Science for Life Laboratory, the national infrastructure SNISS (Uppmax), Daniel Lundin from BILS (Bioinformatics Infrastructure for Life Sciences) for providing assistance in massively parallel sequencing and computational infrastructure. This study was supported by a grant from Swedish Research Council (2008-3027). Field work and subject recruitment were supported by a grant from Digestive Disease Research Center of Tehran University of Medical Sciences (grant 82-603). Case-Control study received support from Intramural funds of the National Cancer Institute at the National Institutes of Health. D.N. was partially supported by KID- funding (Faculty funds for partial financing of doctoral student) from Karolinska Institutet.
Author information
Authors and Affiliations
Contributions
Conception and design: W.Y., D.N., R.M. Development and Methodology: D.N. and W.Y. Acquisition of data: D.N., M.S., R.M., S.N.M., R.S., S.F. and F.I. Laboratory analysis: D.N. and W.Y. Analysis and interpretation of data: D.N., A.P. and W.Y. Drafting Manuscript: D.N. Review, revision of manuscript: W.Y., D.N., B.W., A.P., R.M., S.D., C.A., F.K., P.Bo. and P.Br. Study Supervision: W.Y. and R.M.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Nasrollahzadeh, D., Malekzadeh, R., Ploner, A. et al. Variations of gastric corpus microbiota are associated with early esophageal squamous cell carcinoma and squamous dysplasia. Sci Rep 5, 8820 (2015). https://doi.org/10.1038/srep08820
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep08820
This article is cited by
-
Non-Acid Fluid Exposure and Esophageal Squamous Cell Carcinoma
Digestive Diseases and Sciences (2022)
-
The role of the bacterial microbiome in the treatment of cancer
BMC Cancer (2021)
-
Porphyromonas gingivalis promotes tumor progression in esophageal squamous cell carcinoma
Cellular Oncology (2021)
-
Epidemiologic studies of the human microbiome and cancer
British Journal of Cancer (2016)
-
Enigmas, priorities and opportunities in cancer epidemiology
European Journal of Epidemiology (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.