Abstract
Mifepristone (RU486), a synthetic steroid compound used as an abortifacient drug, has received considerable attention to its anticancer activity recently. To explore the possibility of using mifepristone as a cancer metastasis chemopreventive, we performed a systems pharmacology analysis of mifepristone-related molecules in the present study. Data were collected by using Natural Language Processing (NLP) and 513 mifepristone-related genes were dug out and classified functionally using a gene ontology (GO) hierarchy, followed by KEGG pathway enrichment analysis. Potential signal pathways and targets involved in cancer were obtained by integrative network analysis. Total thirty-three proteins were involved in focal adhesion-the key signaling pathway associated with cancer metastasis. Molecular and cellular assays further demonstrated that mifepristone had the ability to prevent breast cancer cells from migration and interfere with their adhesion to endothelial cells. Moreover, mifepristone inhibited the expression of focal adhesion kinase (FAK), paxillin and the formation of FAK/Src/Paxillin complex, which are correlated with cell adhesion and migration. This study set a good example to identify chemotherapeutic potential seamlessly from systems pharmacology to cellular pharmacology and the revealed hub genes may be the promising targets for cancer metastasis chemoprevention.
Similar content being viewed by others
Introduction
Mifepristone (RU486), an organic chemical used for abortifacient initially, was developed during the early 1980s by a team of researchers working for the French pharmaceutical company1. Although discovered in France, mifepristone is now widely registered for use in 55 countries, including several countries in the European Union, the United States and China for her family-plan policy2. Mifepristone is a progestational and glucocorticoid hormone antagonist. It is mainly used as an abortifacient by interfering with the hormones (progesterone) function in the body3,4. As a glucocorticoid receptor antagonist, mifepristone has been widely used to treat hypercortisolism in patients with refractory Cushing's Syndrome, major depression with psychotic features and glaucoma2.
Mifepristone used in cancer therapy has attracted increasing attention in recent years. Mifepristone could block cell surface receptors, such as progesterone receptor (PR), glucocorticoid receptors (GR) and estrogen receptors (ER), which are overabundant in some tumor cells5,6,7. In PR-positive endometrial adenocarcinoma or sarcoma women, mifepristone given at 200 mg daily could result in a stable disease rate of 25%8,9. In premenopausal women, especially for those ER-positive, mifepristone given at 50 mg on alternate days for 3 months reduced the expression of Ki-67, a marker of cell proliferation10. Furthermore, mifepristone has been clinically used for leiomyoma, uterine fibroids, ovary, prostate cancer, cervical cancer, gastrointestinal tract and cancer chemotherapy2,11,12. Recent studies further showed that mifepristone also inhibited the growth of different cancer cell lines regardless of the expression of hormone responsiveness13.
Although the anticancer activity of mifepristone has been exploited, its exact molecular mechanisms of actions and related pathways and targets towards cancer remain poorly understood. As cancer-related molecular signatures are usually a series, instead of a few, it is necessary to systematically analyze the mifepristone-related pathways and targets, especially those associated with cancer therapy.
Metastases from a primary tumor to secondary locations throughout the body are a major cause of cancer related deaths14. One of the principal requirements for cancer metastasis to the distant organs is the activation, adhesion and motility of circulating tumor cells (CTCs)15,16. Once activated and adhered to the vascular endothelium, the cancer metastasis cascade process starts16,17. Therefore, preventing cancer cells from activation, adhesion and migration as well as intervening with the key proteins in focal adhesion pathway are the main research objectives for us to identify safe and effective cancer metastasis chemopreventives.
To expedite discovery of new mifepristone-related targets for effective cancer metastasis chemoprevention, we established a systems pharmacologgy method to systematically analyze the existing information of mifepristone to pinpoint its potential targets for intervention. By using this method, i.e., systems pharmacology18. The analysis revealed the potential functions, signaling pathways and network of mifepristone-related molecules involved in cancer therapy. The integrative network analysis identified mifepristone-related hub genes, in particular, FAK-the key signal molecule associated with cancer metastasis. To demonstrate the usefulness of systems pharmacology in drug discovery and development, we, under the guidance of the systems pharmacology of mifepristone, investigated the anti-metastatic potential of mifepristone by using the most aggressive metastatic cancer cell lines and then in particular, focused on the effects of mifepristone on FAK and its functional complex “FAK/Src/Paxillin” in vitro. The present study, to the best our knowledge, is the first that revealed the interaction between mifepristone and the FAK/Src/Paxillin complex and provides a new strategy to identify molecular targets for development of cancer metastasis chemopreventives based on the information of systems pharmacology. The detail study designs and results are reported below.
Methods
NLP analysis of mifepristone
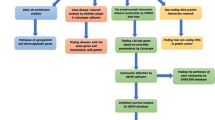
We conducted a search in the PubMed, attempting to cover all papers published between January 1980 and May 2013, with the following combinations of query terms: (“mifepristone” or “RU486”) and (“1980/01/01” [PDAT]: “2013/05/31” [PDAT]). All of studies identified by the computerized search were retrieved, assessed and then downloaded as HTML text without images and converted into XML documents. All the genes and proteins associated with keywords were dug out and added to a list, followed by gene mention tagging using A Biomedical Named Entity Recognizer (ABNER, an open source tool for automatically tagging genes, proteins and other entity names in text, (http://pages.cs.wisc.edu/~bsettles/abner/)19. Furthermore, conjunction resolution was conducted to obtain individual descriptions of the extracted genes. For the extracted genes, such as “STAT3/5 gene”, the analysis will be resolved into the STAT3 and STAT5 gene. In this study, the gene symbols in entrez gene database of NCBI are commonly-used (http://www.ncbi.nlm.nih.gov/gene)20,21. Flow chart of the NLP analysis is shown in Figure 1.
For each gene, the frequency of its occurrence in the literature-based dataset was calculated. The higher the frequency of the genes corresponded to, the greater the likelihood of the association between mifepristone and the certain gene. “N” represents the total number of publications identified from the PubMed database. “m” and “n” represent the frequency of genes and mifepristone, found in the PubMed database. While k denotes the occurrences of gene and mifepristone. Then, by using hypergeometric distribution, we calculated probability of the frequency greater than k simultaneously cited under the completely random conditions:


(The notion of co-citation are defined in the network analysis).
The mifepristone-gene relations with P-value < 0.05 were then summarized and subjected to a relational database for further analysis.
Gene ontology (GO) analysis
GO analysis was conducted with the GSEABase package from R (http://www.r-project.org/) statistical platform22. Genes were classified in three major groups: the biological process, cellular component and molecular function.
Pathway analysis
Genes were mapped to the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database by using GenMAPP v2.1 (http://www.genmapp.org/) and the P-value of the enrichment was calculated for each individual pathway23.
Network analysis of mifepristone-related proteins/genes involved in cancer pathways
Genes that involved in cancer pathway were integrated into three different interaction relationships among: (i) protein interaction, gene regulation, protein modification listed in the KEGG database; (ii) existing high-throughput protein interaction experiments confirmed by yeast two-hybrid; (iii) gene interaction demonstrated in previous reports. In brief, the pathway data were downloaded from the KEGG database and used for analyzing the interaction between genes with the KEGGSOAP package from R statistical analysis platform (http://www.bioconductor.org/packages/2.4/bioc/html/KEGGSOAP.html) that evaluated three kinds of relationships: ECrel (enzyme-enzyme relation, indicating two enzymes catalyzing successive reaction steps), PPrel (protein-protein interaction, such as binding and modification), GErel (gene expression interaction, indicating relation of transcription factor and target gene product)24. The protein-protein interaction data were downloaded from the mammalian protein-protein interaction (MIPS) database (http://mips.helmholtz-muenchen.de/proj/ppi)25.
For interactions that had been previously reported, we used co-citation matrices in PubMed. Using this algorithm, a gene term and all its term variants that co-occur within the sentences of an abstract listed on PubMed are identified and the frequency of the co-cited gene was calculated. Finally, statistical analysis was conducted as described above in equation 1. The resulting network was displayed by using the open-source Medusa software26. Then, the highly connected hub genes, which played an important role in the network stability and maybe the potential mifepristone-related targets involved in cancer therapy, were calculated and decided.
Cell culture, antibodies and reagents
MDA-MB-231 human breast cancer cells were purchased from American Type Culture Collection (ATCC, Manassas, VA) and maintained in ATCC-formulated Leibovitz's L-15 Medium (Catalog No. 30-2008). Cells were supplemented with heat inactivated fetal bovine serum to a final concentration of 10% and incubated at 37°C in a free gas exchange with atmospheric air. Mouse monoclonal anti-FAK (ab72140), rabbit polyclonal anti-Src(ab47405), -paxillin(ab39537) and -β-actin antibodies (ab8227) and goat anti-mouse (ab1500117), goat anti-rabbit IgG (ab150077) antibodies were all obtained from Abcam Corporation.
In vitro cytotoxicity studies
MTT assay was used to investigate the cytotoxicity of mifepristone in vitro as described previously by this lab16,17. Briefly, MDA-MB-231 cells were seeded into 96-well plates at a density of 1 × 104 cells/well and then incubated at 37°C in a humidified atmosphere with 100% air. After overnight incubation, the cells were treated with mifepristone at different concentrations of 0, 20, 40, 60, 80 and 100 μmol/L, respectively. Culture medium was used as a blank control. After 24 hours of incubation, MTT solution (5 mg/ml) was added to each well and the cells were incubated for another 4 hours in the medium without phenol red and serum. The MTT-formazan formed by metabolically viable cells was dissolved in 150 μl of dimethyl sulfoxide (DMSO). The optical density (OD) measured at 565 nm using an infinite M200 Pro microplate reader (Tecan, Switzerland). The absorbance of untreated cells was considered as 100%. Each sample was assayed in triplicate in three independent experiments. Percent growth inhibition of cells exposed to treatments was calculated as follows:
Inhibition (%) = (1 - OD of test group/OD of control group) × 100%
Cell migration assays
Cell migration assay was performed using 24-well transwells (Costar, Coring Incorporated, USA), which allows cells to migrate through a polycarbonate membrane with 8-μm pore size as we described previously27. Cells were seeded 5 × 104 per well on the upper chamber of the transwell apparatus. Migration assay was performed in the presence of 0, 25, 50, 75, 100 μM of mifepristone. DMSO (final concentration: 0.1%) was used as vehicle control. After 24 h incubation, cells in the upper chamber were carefully scraped off using a cotton swab. Cells migrated to the basal side of the membrane were fixed in methanol, stained with 0.1% crystal violet for 20 min and counted the average number from the randomly selected group of 3 pictures.
Cell adhesion assay
The adhesion assay of MDA-MB-231 cells to the HUVEC was assessed according to the method described previously by this lab with minor modifications16,17. Briefly, Human umbilical vein endothelial cells (HUVECs) were isolated and utilized between passages 2 and 5 and grown to confluence in 24-well plates in the M199 medium. Then, TNF-α (final concentration: 10 ng/ml) was used to activate HUVECs for 4 hours. MDA-MB-231 cells labled with Rhodamine 123 were co-cultured with the HUVEC monlayers in each well, followed by treatment with mifepristone for 1 hour. DMSO (0.1%) was used as the vehicle control. Then, nonadherent cells were removed by careful three-time washings with PBS. Ten visual fields for each well were selected randomly and taken pictures using a fluorescence microscope (Zeiss, Germany). Mean inhibition of adhesion for 10 visual fields was calculated by using the equation: % of control adhesion = [the number of adhered cells in treated group/the number of adhered cells in the control group] × 100%.
RNA extraction and Real-time PCR analysis for gene expression
Total RNA was isolated from cells in control group and mifepristone treated groups at different concentrations, using the RiboPureTMkit (Applied Biosystems/Ambion, Austin, TX, USA). Isolated RNA (1 μg) was used to synthesize cDNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Primers were designed to amplify a 240-bp FAK fragment (BC028733.2): forward primer 5′- gattttatcccagcccacagc-3′; reserve primer 5′-cttccatttcctgttgctgtc-3′.
Western blot analysis
Cell lysates were separated by SDS-PAGE and then transferred to polyvinylidene difluoride membranes (Bio-Rad). The membranes were blocked in 5% milk and then separately incubated with primary antibody against human FAK, β-actin at 4°C overnight. After additional washing, membranes were incubated with horseradish peroxidase (HRP)-conjugated secondary antibody (goat anti-mouse or goat anti-rabbit) at 37°C for 1 h. Then, immunodetection was accomplished using enhanced chemiluminescence and data were acquired with a quantitative digital imaging system (Quantity One, Bio-Rad) allowing to check for saturation. Overall emitted photons were quantified for each band, particularly for homogeneously the loading controls.
Immunoprecipitation
The formation of the FAK/Src/Paxillin complex in MDA-MB-231 cells was analyzed by immunoprecipitation and western blot. Cells were lysed with lysis buffer (1% Triton X-100 in 50 mM Tris-HCl [pH 7.4] containing 150 mM NaCl, 5 mM EDTA, 2 mM Na3VO4, 2.5 mM Na4PO7, 100 mMNaF, 200 nM microcystin lysine-arginine and protease inhibitors). Cell lysates (300 μg) were mixed with 10 μg of moused anti-FAK monoclonal antibody. Purified mouse IgG (Sigma) was used as the negative control. The samples were incubated for 4 h, mixed with Protein A/G PLUS-agarose immunoprecipitation reagent. (Pierce, Rockford, IL) and then incubated for an additional 12 h. The beads were washed four times and the bound proteins were released from the beads by boiling in SDS-PAGE sample buffer for 5 min. Samples were analyzed by western blot (described aboved) with rabbit anti-Src polyclonal antibody and rabbit anti-Paxillin polyclonal antibodies.
Statistical analysis
All data were analyzed using SASS software and expressed as the mean ± SD or SE. Statistical comparisons between different groups were performed using Student t-test. A P value of <0.05 was considered to be statistically significant.
Results
NLP analysis of Mifepristone
The initial computerized search through PubMed identified 5617 primary studies reported mifepristone-related genes. As a result, a total of 513 Mifepristone (RU486)-related genes were obtained. The 10 most frequently cited genes were listed in Table 1, including glucocorticoid receptor (NR3C1), progesterone receptor (PR), tumor necrosis factor (TNF), interleukin 6 (IL6), vascular endothelial growth factor A (VEGFA), cytochrome P450 and others.
GO analysis
Each of the 513 genes was categorized in GO according to biological process, cellular component and molecular function. As showed in Table 2, these genes are mainly involved in signal transduction activity, nucleic acid binding activity, transcription regulatory activity, transporter activity and kinase activity.
Integrative analysis, pathway and network of potential targets
Pathway information is required for understanding of gene function28. To better understand the gene function related to mifepristone, we mapped the 513 genes to canonical signaling pathways found in the Kyoto Encyclopedia of Genes and Genomes (KEGG). A total of 134 mifepristone-related pathways were identified, which were assigned into 24 statistically remarkable categories (P value < 0.01), including cytokine-cytokine receptor interaction, Jak-STAT signaling pathway, Toll-like receptor signaling pathway, ErbB signaling pathway, focal adhesion and apoptosis (Table 3).
As the KEGG pathways could map for 14 cancers and summarize different signaling pathways and cancer-related genes involved in different stages of oncogenesis, we analyzed the 513 mifepristone-related genes and mapped 66 genes to the pathways associated with cancer (Figure 2). Then, we constructed a gene network based on the 66 genes.
Construction of gene networks not only directly reflects the physiological situation as a whole, but also the stability of the network. The highly connected hub genes often play important roles in the stability of the network. As the hub genes are at the core of gene regulation and can affect a majority of genes in the network, the hub genes are generally believed to be higher in important than other genes and considered to be potential drug target for therapy. In this study, forty-seven hub genes were identified by network analysis. Among them, CCND1 (cyclin D1), EGFR (epidermal growth factor receptor), JUN (jun oncogene), MYC (v-myc myelocytomatosis viral oncogene homolog), VEGFA (vascular endothelial growth factor A), RELA (v-rel reticuloendotheliosis viral oncogene homolog A) and IGF1R (insulin-like growth factor 1 receptor), showed extremely high connectivity to other genes (Figure 3).
Mifepristone inhibits cell migration and adhesion by regulating focal adhesion pathway
we analyzed the 513 mifepristone-related genes and mapped 33 genes to the pathways associated with focal adhesion. Interestingly, by network analysis performed as described above, a gene network was constructed based on the 33 genes, including FAK, paxillin, ILK and others, implying that mifepristone may have the potential to inhibit cancer cells migration and adhesion, the major steps towards metastasis. Among them, FAK showed extremely high connectivity to other genes (Figure 4).
Genes involved in focal adhesion pathway.
A, focal adhesion-related pathways generated by KEGG analysis. Red indicates mifepristone-related. B, network and connectivity analysis of mifepristone-related therapeutic targets in focal adhesion pathways. The connectivity of FAK (PTK2) is the highest one that has a total of 10-related genes (z-test, P < 0.05).
To explore the metastasis chempreventives function of mifepristone, the cytostatic effect was examined first on human breast cancer cell MDA-MB-231. As showed in Figure 5A, the cytotoxicity of mifepristone was low. Even at concentration of 100 μM, its IC50 was not achieved. As demonstrated in Figure 5B, however mifepristone exhibited significant inhibition on migration of the MDA-MB-231 cells through the transwell membrane at concentrations lower than its IC50, suggesting its specific inhibition on cell migration.
Cellular pharmacology analysis of mifepristone.
A, in vitro activity of mifepristone against MDA-MB-231 cell line. B, dose-dependent inhibition by mifepristone on cell migration. A Corning transwell system was used to assay cell migration as described in methods. The amount of MDA-MB-231 cells migrated through polycarbonate membranes was counted by microscopic observation (10×). Each experiment was carried out at least three times. *, P < 0.05, **, P < 0.01. C, inhibition by mifepristone of MDA-MB-231 cells adhesion to HUVECs. Representative microscopic observation of the inhibition by mifepristone at 0, 50 and 100 μM. DMSO (0.1%) was used as vehicle control (average of 10 independent microscope fields for each of 3 independent experiments).
The adherence of MDA-MB-231 cells to HUVECs was assessed to determine whether mifepristone can regulate cell adhesion. Ten fields of each well were randomly selected and the adhered spots were counted. Compared with the control, the adhesion rate of MDA-MB-231 cells was 86, 83, 67 and 54%, respectively, at 25, 50, 75 and 100 μM of mifepristone (Figure 5C). Mifepristone markedly and in a concentration-dependent manner inhibited the adherence of MDA-MB-231 cells to endothelial monolayers, indicating that it may fit into a new class of therapy for the reduction of risk factors of cancer metastasis.
Molecular analysis of effects of mifepristone on FAK
Since mifepristone had the ability to inhibit cancer cell migration and adhesion (Figure 5B and C), we focused on the key regulator of mifepristone-related proteins involved in focal adhesion pathway. As showed in Figure 4, FAK, also named Protein-tyrosine kinase 2 (PTK2), had extremely high connectivity to other genes in mifepristone-related focal adhesion pathway. Then, FAK expression was tested by Western blot and RT-PCR experiments. Obvious decreases in FAK expression were observed in the MDA-MB-231 cells treated with mifepristone in a concentration-dependent manner both at protein level and mRNA level, while no change in β-actin expression was observed (Figure 6A, 6B), further confirmed that FAK is a critical regulator involved in mifepristone-related focal adhesion pathway.
Concentration-dependent effects of mifepristone on FAK activity.
A. Western blot assay showing that mifepristone inhibits the expression of FAK; B. Real-time PCR assay showing that mifepristone inhibited mRNA expression. Data are the means ± SEM of 3 repeat experiments. N = 3. *, p < 0.05; **, p < 0.01.
Effect of mifepristone on formation of FAK/Src/Paxillin complex
As showed in Figure 4, FAK is an important hub gene connected to other genes in mifepristone-related focal adhesion pathway. FAK mainly acts as a scafford to recruit Src to activate FAK-associated substrate, paxillin. In addition, the formation of FAK/Src/Paxillin complex is required for the activation of integrins, which plays an important role in cell migration and adhesion. To determine whether or not mifepristone plays a role in regulating FAK/Src/Paxillin complex formation, the levels of FAK, Src and Paxillin were analyzed in the presence and absence of mifepristone with western blot or immunoprecipitation. Mifepristone decreased FAK and Paxillin expression but did not change Src expression in cell lysate. However, mifepristone decreased the formation of the FAK/Src/Paxillin complex as shown in immunoprecipitation experiment (Figure 7 ).
Mifepristone-mediated formation of FAK/Src/Paxillin complex.
Cells were pretreated in the absence and presence of mifepristone (50 μM) for 1 hour. Cell lysates were analyzed by western blot with antibodies that recognize FAK, Src and Paxillin. Immunoprecipitation (IP) with the anti-FAK antibody or control IgG antibody followed by blotting with the anti-Src or anti-Paxillin antibody was performed. Each of the examples is representation of three independent experiments. The lower part depicting the bars denotes the mean ± SE of three independent experiments for each condition determined from densitometry relative to control. *, P < 0.05.
Discussion
Metastatic spread of cancers accounts for the lethality of the disease and therefore there is a great need to develop new chemopreventives to inhibit cancer cell metastasis. Mifepristone has been used for termination of early pregnancy because of its capacity to work as an antiprogestin and antiglucocorticoid. However, the study of mifepristone has been expanded to the field of oncology over the past 30 years. Despite its importance as a potential anticancer agent29, studies on the pathways and molecular mechanism of action involved in mifepristone-related cancer therapy are scarce and it still remains unclear what the potential mifepristone therapeutic targets are in terms of cancer treatment.
In this work, we identified 513 genes related to the response of mifepristone by using the NLP analytic approach. Followed by GO analysis and pathway analysis, we established their potentially functional classification for the first time: there are 134 pathways and among them 24 have a P value less that 0.01. Examples are cytokine-cytokine receptor interaction30, Jak-STAT signaling pathway31, Toll-like receptor signaling pathway32, apoptosis33, focal adhesion34, VEGF signaling pathway35 and p53 signaling pathway36. They were reported to be involved in cancer carcinogenesis, prognosis and therapeutics. For example, the cytokine-cytokine receptor interaction signaling pathway has attracted more attention of many investigators in recent years, in part because their structures, biophysical basis of their binding and their mode of biological activation are important for small molecule drug design37. In this work, we identified 61 genes related to cytokine-cytokine receptor interaction responding to mifepristone treatment, such as TNF38, IL639 and VEGFA40 (Table 3 and Figure 2). Furthermore, mifepristone has been reported to induce apoptosis through reduction in the mitochondrial membrane potential and activation of p38 MAPK in U937 human leukemia cells41. In this study, 22 mifepritone-related genes were found to be involved in apoptosis pathway, such as AKT1, BAD and CYCS.
Since the KEGG offers important signaling pathways and cancer-related genes involved in different stages of oncogenesis, we analyzed the 513 mifepristone-related genes and mapped 66 genes to the pathways associated with cancer (Figure 2). In order to find out the potential proteins/genes that may be key therapeutic targets of mifepristone, network analysis was conducted in the present study and reveal 47 hub genes, most of which were transcription factors or kinases, such as MAPK system (Figure 2 and 3). The highly connected hub genes regulate and affect a majority of genes in the collective dataset and are generally believed to be more important than other normal genes and used as key therapeutic targets. The present gene network and connectivity analyses showed that the hub genes, EGFR and CCND1, had the highest connectivity rate related to 12 mifepristone-related genes (z-test, P < 0.01). Other hub genes such as JUN and MYC were also associated with cancer therapy (z-test, p < 0.05) (Figure 3). EGFR (epidermal growth factor receptor, also named ErbB1 or HER1)42, has been one of the most targeted receptors in the field of oncology. EGFR ligand binding and the subsequent formation of ErbB family dimers could promote the cross-phosphorylation of the dimmer partner and generate a network of intracellular signals that control numerous biological processes, especially, activating MAPK cascade33 and phosphatidylinositol 3-kinase (PI3-K)/Akt signaling43, which are important in gene transcription, cell proliferation, migration, or differentiation. NLP analysis of mifepristone related targets showed that mifepristone could inhibit EGFR mRNA levels in breast cancer cells44 and astrocytomas cells45 via abrogating progesterone receptors. Inhibiting EGFR mRNA levels could turn off EGFR signals and block it from sending messages and eventually result in cell death. So, EGFR may be an important mifepristone's target in cancer therapeutic.
Over the past two decades, many striking similarities have been revealed between implanted embryos and circulating tumor cells (CTCs) in terms of their proliferative, migratory and invasive properties46,47. Similar to vascular endothelial cells that involved in regulating the migration and invasion of extravillous trophoblast (EVT) cells to uterus, the activated CTCs could adhere and interact with vascular endothelium cells before they extravasate in the distant metastatic organs. We proposed that the initiation of adhesion of CTCs to vascular endothelial cells is the first and important step for CTCs to start the metastastic cascade. Inhibition of the initial step may thus prevent consequential formation of the metastasis foci. A previous study on a rabbit laryngeal wound-healing model had reported that mifepristone impaired post-surgical wound healing of the larynx by increasing the presence of granulation tissue and polyps and appeared to delay re-epithelialization48. It was also found that cytostatic concentrations of mifepristone caused morphological changes in SKOV-3, MDA-MB-231 and U87MG, but not in LNCaP cells that underwent cellular senescence. Interestingly, mifepristone-pretreated SKOV-3, MDA-MB-231, U87MG and LNCaP cells delayed adhesion to extracellular matrix proteins49. These reports indicated that mifepristone could inhibit migration and adhesion in cancer cells.
In this study, we performed a systematic meta-analysis of mifepristone-related molecules and found that there are 33 mifepristone-related proteins involved in the focal adhesion pathway (Figure 4). The cell migration test and adhesion assay further demonstrated that mifepristone had a potential ability to suppress metastatic activity of cancer cells (Figure 5B and C). More importantly, FAK has the extremely high connectivity to other genes involved in focal adhesion pathway (Figure 4), indicating that FAK is a key mifepristone's target. FAK is the primary enzyme involved in the engagement of integrins and assembly of focal adhesion by interaction with many adaptor proteins, including Src and paxillin among others. This scaffolding function of FAK is needed for cell motility and the increased expression of FAK is often correlated with cacinogenesis and tumor metastasis50. Herein, we demonstrated that mifepristone decrease the expression of FAK in the MDA-MB-231 cells in a concentration-dependent manner both at protein level and mRNA levels (Figure 6). Mifepristone also decreased Paxillin expression but did not change Src expression in cell lysate (Figure 7).
In summary, by using computational and bioinformatic methods, we identified the potential functions, signaling pathways and network of mifepristone-related molecules involved in cancer therapy. Particularly, the present cellular pharmacology studies further demonstrated that the potential of mifepristone for preventing MDA-MB-231 cell migration and adhesion and its regulating FAK/Src/Paxillin complex formation. Overall, the present study demonstrates, for the first time, the relationship between mifepristone and the formation of FAK/Src/Paxillin complex. Our data provide more details in understanding anticancer mechanism of mifepristone, offer a cache of potential therapeutic targets and more importantly, provide a molecular framework for clinical evaluation of mifepristone as a potential cancer metastatic chemopreventive agent.
References
Herrmann, W. et al. Effects of an anti-progestin steroid in women: interruption of the menstrual cycle or early pregnancy (author's transl)]. Contracept Fertil Sex (Paris). 10, 389–393 (1982).
Chen, J. Z. et al. The Unique Pharmacological Characteristics of Mifepristone (RU486): from Terminating Pregnancy to Preventing Cancer Metastasis. Medicinal Research Reviews. 34, 979–1000 (2014).
Lalitkumar, S., Bygdeman, M. & Gemzell-Danielsson, K. Mid-trimester induced abortion: a review. Hum Reprod Update. 13, 37–52 (2007).
Winikoff, B. Acceptability of medical abortion in early pregnancy. Fam Plann Perspect. 27, 142–148, 185 (1995).
Wu, J., Liang, Y., Nawaz, Z. & Hyder, S. M. Complex agonist-like properties of ICI 182,780 (Faslodex) in human breast cancer cells that predominantly express progesterone receptor-B: implications for treatment resistance. Int J Oncol. 27, 1647–1659 (2005).
Sommer, P. et al. Glucocorticoid receptor over-expression promotes human small cell lung cancer apoptosis in vivo and thereby slows tumor growth. Endocrine-related cancer. 17, 203–213 (2010).
Vladusic, E. A., Hornby, A. E., Guerra-Vladusic, F. K., Lakins, J. & Lupu, R. Expression and regulation of estrogen receptor beta in human breast tumors and cell lines. Oncol Rep. 7, 157–167 (2000).
Ramondetta, L. M. et al. Phase 2 trial of mifepristone (RU486) in advanced or recurrent endometrioid adenocarcinoma or low-grade endometrial stromal sarcoma. Cancer. 115, 1867–1874 (2009).
Mei, L. et al. A novel mifepristone-loaded implant for long-term treatment of endometriosis: in vitro and in vivo studies. Eur J Pharm Sci. 39, 421–427 (2010).
Walker, K. J., Price-Thomas, J. M., Candlish, W. & Nicholson, R. I. Influence of the antioestrogen tamoxifen on normal breast tissue. Br J Cancer. 64, 764–768 (1991).
Llaguno-Munive, M., Medina, L. A., Jurado, Romero-Piña, M. & Garcia-Lopez, P. Mifepristone improves chemo-radiation response in glioblastoma xenografts. Cancer Cell Int. 13, 29 (2013).
Engman, M. et al. GSTM1 gene expression correlates to leiomyoma volume regression in response to mifepristone treatment. PLoS One 8, e80114 (2013).
Tieszen, C. R., Goyeneche, A. A., Brandhagen, B. N., Ortbahn, C. T. & Telleria, C. M. Antiprogestin mifepristone inhibits the growth of cancer cells of reproductive and non-reproductive origin regardless of progesterone receptor expression. BMC Cancer. 11, 207 (2011)
Leong, S. P & Tseng, W. W. Micrometastatic cancer cells in lymph nodes, bone marrow and blood: Clinical significance and biologic implications. CA Cancer J Clin. 64, 195–206 (2014).
Nguyen, D. X., Bos, P. D. & Massagué, J. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer. 9, 274–284 (2009).
Lu, Y. S. et al. Nitric Oxide Inhibits Hetero-adhesion of Cancer Cells to Endothelial Cells: Restraining Circulating Tumor Cells from Initiating Metastatic Cascade. Sci Rep. 4, 4344 (2014).
Wang, J. C. et al. Synthesis, spectral characterization and in vitro cellular activities of metapristone, a potential cancer metastatic chemopreventive agent derived from mifepristone (RU486). AAPS J. 16, 289–298 (2014).
Berg, J. M., Rogers, M. E. & Lyster, P. M. Systems biology and pharmacology. Clin. Pharmacol. Ther. 88, 17–19 (2010).
Settles, B. ABNER: an open source tool for automatically tagging genes, proteinsand other entity names in text. Bioinformatics. 21, 3191–3192 (2005).
Smith, L. et al. Overview of BioCreative II gene mention recognition. Genome Biol. 9 Suppl 2S2 (2008).
Morgan, A. A. et al. Overview of BioCreative II gene normalization. Genome Biol. 9 Suppl S3 (2008).
Bauer, S., Robinson, P. N. & Gagneur, J. Model-based gene set analysis for Bioconductor. Bioinformatics. 27, 1882–1883 (2011).
Dahlquist, K. D., Salomonis, N., Vranizan, K., Lawlor, S. C. & Conklin, B. R. GenMAPP, a new tool for viewing and analyzing microarray data on biological pathways. Nat Genet. 31, 19–20 (2002).
Ogata, H., Goto, S., Fujibuchi, W. & Kanehisa, M. Computation with the KEGG pathway database. Biosystems. 47, 119–128 (1998).
Mewes, H. W., Albermann, K., Heumann, K., Liebl, S. & Pfeiffer, F. MIPS: a database for protein sequence, homology data and yeast genome information. Nucleic Acids Res. 25, 28–30 (1997).
Hooper, S. D., & Bork, P. Medusa: a simple tool for interaction graph analysis. Bioinformatics. 21, 4432–4433 (2005).
Jia, L., Wong, H., Wang, Y., Garza, M., & Weitman, S. D. Carbendazim: disposition, cellular permeability, metabolite identification and pharmacokinetic comparison with its nanoparticle. J Pharm Sci. 92, 161–72 (2003).
Wang, K., Li, M. & Bucan, M. Pathway-based approaches for analysis of genomewide association studies. Am J Hum Genet. 81, 1278–1283 (2007).
Chabbert-Buffet, N., Meduri, G., Bouchard, P. & Spitz, I. M. Selective progesterone receptor modulators and progesterone antagonists: mechanisms of action and clinical applications. Hum Reprod Update. 11, 293–307 (2005).
Hernández, J. et al. Metallothionein induction by restraint stress: role of glucocorticoids and IL-6. Cytokine. 12, 791–796 (2000).
Tayel, S. S. et al. Progesterone suppresses interferon signaling by repressing TLR-7 and MxA expression in peripheral blood mononuclear cells of patients infected with hepatitis C virus. Arch Virol. 158, 1755–1764 (2013).
Shibata, M. et al. Glucocorticoids enhance Toll-like receptor 2 expression in human keratinocytes stimulated with propionibacteriumacnes or proinflammatory cytokines. J Invest Dermatol. 129, 375–382 (2009).
Jang, J. H. et al. RU486, a glucocortcoid receptor antagonist, induces apoptosis in U937 human lymphoma cells through reduction in mitochondrial membrane potential and activation of p38 MAPK. Oncol Rep. 30, 506–512 (2013).
Meyer, G., Leipprandt, J., Xie, J., Aupperlee, M. D. & Haslam, S. Z. A potential role of progestin-induced laminin-5/α6-integrin signaling in the formation of side branches in the mammary gland. Endocrinology. 153, 4990–5001 (2012).
Ebrahem, Q., Minamoto, A., Hoppe, G., Anand-Apte, B. & Sears, J. E. Triamcinolone acetonide inhibits IL-6 and VEGF-induced angiogenesis downstream of the IL-6 and VEGF receptors. Invest Ophthalmol Vis Sci. 47, 4935–4941 (2006).
Zhou, R. et al. Blockage of progesterone receptor effectively protects pancreatic islet beta cell viability. Steroids. 78, 987–995 (2013).
Schreiber, G. & Walter, M. R. Cytokine-receptor interactions as drug targets. Curr Opin Chem Biol. 14, 511–519 (2010).
Eid, M. A., Lewis, R. W. & Kumar, M. V. Mifepristone pretreatment overcomes resistance of prostate cancer cells to tumor necrosis factor alpha-related apoptosis-inducing ligand (TRAIL). Mol Cancer Ther. 1, 831–840 (2002).
Verhoog, N. J., Du-Toit, A., Avenant, C. & Hapgood, J. P. Glucocorticoid-independent repression of tumor necrosis factor (TNF) alpha-stimulated interleukin (IL)-6 expression by the glucocorticoid receptor: a potential mechanism for protection against an excessive inflammatory response. J Biol Chem. 286, 19297–19310 (2011).
Hyder, S. M., Chiappetta, C., Stancel, G. M. Pharmacological and endogenous progestins induce vascular endothelial growth factor expression in human breast cancer cells. Int J Cancer. 92, 469–473 (2001).
Herbst, R. S. Review of epidermal growth factor receptor biology. Int J Radiat Oncol Biol Phys. 59, 21–26 (2004).
Zhou, Y. et al. Blockade of EGFR and ErbB2 by the novel dual EGFR and ErbB2 tyrosine kinase inhibitor GW572016 sensitizes human colon carcinoma GEO cells to apoptosis. Cancer Res. 66, 404–411 (2006).
Grasso, A. W. et al. ErbB kinases and NDF signaling in human prostate cancer cells. Oncogene. 15, 2705–2716 (1997).
Ewing, T. M. et al. Regulation of epidermal growth factor receptor by progestins and glucocorticoids in human breast cancer cell lines. Int J Cancer. 44, 744–752 (1989).
Hernández-Hernández, O. T., González-García, T. K. & Camacho-Arroyo, I. Progesterone receptor and SRC-1 participate in the regulation of VEGF, EGFR and Cyclin D1 expression in human astrocytoma cell lines. J Steroid Biochem Mol Biol. 132, 127–134 (2012).
Janneau, J. L. et al. Transcriptional expression of genes involved in cell invasion and migration by normal and tumoral trophoblast cells. J Clin Endocrinol Metab. 87, 5336–5339 (2002).
Ferretti, C., Bruni, L., Dangles-Marie, V., Pecking, A. & Bellet, D. Molecular circuits shared by placental and cancer cells and their implications in the proliferative, invasive and migratory capacities of trophoblasts. Hum Reprod Update. 13, 121–141 (2007).
Tcacencu, I. Mifepristone (RU-486) impairs post-surgical wound healing of the larynx. Med. Sci. Monit. 8, BR397–400 (2002).
Brandhagen, B. N. et al. Cytostasis and morphological changes induced by mifepristone in human metastatic cancer cells involve cytoskeletal filamentous actin reorganization and impairment of cell adhesion dynamics. BMC Cancer. 13, 35 (2013).
Parri, M. & Chiarugi, P. Rac and Rho GTPases in cancer cell motility control. Cell Commun Signal. 8, 23 (2010).
Acknowledgements
This work was supported by the Grant of Ministry of Science & Technology of China (2015CB931804), Fujian Development and Reform Commission (2014/168), National Scientific Fund of China (No.81272548) and Fuzhou University Start-up fund. We also wish to express our gratitude to Shanghai Sensichip Co Ltd for bioinformatics analysis.
Author information
Authors and Affiliations
Contributions
L.J., S.Y. and P.J.S. conceived and designed the experiments. C.Y. and F.M. performed cell culture experiments. X.Y., Y.Z. and G.C. carried out the cell migration and adhesion experiments, Y.L. and T.Y. performed the Real-time PCR and western blot experiments, S.Y. and Y.G. conducted the immunoprecipitation experiments. S.Y. and X.Y. acquired and analyzed the experimental data. F.X. and J.S. conducted some experiments and discussed the results. L.J. and S.Y. wrote the manuscript. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Systems pharmacology of mifepristone (RU486) reveals its 47 hub targets and network: Comprehensive analysis and pharmacological focus on FAK-Src-Paxillin complex
Supplementary Information
Supplementary Dataset 1
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Yu, S., Yang, X., Zhu, Y. et al. Systems pharmacology of mifepristone (RU486) reveals its 47 hub targets and network: Comprehensive analysis and pharmacological focus on FAK-Src-Paxillin complex. Sci Rep 5, 7830 (2015). https://doi.org/10.1038/srep07830
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep07830
This article is cited by
-
The traditional Chinese medicine Achyranthes bidentata and our de novo conception of its metastatic chemoprevention: from phytochemistry to pharmacology
Scientific Reports (2017)
-
Pharmacoproteomic analysis reveals that metapristone (RU486 metabolite) intervenes E-cadherin and vimentin to realize cancer metastasis chemoprevention
Scientific Reports (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.