Abstract
Head and neck squamous cell carcinoma (HNSCC) is the sixth most prevalent cancer in the world. HNSCC remains difficult to treat and despite advances in treatment, overall survival rate has modestly improved over the past several years. Poor survival rate is attributed to high frequency of local recurrence and distant metastasis. Cancer stem-like cells (CSCs) have been implicated in tumor recurrence and confer resistance to anti-cancer therapy treatment. In this study, we have characterized genes that are modulated in HNSCC-CSCs and can be targeted in future as potential therapeutics. CSCs were isolated from HNSCC cells (oralspheres) and examined for tumorigenicity in immunodeficient mice. We observed aggressive tumor growth with oralspheres as compared to parental cells. The CSC-derived tumors were grossly extremely vascularized and expressed VEGFR1. We next analyzed the molecular determinant of oralspheres. In addition to CD133 and Nanog, we observed significant higher expression of Notch1 protein in the oralspheres. There was differential expression of angiogenesis and invasive marker genes such as angiopoietin1, integrin β3, MMP9 and THBS1. Interestingly, c-Fos was upregulated in oralspheres as compared to parental cells. Our observations suggest that understanding the molecular determinant of oralspheres will help in developing future therapeutic modalities against treatment resistant HNSCC.
Similar content being viewed by others
Introduction
Head and neck squamous cell carcinoma (HNSCC) refers to a group of biologically similar cancers that start in the lip, tongue, oral cavity (mouth), nasal cavity (inside the nose), paranasal sinuses, pharynx and larynx. HNSCC is the sixth most common cancer worldwide and accounts for about 3–5% of all cancers in the United States. The American Cancer Society estimates 55,070 people will develop head and neck cancers in 2014. HNSCC patients often present with late stage tumors and the five year survival rate is less than 50%1. Poor survival rate is attributed to high frequency of local recurrence and distant metastases2.
Many types of solid tumors originate from a small population of cancer stem-like cells (CSCs) or tumor-initiating cells (from here CSCs) that are able to initiate and maintain tumor growth and progression. Subpopulations of CSCs have been identified in most tumors, including HNSCC. These CSCs are involved in cell growth, migration/invasion and apoptosis resistance, attributing to treatment resistance and metastasis leading to a poor clinical outcome3. However, the pathogenesis and biological significance of CSCs in HNSCC has not been well characterized. CSCs can offer new insights into primary tumor growth and metastatic progression. Targeting CSCs in HNSCC may lead to more effective therapies to reduce metastasis.
Several lines of evidence suggest that the process of epithelial-to-mesenchymal transition (EMT) generate cells with stem-like properties, CSCs4. These CSCs have the ability to self-renew and generate secondary tumors. These observations provide a critical connection between the induction of metastasis and the acquisition of stem-like properties in cancer cells undergoing EMT5,6. Overexpression of HIF-1α, Twist1 or Bmi1 confers stem-like properties and induces EMT in head and neck cancer cell lines7. Notch1 also plays an important role in EMT and CSCs8. Notch signaling is required for the maintenance of the CSC phenotype in breast cancer. Inhibition of Notch signaling has been shown to prevent the formation of secondary mammospheres from cell lines and primary patient samples9. It has been proposed that the Notch pathway has a role in tumor metastasis10,11. Nam and colleagues12 reported activation of the Notch pathway in a xenograft model of brain metastasis. Notch1 mutations occur in approximately 15% of patients with HNSCC, implicating a critical role of the Notch signaling pathway in CSCs and in the etiology of head and neck cancer13,14,15.
In the present study, we isolated and characterized sphere forming cells from human HNSCC cells (OSC19, Cal27 and JHU29) as a population of CSCs. The oralspheres form tumors in immunodeficient mice and express CSC marker genes such as CD133, Nanog and Notch1. The pathway specific gene expression profiling analysis indicated that oralspheres from OSC19 cells display differential expressions of several genes, including MMP9, angiopoietin1, integrinβ3, c-Fos and THBS1. These genes are involved in cell cycle, angiogenesis and EMT pathways. Our data suggests that the cellular pathways that are activated in oralspheres could potentially be targeted as novel therapies against head and neck cancer.
Results
Presence of cancer stem-like cells (CSCs) in HNSCC
CSCs are well established as being ‘migrating tumor initiating cells’ which contribute to metastatic spread. Several different techniques have been used to identify CSCs. We have used the in vitro spheroid colony formation method for separation of CSCs from the parental populations. We initially investigated whether NOK, OSC19, Cal27 and JHU29 cell lines grow as spheroid cultures. Single cell suspensions of cells (5,000 cells/well) were seeded on ultra-low adherent plate. After 10 days, the spheres were counted. Successful oralsphere populations were determined by >75 micron in size (Fig. 1a). The percentage of the cancer-like stem cells in parental cells was calculated by the number of oralspheres, equal to or larger than 75 microns, divided by the total number of cells plated. In our hands, we have observed OSC19 cells possess ~1.7%, Cal27 cells possess ~1.6% and JHU29 possess 0.6% of CSC population in total no. of parental cells. We did not observe any spheroid formation from NOK cell lines (Fig. 1b).
HNSCC possess cancer stem-like cell (CSC) populations.
(a) OSC19, Cal27 and JHU29 cells are seeded in serum-free media on ultra-low attachment plates and Oralspheres were formed. Scale bars represent 100 μm. (b) Spheres >75 μm diameter was counted after 10 days. The percentage of sphere forming cells was calculated by dividing the number of oralspheres by the number of cells seeded. The experiments were performed at least three times and data are presented here as mean ± standard errors.
Enhanced expression of CSC markers in oralspheres
We next examined CSC marker expression in oralspheres and compared with the expression levels in parental cells. CD133 is a glycosylated, ~120-kDa protein with five transmembrane domains and two large extracellular loops16. CD133+ phenotype is highly expressed on the tumor-initiating population of brain neoplasms17. Nanog is one of the key transcription factors that is involved in the maintenance of pluripotency and self-renewal in undifferentiated embryonic stem cells and are highly expressed in several CSCs18,19. We observed increased mRNA expression of CD133 and Nanog in oralspheres in comparison to parental cells in OSC19 and Cal27 (Fig. 2a and 2b). We also observed higher expression of Nanog in oralspheres as compared to parental cells by Western blot analysis (Fig. 2c). We could not detect Sox2 and Oct4 in the oralspheres.
HNSCC express CSC markers.
(a) qRT-PCR analyses of CD133 in parental and oralspheres from OSC19 and Cal27. Expression of CD133 was normalized to GAPDH. The results are presented as means of three different experiments with standard errors (**, p < 0.001). (b) qRT-PCR analyses of Nanog in parental and oralspheres from OSC19 and Cal27. Expression of Nanog was normalized to GAPDH. The results are presented as means of three different experiments with standard errors (*, p < 0.05). (c) Cell lysates from parental and oralspheres derived from OSC19 and Cal27 were subjected to Western blot analysis for Nanog using specific antibody. The blot was reprobed with an antibody to GAPDH for comparison of equal protein load. Note that cropped gel images are used in this figure and the gels were run under the same experimental conditions.
Oralspheres form tumors in NOD-SCID IL2Rgammanull (NSG) mice
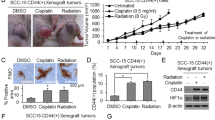
We, next, examined the ability to form tumor from parental and oralspheres of OSC19 cells in NSG mice. We injected 3 × 106 OSC19 cells and 1 × 104 OSC19 sphere forming cells into NSG mice (6 mice/gr). Tumor formation was observed from both cell lines ~16 days post-injection and tumor volume was measured. Tumor volume was significantly larger in oralspheres implantation group with buildup tumor interstitial fluid, although tumor size was similar between two groups (Fig. 3a and 3b). The tumors from oralspheres implantation group were found to be vascular and due to tumor condition, mice were sacrificed prematurely despite tumor volumes of ~350 mm3. Histopathology displayed poorly differentiated with areas of glandular differentiation in the tumors of both groups (Fig. 3c). Grossly, vascularization appeared enhanced in oralsphere tumors as compared to tumors of parental OSC19 cells. We also observed higher expression of VEGFR1 expression from oralspheres implanted tumors and the localization of VEGFR1 was found both in tumor and endothelial cells (Fig. 3d).
Oralspheres display higher tumorigenicity in NSG mice xenograft model.
(a) Parental and oralspheres from OSC19 cells were implanted subcutaneously into the flank of NSG mice. After ~16 days post injection, tumor formation was observed. Volume of tumor growth was monitored once a week. Data are presented here as the means and standard deviations (*p < 0.01). (b) Representative images of tumors extracted from control and oralspheres injected into NSG mice. (c) A representative tumor section from mice injected with parental and oralspheres from OSC19 cells, stained with hematoxylin and eosin (magnification, 40×) are shown. (d) A representative tumor section from mice injected with parental and oralspheres from OSC19 cells, stained with VEGFR1 (magnification, 40×) are shown.
Elevated Notch signaling in oralsphere
Emerging evidence suggests that cancer progression is often associated with the acquisition of EMT phenotype that is reminiscent of CSCs, which is partly responsible for the ability of HNSCC cells to acquire aggressiveness, contributing to tumor metastasis6,20. It has been postulated that Notch1 also plays an important role in EMT and CSCs8. We observed higher expression of Notch1 in oralspheres as compared to parental OSC19 cells (Fig. 4a). A higher level of Notch1 expression in patients with oral cancer (tongue origin) tissues as compared to adjacent non-tumor tissues was also observed (Fig. 4b). We also observed increased expression of JAG1, receptor ligand for Notch1, at mRNA level in oralspheres derived from OSC19 as compared to parental cells, although the upregulation was not significant in Cal27 oralspheres (Fig. 4c). To further confirm the Notch1 activation in oralspheres, we compared downstream activation of Notch1 signaling pathway driven by transcriptional changes between oralspheres and parental cells by qRT-PCR. Hes1 expression was upregulated in oralspheres derived from OSC19 as compared to parental cells (Fig. 4d).
Activation of Notch signaling in HNSCC.
(a) Cell lysates from parental and oralspheres from OSC19 and Cal27 were subjected to Western blot analysis for Notch1 using specific antibody. The blot was reprobed with an antibody to GAPDH for comparison of equal protein load. (b) Cell lysates were prepared from adjacent squamous normal epithelia and tumor sections of HNSCC derived patients. The expression level of Notch1 was examined by Western blot analysis using specific antibody. The blot was reprobed with an antibody to GAPDH for comparison of equal protein load. (c) qRT-PCR analyses of JAG1 in parental and oralspheres from OSC19 and Cal27 cells. Expression of JAG1 was normalized to GAPDH. The results are presented as means of three different experiments with standard errors (*, p < 0.05). (d) qRT-PCR analyses of Hes1 in parental and oralspheres from OSC19 cells. Expression of Hes1 was normalized to GAPDH. The results are presented as means of three different experiments with standard errors (*, p < 0.05). Note that cropped gel images are used in this figure and the gels were run under the same experimental conditions.
Oralspheres activate signaling pathways to promote cancer progression
CSCs play a critical role in tumorigenesis, chemotherapy resistance and metastasis. During oncogenesis, several biological pathways, such as apoptosis, cell cycle, DNA damage repair, signal transduction, angiogenesis and EMT are deregulated. We wanted to examine whether these pathways are further modulated in oralspheres as compared to parental cells. To accomplish this, a human cancer pathway profiler array was performed for identifying the modulation of cancer related genes in oralspheres. Several genes related to signal transduction, angiogenesis, migration and invasiveness were significantly upregulated in oralspheres as compared to parental OSC19 cells (Fig. 5a). Among them, angiopoietin1 (ANG1) plays an important role in vascular stabilization and angiogenesis in endothelial cells. ANG1 expression was 12 fold higher in oralspheres as compared to parental OSC19 cells. Integrin β3 (ITGB3) participates in cell adhesion and cell-surface mediated signaling. ITGB3 expression was 36 fold higher in oralspheres as compared to parental OSC19 cells. Positive correlation of ITGB3 and breast cancer stem cells has been reported21. Matrix metalloproteinases (MMPs) contribute to extracellular matrix (ECM) degradation and cancer cell migration22. MMP9 plays a key role in cell migration, invasion and metastasis. From the array, MMP9 expression was found to be 45 fold higher in oralspheres as compared to parental OSC19 cells. We validated the expression of MMP9 in control and oralspheres generated from two different cell lines, OSC19 and Cal27. Increased expression of MMP9 in oralspheres as compared to parental cells was observed (Fig. 5b). Slug plays a key role in EMT and cell migration in human head and neck squamous cell carcinoma23. Higher expression of Slug was also observed in oralspheres in comparison to parental cells (Fig. 5c). We observed downregulation of angiogenic suppressor gene THBS1 (also called TSP1). THBS1 overexpression in breast cancer cells suppresses metastatic outgrowth in lung24. Low expression of THBS1 has also been reported in prostate cancer CSCs25. Interestingly, c-Fos expression is ~20 fold higher in oralspheres as compared to parental OSC19 cells. Early oncogene c-Fos plays a pivotal role in cell growth regulation in association with c-JUN by forming AP-1 complex. We also observed higher expression of c-Fos in oralspheres as compared to parental cells at the protein level (Fig. 5d). Next, we used String Analysis to analyze the functional network of MMP9 and c-Fos signaling pathways (Fig 6). MMP9 play an essential role in local proteolysis of extracellular matrix and in cancer cell migration. String analysis suggest that Jun oncogene, N-cadherin (CDH2), Fos-like antigen1 (FOSL1) and THBS1 activate MMP9 expression by functional interactions. In general, c-Fos is known to be involved in signal transduction and cell proliferation in cancer cells. Jun oncogene, cAMP responsive element binding protein1 (CREB1), mitogen-activated protein kinase 8 (MAPK8), serum response factor (SRF) and prostaglandin-endoperoxide synthase 2 (PTGSS2) are suggested to be involved in activation of c-Fos signaling network. Taken together, genes identified in our human cancer pathway array indicated increased expression of highly metastatic genes in oralspheres in comparison to parental cells, implicating the CSCs characteristics and tumorigenic potential of oralspheres.
Modulation of genes related to cancer specific pathway in oralspheres.
(a) Gene expression profiling of cancer pathway finder specific array was performed using RNA from parental and oralspheres derived from OSC19 cells. OSC19 spheres displayed an increased expression of genes related to cell migration and metastasis (e.g., ANG1, ITGB3, MMP9 and c-Fos). (b) qRT-PCR analyses of MMP9 in control and oralspheres from OSC19 and Cal27. Expression of MMP9 is normalized to GAPDH. The results are presented as means of three different experiments with standard errors (**, p < 0.001). (c) Cell lysates were prepared from parental and oralspheres of OSC19 and Cal27. Expression of Slug was examined by Western blot using specific antibody. The blot was reprobed with an antibody to GAPDH for comparison of protein load. (d) Cell lysates were prepared from parental and oralspheres of OSC19 and Cal27. Expression of c-Fos was examined by Western blot using specific antibody. The blot was reprobed with an antibody to GAPDH for comparison of protein load. Note that cropped gel images are used in this figure and the gels were run under the same experimental conditions.
Discussion
CSCs play a critical role during tumor initiation and progression and therefore, the existence of CSCs has significant clinical implications; however, little is known about gene expression profiles of CSCs derived from oral cancers. In the present study, we found that a small proportion of HNSCC cells behave like CSCs. This subset of cells display aggressive phenotypes with increased expression of genes involved in angiogenesis and tumor invasiveness, which is consistent with overexpression of CSC markers. Therefore, understanding the pathway will help in developing future therapeutic approaches that selectively induce death of CSCs and may reduce the risk of metastasis in HNSCC.
One potential confounding factor of this study was the common expression of CD44 in both CSCs and the parental cells. Although some studies suggested CD44 positive cells may be a marker for HNSCC alone, we have observed a similar expression of CD44 in CSC and non-CSC forming cells (data not shown). While our manuscript was in preparation, Chinn et al3 reported similar observation regarding the status of CD44 expression in both cell types. CSCs did demonstrate differences in other important signaling pathways. We have observed higher expression of CD133 and Nanog in oralspheres in comparison to parental cells. Increased levels of CD133 could be a survival mechanism since it has been reported that CD133 expression is associated with activation of c-Src and required for maintenance of CSC phenotype through EMT modulation in head neck cancer26. Moreover, elevated expression of Nanog and CD133 has been shown to be positively associated with late-stage progression and worse prognosis in patients with oral cancer27. EMT association with colon cancer CSC has been reported recently28. HNSCC is a heterogeneous tumor and CSCs from HNSCC may have potential for metastasis, since we observed the migratory or invasive genes are overexpressed in oralspheres. Notch1 mutations occur in approximately 15% of patients with HNSCC, implicating a critical role of Notch signaling pathways in HNSCC tumors15. It has been suggested that Notch1 plays an important role in EMT and CSCs8. Development of Notch1 inhibiting therapies may be a potential therapeutic target in Notch1 activated HNSCC tumors. Notch 1 expression was significantly increased in the oralspheres in vitro and the HNSCC tumors in vivo as compared to adjacent non-tumor tissues. To date, the best characterized gene targets indicating Notch signaling pathway activation are members of the hairy and enhancer of split (HES) and the Hes related-repressor protein families of bHLH transcriptional repressors such as Hes1 and Hey115. We also observed activation of Hes1 in oralspheres as compared to parental cells. Together these results further suggested the activation of Notch1 signaling in oralspheres.
In our study, we observed higher expression of genes related to cell migration, angiogenesis and invasiveness such as, angiopoietin 1, Integrin β3 and MMP9 in oralspheres as compared to parental cells. Slug expression was found to be higher in oralspheres as compared to parental cells, implicating activation of EMT in oralspheres. Enhanced EMT characteristic is associated with a poor prognosis overall and metastatic free survival in patients with HNSCC7,29. High MMP9 protein levels have been observed in human oral cancers and MMP9 is a marker of malignant human oral cancer30,31. Enhanced expression of MMP9 was observed in oralspheres suggesting the possibility of higher rate of malignancy in HNSCC. Although a soluble secreted enzyme, MMP9 has been reported to localize at the cell surface via CD44 in murine mammary carcinoma cells and facilitate tumor invasion and angiogenesis by proteolytic activation of latent TGF-β32,33. We observed that tumors from oralspheres display higher level of VEGFR1. VEGFR1 expression and activation is associated with increased malignant potential in osteosarcoma cells34. We also observed downregulation of angiogenic suppressor gene, THBS1. Interestingly, it has been shown that absence of THBS1 resulted in increased association of VEGF with its receptor VEGFR2 and higher levels of active MMP9, to facilitate both angiogenesis and tumor invasion35. In addition, THBS1 suggested being a negative regulator of tumor vasculature which is supported by higher tumor vascularization in tumors from oralspheres by our gross observation. Surprisingly, we observed the overexpression of c-Fos in oralspheres. AP-1 complex formed by association between c-Fos and c-Jun is recently implicated in maintenance of colon cancer stem cells36. The transcription of MMP9 gene is regulated by upstream regulatory elements, such as AP-1, STAT3 and NF-κB binding sites37,38,39. It is possible that c-Fos can associate with MMP9 promoter through AP1 transcription factor to promote tumor cell invasion in oralspheres. Further studies need to be performed to elucidate the role of these factors in HNSCC CSCs. In conclusion, our studies suggest the enhanced expression of molecular signatures associated with tumor progression, invasion and metastasis in CSCs derived from HNSCC and development of drugs targeting CSCs needs further investigation.
Methods
Cell Culture
Cal27 cells (tongue) were recently obtained from ATCC. Cal27 cells were maintained in Dulbecco's Modified Eagle Medium (Sigma) supplemented with 10% FBS and 1% penicillin/streptomycin (Sigma). JHU29 (tongue) cells were recently procured from the Johns Hopkins University40. JHU29 cells were maintained in RPMI-1640 Medium (Sigma) supplemented with 10% FBS and 1% penicillin/streptomycin. NOK cells (kindly provided by K. Mugner) are grown in Keratinocyte SFM medium supplemented with EGF and bovine pituitary extract (Gibco, Life technologies). OSC19 cells (kindly provided by J. Myers) were maintained in G-DMEM containing high glucose (Life technologies) supplemented with 1 × NEAA, 1 × Pyruvate, 1 × Vitamin, 2 mM Glutamine (Life technologies) and 10% FBS with penicillin (100 U/ml) and streptomycin (100 μg/ml) (Sigma-Aldrich).
Sphere formation assay
Single cell suspensions of OSC19, Cal27 and JHU29 cells (5000 cells/well) were cultured on ultralow attachment plates (Corning, Lowell, MA). Cells were grown in a serum-free growth medium supplemented with EGF and FGF. After 10 days of incubation, percentage of sphere forming cells were calculated by dividing the number of oralspheres by the number of cells seeded. Spheres >75 μm diameter were counted after 10 days. Primary oralspheres were dissociated mechanically and enzymatically using Accutase (Gibco, life technologies) to break up sphere clusters and generate single cell suspension, counted and re-seeded to generate secondary and tertiary spheres.
Real time RT-PCR
TRIzol reagent (Life technologies) was used to extract total RNA from control or oralspheres according to the manufacturer's instructions. 1 μg of RNA was reverse-transcribed using superscript III reverse transcription (Invitrogen, Life technologies). Power SYBR Green Assay kit (Applied Biosystems) was used for real-time PCR reaction, following manufacturer's protocol. Sequences of PCR primers were listed in Table 1. Data were analyzed using Ct method and normalized by glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression in each sample.
Western blot analysis
Cell lysates were prepared from parental and oralspheres derived from each of Cal27 and OSC19 cell lines. Proteins were separated by SDS-PAGE and transferred onto 0.45 μM nitrocellulose membrane. Membranes were blocked using 5% low fat dry milk in TBST and probed with the following primary antibodies. Proteins were detected using ECL Western Blotting Substrate (Thermo Scientific) and autoradiography. Protein loads were normalized using antibody for GAPDH (Cell Signaling Technologies). The following antibodies were used in this study: Nanog, Notch1, Slug (Cell Signaling Technologies) and c-Fos (Santa Cruz Biotechnology).
Human Cancer Pathway Finder Array Profiling
Total RNA was extracted from control and oralspheres isolated from OSC19 cells. A RT2 profiler PCR Array for human cancer pathway finder (Qiagen) to analyze the modulation of 84 cancer related genes was performed as described previously40. Array data was analyzed using free web based software (http://pcrdataanalysis.sabiosciences.com/pcr/arrayanalysis.php) and automatically perform all ΔΔCt fold change calculations.
Tumor xenograft in NSG mice
All animal experiments were performed in accordance with a protocol approved by the Institutional Animal Care and Use Committee of Saint Louis University. A total of 3 × 106 parental OSC19 cells and 1 × 104 OSC19 oralspheres were resuspended in PBS with 40% Matrigel composition for subcutaneous injections. Tumor xenografts were established in five week old female NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ mice, commonly known as NOD SCID gamma (NSG) mice (Jackson Laboratory) by direct injection of cells into the flanks. Tumor growth was measured weekly till the end of experiments. A caliper was used for tumor measurements and tumor volume was calculated using the formula (L × W2) × 0.5. Mice were sacrificed and tumors were harvested for histopathology and biochemical assays.
String analysis
Protein-protein interactions were predicted using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database v9.0 (http://www.string-db.org/). Proteins were linked based on the following six criteria; neighborhood, gene fusion, co-occurrence, co-expression, experimental evidence and existing databases41.
References
Siegel, R., Ward, E., Brawley, O. & Jemal, A. Cancer statistics, 2011: the impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA. Cancer J. Clin. 61, 212–36 (2011).
Sahu, N. & Grandis, J. R. New advances in molecular approaches to head and neck squamous cell carcinoma. Anticancer Drugs. 22, 656–64 (2011).
Chinn, S. B. et al. Cancer stem cells: Mediators of tumorigenesis and metastasis in head and neck squamous cell carcinoma. Head Neck (2014).
Mani, S. A. et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 133, 704–715 (2008).
Scanlon, C. S., Van Tubergen, E. A., Inglehart, R. C. & D'Silva, N. J. Biomarkers of epithelial-mesenchymal transition in squamous cell carcinoma. J. Dent. Res. 92, 114–121 (2013).
Chen, C., Zimmermann, M., Tinhofer, I., Kaufmann, A. M. & Albers, A. E. Epithelial-to-mesenchymal transition and cancer stem(-like) cells in head and neck squamous cell carcinoma. Cancer Lett. 338, 47–56 (2013).
Yang, M. H. et al. Direct regulation of TWIST by HIF-1alpha promotes metastasis. Nat Cell Biol. 10, 295–305 (2008).
Takebe, N., Harris, P. J., Warren, R. Q. & Ivy, S. P. Targeting cancer stem cells by inhibiting Wnt, Notch and Hedgehog pathways. Nat. Rev. Clin. Oncol. 8, 97–106 (2011).
Grudzien, P. et al. Inhibition of Notch signaling reduces the stem-like population of breast cancer cells and prevents mammosphere formation. Anticancer Res. 30, 3853–67 (2010).
Bin Hafeez, B. et al. Targeted knockdown of Notch1 inhibits invasion of human prostate cancer cells concomitant with inhibition of matrix metalloproteinase-9 and urokinase plasminogen activator. Clin. Cancer Res. 15, 452–459 (2009).
Hu, X. B. et al. Blockade of Notch signaling in tumor-bearing mice may lead to tumor regression, progression, or metastasis, depending on tumor cell types. Neoplasia. 11, 32–38 (2009).
Nam, D. H. et al. Activation of notch signaling in a xenograft model of brain metastasis. Clin. Cancer Res. 14, 4059–4066 (2008).
Agrawal, N. et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 333, 1154–1157 (2011).
Stransky, N. et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 333, 1157–1160 (2011).
Sun, W. et al. Activation of the NOTCH pathway in head and neck cancer. Cancer Res. 74, 1091–1104 (2014).
Keysar, S. B. & Jimeno, A. More than markers: biological significance of cancer stem cell-defining molecules. Mol. Cancer Ther. 9, 2450–2457 (2010).
Singh, S. K. et al. Identification of human brain tumour initiating cells. Nature. 432, 396–401 (2004).
Wang, J. et al. A protein interaction network for pluripotency of embryonic stem cells. Nature. 444, 364–368 (2006).
Prud'homme, G. J. Cancer stem cells and novel targets for antitumor strategies. Curr Pharm Des. 18, 2838–2849 (2012).
Fessler, E., Dijkgraaf, F. E., De Sousa, E., Melo, F. & Medema, J. P. Cancer stem cell dynamics in tumor progression and metastasis: is the microenvironment to blame? Cancer Lett. 341, 97–104 (2013).
Liu, C. & Tang, D. G. MicroRNA regulation of cancer stem cells. Cancer Res. 71, 5950–5954 (2011).
Munshi, H. G. & Stack, M. S. Reciprocal interactions between adhesion receptor signaling and MMP regulation. Cancer Metastasis Rev. 25, 45–56 (2006).
Smith, A., Teknos, T. N. & Pan, Q. Epithelial to mesenchymal transition in head and neck squamous cell carcinoma. Oral Oncol. 49, 287–292 (2013).
Ghajar, C. M. et al. The perivascular niche regulates breast tumour dormancy. Nat. Cell. Biol. 15, 807–817 (2013).
Oktem, G. et al. Cancer stem cell differentiation: TGFβ1 and versican may trigger molecules for the organization of tumor spheroids. Oncol Rep. 32, 641–649 (2014).
Chen, Y. S. et al. CD133/Src axis mediates tumor initiating property and epithelial-mesenchymal transition of head and neck cancer. PLoS One. 6, e28053 (2011).
Chiou, S. H. et al. Positive correlations of Oct-4 and Nanog in oral cancer stem-like cells and high-grade oral squamous cell carcinoma. Clin Cancer Res. 14, 4085–95 (2008).
Han, X. Y. et al. Epithelial-mesenchymal transition associates with maintenance of stemness in spheroid-derived stem-like colon cancer cells. PLoS One. 8, e73341 (2013).
Mandal, M. et al. Epithelial to mesenchymal transition in head and neck squamous carcinoma: association of Src activation with E-cadherin down-regulation, vimentin expression and aggressive tumor features. Cancer. 112, 2088–2100 (2008).
Fan, H. X., Li, H. X., Chen, D., Gao, Z. X. & Zheng, J. H. Changes in the expression of MMP2, MMP9 and ColIV in stromal cells in oral squamous tongue cell carcinoma: relationships and prognostic implications. J. Exp. Clin. Cancer Res. 31, 90 (2012).
Patel, B. P., Shah, S. V., Shukla, S. N., Shah, P. M. & Patel, P. S. Clinical significance of MMP-2 and MMP9 in patients with oral cancer. Head Neck. 29, 564–572 (2007).
Bourguignon, L. Y. et al. CD44v (3, 8–10) is involved in cytoskeleton-mediated tumor cell migration and matrix metalloproteinase (MMP9) association in metastatic breast cancer cells. J. Cell Physiol. 176, 206–215 (1998).
Yu, Q. & Stamenkovic, I. Cell surface-localized matrix metalloproteinase-9 proteolytically activates TGF-beta and promotes tumor invasion and angiogenesis. Genes Dev. 14, 163–176 (2000).
Ohba, T. et al. Autocrine VEGF/VEGFR1 Signaling in a subpopulation of cells associates with aggressive asteosarcoma. Mol. Cancer Res. 12, 1100–1111 (2014).
Rodriguez-Manzaneque, J. C. et al. Thrombospondin-1 suppresses spontaneous tumor growth and inhibits activation of matrix metalloproteinase-9 and mobilization of vascular endothelial growth factor. Proc. Natl. Acad. Sci. U. S. A. 98, 12485–12490 (2001).
Apostolou, P. et al. AP-1 Gene Expression Levels May Be Correlated with Changes in Gene Expression of Some Stemness Factors in Colon Carcinomas. Signal Transduct. 2013, 497383 (2013).
Troussard, A. A. et al. The integrin linked kinase (ILK) induces an invasive phenotype via AP-1 transcription factor-dependent upregulation of matrix metalloproteinase 9 (MMP9). Oncogene. 19, 5444–5452 (2000).
Schröer, N., Pahne, J., Walch, B., Wickenhauser, C. & Smola, S. Molecular pathobiology of human cervical high-grade lesions: paracrine STAT3 activation in tumor-instructed myeloid cells drives local MMP9 expression. Cancer Res. 71, 87–97 (2011).
Tai, K. Y., Shieh, Y. S., Lee, C. S., Shiah, S. G. & Wu, C. W. Axl promotes cell invasion by inducing MMP9 activity through activation of NF-kappaB and Brg-1. Oncogene. 27, 4044–4055 (2008).
Rajamoorthi, A. et al. Bitter melon reduces head and neck squamous cell carcinoma growth by targeting c-Met signaling. PLoS One. 8, e78006 (2013).
Franceschini, A. L. et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 41 (2013).
Acknowledgements
This work was supported by Cancer Center seed grant and Doisy Research Fund from Saint Louis University Pathology Department. The authors thank Dr. Karl Mugner for providing NOK cell line, Dr. Jeffrey Myers for providing OSC19 cell line and Philip Fitchev for technical assistance.
Author information
Authors and Affiliations
Contributions
S.S., R.S., M.S. and R.R. performed the experiments and all authors wrote the manuscript. S.C. performed the histopathology and M.V. provided us the HNSCC samples. All the authors read and approved the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Shrivastava, S., Steele, R., Sowadski, M. et al. Identification of molecular signature of head and neck cancer stem-like cells. Sci Rep 5, 7819 (2015). https://doi.org/10.1038/srep07819
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep07819
This article is cited by
-
Cancer stem cells and oral cancer: insights into molecular mechanisms and therapeutic approaches
Cancer Cell International (2020)
-
Cannabidiol enhances cytotoxicity of anti-cancer drugs in human head and neck squamous cell carcinoma
Scientific Reports (2020)
-
Depletion of PCAT-1 in head and neck cancer cells inhibits tumor growth and induces apoptosis by modulating c-Myc-AKT1-p38 MAPK signalling pathways
BMC Cancer (2019)
-
Prognostic implication of NOTCH1 in early stage oral squamous cell cancer with occult metastases
Clinical Oral Investigations (2018)
-
Induction of artificial cancer stem cells from tongue cancer cells by defined reprogramming factors
BMC Cancer (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.