Abstract
High-risk human papillomavirus (HPV) type 16, which is responsible for greater than 50% of cervical cancer cases, is the most prevalent and lethal HPV type. However, the molecular mechanisms of cervical carcinogenesis remain elusive, particularly the early steps of HPV infection that may transform normal cervical epithelium into a pre-neoplastic state. Here, we report that a group of microRNAs (microRNAs) were aberrantly decreased in HPV16-positive normal cervical tissues and these groups of microRNAs are further reduced in cervical carcinoma. Among these miRNAs, miR196a expression is the most reduced in HPV16-infected tissues. Interestingly, miR196a expression is low in HPV16-positive cervical cancer cell lines but high in HPV16-negative cervical cancer cell lines. Furthermore, we found that only HPV16 early gene E5 specifically down-regulated miRNA196a in the cervical cancer cell lines. In addition, HoxB8, a known miR196a target gene, is up-regulated in the HPV16 cervical carcinoma cell line but not in HPV18 cervical cancer cell lines. Various doses of miR196a affected cervical cancer cell proliferation and apoptosis. Altogether, these results suggested that HPV16 E5 specifically down-regulates miR196a upon infection of the human cervix and initiates the transformation of normal cervix cells to cervical carcinoma.
Similar content being viewed by others
Introduction
Cervical cancer is the third most commonly diagnosed cancer and the fourth leading cause of cancer death in females worldwide, accounting for 9% of total new cancer cases and 8% of total cancer deaths among females in 20081. Similar to other malignant tumors, cervical cancer results from a multistep process that includes the transformation of the normal cervical epithelium to a preneoplastic state followed by cervical intraepithelial neoplasia (CIN) and ultimately invasive cervical cancer2. The transformation of the normal cervical epithelium to cervical carcinoma is most likely caused by human papillomavirus (HPV)s, which are double-stranded DNA viruses that induce epithelial lesions. Low-risk HPV types only induce benign genital warts, whereas high-risk HPV types are highly associated with cervical cancers. High-risk HPV16 is the most prevalent type, followed by types 18, 31, 33 and 353,4. According to their expression in the epithelial differentiation stage of epithelium, E1, E2, E5, E6 and E7 viral genes are classified as exhibiting early stage expression3. E1 and E2 regulate viral gene expression via their enzymatic activities in replication and transcriptions5,6. The oncogenic potential of high risk HPV is mostly attributed to three early genes: E5, E6 and E7. E6 and E7 exert their oncogenic effect by destabilizing and degrading pRB and p537,8,9,10,11,12. E5 encodes a hydrophobic protein that localizes to host cell membranes. E5 is believed to play a role during the early steps of transformation in the basal layers of the epithelium and enhance the oncogenic effect of E6 and E7. However, its mechanistic role is not clear during cervical carcinogenesis13,14,15. Interestingly, some cervical cancer cell lines, including HPV18-infected HeLa cells, exhibited a deleted E5 DNA region in the host cell's genome. This observation suggests that different HPVs potentially promote cervical cancer development through different signaling pathways.
Recent studies have indicated that miRNAs are aberrantly expressed in different tumor tissues at various developmental stages16,17,18,19,20 and are considered to be master regulators of many important biological processes18,21,22,23,24,25, suggesting that miRNAs might serve as promising biomarkers for outcome prediction or potential therapeutic targets of a wide array of human cancers26,27,28. Aberrant miRNA expression has been reported in cervical cancer tissues or cell lines29,30,31, but the role of miRNAs in cervical cancer remains unclear due to inconsistencies in miRNA expression reported in different studies. miR196a has recently been reported to be aberrantly expressed in cervical carcinoma and cervical cancer cell lines, but heterogeneous expression levels were reported in several studies32,33. It is likely that these studies did not separate HPV types during their analysis. Moreover, studies regarding the relationship between the premalignant state of cervical tissues infected by specific types of HPVs and miRNA expression are necessary to understand whether miRNA expression is affected during the initial stage of cellular transformation.
In this study, we first analyzed miRNA expression in cervical tissues collected from patients. We focused on the most prevalent HPV type (HPV16) to eliminate potentially heterogeneous expression due to different HPV subtypes. We identified a group of miRNAs with reduced expression specifically in HPV16-positive normal cervices and further reduced in HPV16 squamous cervical carcinoma. Surprisingly, we found that miR196a expression was specifically regulated by HPV early gene E5 and our further analysis indicates that miR196a is involved in regulating cell proliferation, growth and apoptosis.
Results
Aberrant microRNA expression in normal cervix, HPV16-positive normal cervix and HPV16-positive cervical carcinoma
Forty-one pathologically normal cervices were collected from patients who underwent hysterectomy for uterine leiomyoma or adenomyosis. Ten normal HPV16-positive cervix samples and ten normal HPV-negative cervix samples were randomly selected for the following analysis. Cervical cancer tissues were collected from patients who underwent a radical hysterectomy for squamous cell carcinoma of cervix (FIGO Ib stage). Nine of the cervical carcinoma tissues were found to be infected with high-risk HPV16. To determine miRNA expression in these samples, we performed microarray hybridization on microchips containing 435 mature human miRNAs. Ten HPV-negative normal cervix samples (normal control) were divided into two pools of five samples to eliminate individual differences in miRNA expression. Hence, microarray hybridization was performed on miRNA samples from two pools of HPV-negative normal cervix samples, ten HPV16-positive normal cervix samples and nine HPV16-positive cervical cancer samples. No significant differences in age were noted among these three groups.
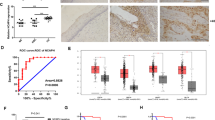
The hybridization intensity of HPV16-positive normal cervix (HPV) and cancerous (CA) samples was compared to that of HPV-negative normal pools (Np). Differential expression profiles were obtained using SAM program34. Only one miRNA, miR-193a, was significantly increased in cervical cancer tissues compared with normal cervices (P < 0.01; Fig. 1A). In contrast, eighteen miRNAs (133b, 154, 218,127, 503, 204, 369-5p, 128a, 196a, 329, 33, 382, 133a, 379, 450, 376b, 299-5p and 495) were significantly decreased in the cancer samples (P < 0.05 all; Fig. 1A). Among them, nine miRNAs (133b, 154, 196a, 329, 369-5p, 127, 133a, 495 and 503) were decreased in every cancer sample examined (Fig. 1A). When normal cervix samples were compared with HPV16-positive normal cervix samples, only three microRNAs (miR196a, 133a and 133b) were identified as significantly decreased (Fig. 1A). Interestingly, all three of these microRNAs were also decreased in all cervical cancer tissues collected. To validate these results using an independent assays, we performed quantitative real-time PCR to determine miR-133b, 154, 196a, 329, 369-5p, 127, 133a, 495 and 503 expression by pooling the same sets of samples: HPV-negative normal cervices (n = 10), HPV16-positive normal cervices (n = 10) and HPV16-positive cervical cancer cervices (n = 9). Consistent with the microarray results, all nine miRNAs were significantly decreased in the cervical cancer group compared with the HPV-negative normal cervix group and only three miRNAs were decreased in HPV16-positive cervices compared with HPV-negative normal cervices (Figure 1B). Among the three miRNAs, miR196a exhibited the most significant decreased in expression in normal cervices compared with HPV16-positive infected cervices and cervical cancer cervices. We chose to further investigate the role of miR196a because it exhibited the highest reduction in both HPV16-positive normal cervices and cervical carcinoma cervices compared with HPV-negative normal cervices.
Aberrant miRNA expression in HPV16 cervical tissues.
(A) A heatmap of the microarray analysis indicating miRNAs that were differentially expressed in ten HPV16-positive cervix samples (HPV) or nine HPV16-positive cervical carcinoma samples (CA) compared with two pools of HPV-negative normal cervix samples (NP). The average fold change in miRNAs that were significantly differentially expressed in HPV16-positive cervices compared with normal cervices or HPV16-positive cervical carcinoma samples compared with normal cervices in microarray analysis (P < 0.05). (B) Relative expression of miRNAs in HPV-negative normal cervix, HPV16-positive cervix and HPV16-positive cervical carcinoma samples using qRT-PCR of pooled samples.
Low expressions of miR-196a in HPV16-positive cervical cancer cell lines
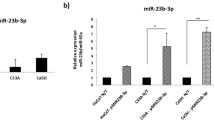
To determine whether the differential expression of miR196a also occurred in cervical cancer cell lines, we examined its expression in two HPV16-positive cell lines (CaSki and SiHa), one HPV18-positive cell line (HeLa) and one HPV-negative cell line (C33-a). miR196a expression in both CaSki and SiHa cell lines were significantly reduced compared with HeLa and C33-a (P < 0.001; Fig. 2). When the expression of miR196a was normalized to C33-a cells, the expression was increased 4.3-fold in HeLa cells. In contrast, miR196a was 4.2-fold and 19.2-fold reduced in CaSki and SiHa cells, respectively. Hence, miR196a expression was specifically reduced in both HPV16-positive cell lines examined. This finding is consistent with the reduced miR196a expression noted in our HPV16-positive cervical carcinoma samples.
HPV16 E5 specifically reduces miR196a levels
The data presented above indicate that miR196a is specifically down-regulated in HPV16 cervical carcinoma samples and HPV16-positive cell lines, but the mechanism by which HPV16 infection affects miR196a levels is unclear. To test whether the HPV16 early genes E5, E6, or E7 participate in this process, we examined miR196a expression upon modulation of E5, E6, or E7 levels in HeLa or SiHa cells. Although both cell lines contain integrated HPV DNA, the E5, E6 and E7 sequences vary between HPV16 and HPV18. Moreover, a segment of HPV18, including E5, is deleted from the integrated HPV18 DNA in the HeLa cell genome35,36. Thus, the effect of HPV16-E5 can be evaluated in isolation when HPV16-E5 is expressed in HeLa cells, whereas the effects of HPV16-E6 and HPV16-E7 are evaluated in the presence of their HPV18-homologs. We constructed vectors to overexpress each HPV16 gene and each gene is expressed separately from the corresponding ectopic vector. Therefore, the potential effect on miR196a expression levels can be evaluated independently. HPV-E2 and HPV-E5 are bicistronic, but previous study indicated that HPV-E2 mainly functions in regulating the transcription of viral genes5. Nevertheless, we also examined the effect of HPV16-E2 in the overexpression experiments. When HPV16-E5, -E6 and -E7 were overexpressed in HeLa cells, E5 exclusively exhibited a significant reduction in miR196a expression in HeLa cells (P < 0.01; Fig. 3A and B). As measured by RT-PCR, RNA transcript levels were not significantly altered by adding DNase before the cDNA preparation step (+gDNA Eraser) and no transcript can be detected without addition of reverse transcriptase during cDNA preparation (No RT), indicating the purity of the RNA we collected from the cells (Supplementary Fig. 1A). In addition, we confirmed that HPV16-E2 did not cause a significant reduction of miR196a expression (Supplementary Fig. 1B, 1C), indicating the independent roles of HPV16-E2 and HPV16-E5.
E5 down-regulates miR196a in HeLa cells and SiHa cells.
(A) Relative expression of E5, E6 and E7 in HeLa cells with and without overexpression of these genes. (B) Normalized expression level of miR196a in HeLa cells overexpressing E5, E6, or E7. (C) Relative expression levels of E5, E6 and E7 in SiHa cells upon shRNA-mediated silencing of these genes. (D) Normalized expression levels of miR196a upon E5, E6, or E7 silencing by shRNAs. GAPDH was used to normalize relative mRNA expressions.
To verify the effects of E5, E6 and E7 in SiHa cells, we constructed shRNA vectors that can efficiently silence each gene. Three shRNAs significantly down-regulated E6 and E7, whereas only two shRNAs were identified that exhibited significant silencing due to the short E5 coding region (Supplementary Fig. 2). We used E5 shRNA-2, E6 shRNA-1 and E7 shRNA-2 in the subsequent experiments. Given that E2 and E5 as well as E6 and E7 are bicistronic, the silencing of one gene might also silence the other gene on the same transcript. Indeed, shRNA targeting E5 transcripts silenced both E2 and E5 and shRNA targeting E6 or E7 silenced both E6 and E7 (Supplementary Fig. 3A, 3C, 3E). The silencing effects of these shRNAs persisted for at least 72 hours after transfection (Supplementary Fig. 3B, 3D, 3F). Hence, the effects of shRNAs targeting E5, E6 and E7 should be evaluated with their bicistronic genes. When E5, E6 and E7 were 7.4-fold, 4.9-fold and 5.6 fold reduced, respectively, E5 silencing up-regulated miR196a levels 7.0-fold compared with the control in SiHa cells (P < 0.01; Fig. 3C and D). These results suggested that silencing of both E2 and E5 genes potentially increases miR196a expression, but silencing of both E6 and E7 did not alter miR196a levels.
HPV16 E5 and miR196a regulate HOXB8 expression
If HPV16 E5 specifically reduces miR196a expression, the downstream target of miR196a may also be affected. HOXB8 was reported to be one of the targets of miR196a37 and the aberrant expression of HOXB8 genes is highly associated with various cancers38,39. To determine whether HOXB8 expression is regulated by miR196a, we examined HOXB8 expression after altering miR196a levels in HeLa or SiHa cells. To confirm the specificity of the miR196a measurements and to modulate miR196a levels, we transfected a miR196a inhibitor and mimic into HeLa cells and SiHa cells. miR196a levels were significantly decreased when HeLa cells were treated with the inhibitor and slightly increased when HeLa cells were treated with the mimic due to the relatively high level of miR196a expression in these cells (Supplementary Fig. 4A). In contrast, miR196a expression was significantly increased in mimic-treated SiHa cells, whereas miR196a levels were slightly decreased upon inhibitor treatment due to the low level of miR196a expression in these cells (Supplementary Fig. 4A). HOXB8 expression was significantly increased 7.7-fold after miR196a was down-regulated in HeLa cells (P < 0.01; Fig. 4A), whereas HOXB8 expression was reduced by 2.9-fold when miR196a was up-regulated in SiHa cells (P < 0.01; Fig. 4A). HPV16 E5 up-regulation also increased HOXB8 expression in HeLa cells, whereas E6 and E7 had no effect (P < 0.01; Fig. 4B). Furthermore, increased HPV16 E2 expression also did not affect HOXB8 levels in HeLa cells (Supplementary Fig. 1D). In contrast, HPV16 E5 silencing exclusively reduced HOXB8 expression in SiHa cells (P < 0.01; Fig. 4C). This silencing experiment could not exclude the effect of HPV16 E2 in SiHa cells given the bicistronic nature of the E2 and E5 genes. However, considering the results of the overexpression and silencing experiments, it is most likely that up-regulation of HPV16 E5 decreased the expression of miR196a and increased the expression of HOXB8.
HOXB8 is specifically up-regulated when miR196a is silenced or when E5 is overexpressed.
(A) Relative expression of HOXB8 in HeLa or SiHa cells upon miR196a silencing or overexpression. (B) Relative HOXB8 expression level in HeLa cells upon E5, E6, or E7 overexpression. (C) Relative HOXB8 expression in SiHa cells upon E5, E6, or E7 silencing. GAPDH was used to normalize relative mRNA expression.
miR196a expression affects cell proliferation, cell growth and apoptosis
To investigate the potential cellular function of miR196a, we examined its effect on cell proliferation, cell growth and apoptosis by modulating miR196a expression in HeLa and SiHa cells. First, we determined the effect of miR-196a on cell proliferation by conducting EdU assays. In HeLa cells, inhibition of miR-196a expression increased the percentage of EdUincorporated cells to 27% compared with the control, which exhibited 15% incorporation (P = 0.003, Fig. 5A and B, Supplementary Fig. 5A). However, EdU incorporation decreased to 38% of cells when miR196a was overexpressed compared with 46% in the control in SiHa cells (P = 0.0133, Fig. 5C and D, Supplementary Fig. 5B). Thus, lower levels of miR196a increased HeLa cell proliferation, whereas higher levels of miR196a decreased SiHa cell proliferation. The effect of miR-196a on cell growth was evaluated using the CCK-8 assay. The cell growth curves indicated that miR-196a down-regulation significantly increased HeLa cell growth, whereas miR-196a up-regulation reduced SiHa cell growth (Fig. 5E and F). Hence, EdU and CCK-8 assays indicated that HeLa and SiHa cell growth and proliferation, respectively, were affected when miR196a levels were modulated.
miR196a regulates cell proliferation and cell growth.
(A) EdU staining indicating HeLa cell proliferation increases when miR196a is silenced. (B) Percentage of EdU-positive HeLa cells upon miR196a silencing. (C) EdU staining indicating increased SiHa cell proliferation when miR196a is overexpressed. (D) Percentage of EdU-positive SiHa cells upon miR196a overexpression. (E) Growth rate of HeLa cells when miR196a is silenced as measured by CCK8 assays. (F) Growth rate of SiHa cells when miR196a is overexpressed as measured by CCK8 assays.
In addition to cell growth and proliferation, miR196a levels may also affect apoptosis. To evaluate the effect of miR-196a on cell apoptosis, we performed Annexin V/propidium iodide (PI) and caspase 3/7 assays. Annexin V+/PI− stained cells represent early apoptotic cells, whereas Annexin V+/PI+ stained cells represent late apoptotic cells or necrotic cells. Down-regulation of miR-196a significantly decreased the percentage of early apoptotic and late apoptotic HeLa cells, whereas miR196a up-regulation did not significantly affect SiHa cell apoptosis (early apoptotic cells: 13.6% ± 2.3% vs. 5.9% ± 0.3%, P = 0.028; late apoptotic cells: 8.2% ± 1.2% vs. 4.3% ± 0.7%, P = 0.045, 48 hours post-transfection; Fig. 6A, B and C). However, miR-196a down-regulation decreased caspase 3/7 activity in HeLa cells and miR-196a up-regulation increased caspase 3/7 activity in SiHa cells (Fig. 6D). In short, miR-196a down-regulation significantly affected both the percentage of cells undergoing apoptosis and caspase 3/7 levels in HeLa cells, whereas miR-196a up-regulation only slightly increased caspase 3/7 levels in SiHA cells.
miR196a modulates apoptosis.
(A) Representative flow cytometry analysis of annexin V and propidium iodide-stained HeLa cells or SiHa cells treated with and without miR196a inhibitors (left panels) or miR196a mimics (right panels). (B) Quantification of early apoptosis cells (annexin V+/PI−) using three independent cell populations. (C) Quantification of early apoptosis cells (annexin V+/PI+) using three independent cell populations. (D) Caspase 3/7 assay indicating Caspase3/7 levels detected in HeLa or SiHa cells treated with and without miR196a inhibitors or miR196a mimics.
Discussion
We began our studies regarding the differential expression of miRNAs in cervical cancers by collecting HPV-negative normal cervix, HPV16-positive normal cervix and HPV16-positive squamous cervical carcinoma tissues from patients. Similar studies on miRNAs mostly utilized cell lines and did not separately analyze HPV subtypes. The identification of three miRNAs (miR196a, miR133a and miR133b) that exhibited significant differential expression in HPV16-positive normal cervices versus HPV-negative normal cervices indicated that the expression of some miRNAs is altered upon HPV16 infection. These three miRNAs were further down-regulated in all of the HPV16-positive cervical carcinoma samples we examined, suggesting that these miRNAs are further altered when the infected cervical tissues are transformed into malignant state. In contrast, six miRNAs (miR154, miR329, miR369-5p, miR127, miR495 and miR503) were down-regulated in HPV16-positive cervical carcinoma versus normal cervix samples but not in normal cervix versus HPV16-positive cervix samples. Thus, there are two groups of miRNAs that are down-regulated in cervical carcinoma: one group demonstrates a stepwise down-regulation from normal cervix samples to HPV16-infected cervix samples, whereas the second group is exclusively down-regulated when the cervix is transformed to a cancerous state. This finding suggests that the first group of miRNAs represent the early targets of HPV16 infection when the cervical tissues still appear normal and are not malignant. The aberrant expression of this group of miRNAs might gradually transform the normal cervix to a cancerous state. When the infected cervix becomes cancerous, the expression of a second group of miRNAs is altered, accompanying or causing abnormal cellular process. To our knowledge, this is the first report demonstrating a stepwise down-regulation of miRNAs in cervical cancer development. Therefore, our studies have potentially identified an early alteration of HPV16 infection at the miRNA level that could be used as a marker or therapeutic target of cellular transformation. Interestingly, four miRNAs (miR133a, miR133b, miR218 and miR495), were down-regulated in HPV16-positive cervical cancer versus HPV-negative normal cervix samples in our studies and these miRNAs were also down-regulated in an independent study comparing miRNA levels in normal cervical samples and HPV16-positive cervical cancer cell lines30. Many miRNAs reported to be reduced in that study were not significantly down-regulated in our study and this difference is likely due to differences in comparing normal cervix to cervical carcinoma versus cervical cancer cell lines. Further studies should focus on miRNAs that are commonly reduced in both studies, such as miR133a and miR133b.
Consistent with data collected from patient samples, miR196a expression was significantly reduced in the HPV16-positive cell lines but not in the HPV-negative cell line(C33-a) or HPV18-positive cell line(HeLa) we examined. However, more cell lines should be examined to determine whether all HPV18 and HPV negative cell lines express higher miR196a expression than HPV16 cell lines. In contrast to cervical tissues, cervical cancer cell lines have been passaged. Hence, the HPV16-specific reduction of miR196a in both cervical tissues and cell lines indicates that its aberrant expression pattern occurs in both states. Many studies have associated miR196a with cancer incidence and development36. miR196a is thought to be oncogenic due to its high expression in numerous cancer cells, including leukemia and pancreatic cancer40,41. However, other studies have suggested that miR196a might function as a tumor suppressor in other malignant cells, such as melanoma and breast cancer cells42,43. Mueller et al. reported that miR-196a-overexpressing cell clones exhibited an approximately 40% reduction in their invasive potential compared with parental cells or empty vector controls44. miR196a expression was down-regulated in malignant melanoma cells compared with normal melanocytes and low miR196a expression up-regulated HOXB742. Moreover, miR196a was suggested to be oncogenic in adenocarcinomas and esophageal cancers but potentially plays a tumor suppressive role in esophageal squamous carcinoma. These opposite roles of miR196a in different types of the same cancer are analogous to our observation. We observed increased miR196a levels in HeLa adenocarcinoma cells and significantly low miR196a levels in SiHa squamous carcinoma cells. Moreover, we confirmed that down-regulation of miR196a increased HOXB8 expression in HeLa cells and that the up-regulation of miR196a decreased HOXB8 levels in SiHa cells. Previous HOXB8 studies demonstrated that miR196a binds to the 3′UTR of HOXB8 mRNA with full complementarity, thus triggering the silencing of its HOXB8 transcripts37.
The most striking result we observed involves the specificity of the HPV16 early gene E5 for down-regulating miR196a. Among the three early genes exhibiting oncogene properties, E5 is regarded as playing a supplementary role in transforming cervical cells compared with E6 and E7. Consistent with the above results, the effects of HPV16-E5-mediated HOXB8 up-regulation through down-regulating miR196a were also supported in our results. E5 has been detected in precancerous HPV16-positive cervical lesions, suggesting its role in malignant development14. Although E5, E6 and E7 are all expressed in the basal/suprabasal epithelia of cervical tissues, the modes of transformation potentially vary. E5 potentially independently targets a group of miRNAs, acting as tumor suppressor and altering the cellular process; in contrast, E6 and E7 target tumor suppressor factors, such as p53. Acting together, the early genes gradually transform the normal cervix to a cancerous state.
Our study demonstrated that modulating miR-196a levels affects cell proliferation, cell growth and apoptosis in the cervical cancer cell lines. The results of EdU experiments suggested that miR196a plays a role in regulating proliferation, but the results of CCK8 assays strengthened this conclusion even more. Taken together, EdU and CCK8 assays revealed increased proliferation and cell growth when miR196a was down-regulated. In our studies, the effect on apoptosis was significantly and exclusively noted in HeLa cells upon miR196a inhibition as assessed by propidium iodide and Annexin V staining. However, we found that miR196a levels significantly regulated apoptosis in both cell lines as measured by Caspase3/7 expression. Altogether, the results suggested that miR196a down-regulation reduces cervical cancer cell apoptosis and the Caspase3/7 assay might be more sensitive in detecting small changes in apoptosis in cells. By promoting cellular proliferation and simultaneously reducing apoptosis in infected cells, HPVs promote the transformation of the normal epithelium to a neoplastic or malignant state45. Hence, our results indicate that miR196a is involved in regulating these processes, perhaps by maintaining these cellular activities at normal levels in normal cervical tissues. Given that miR196a is specifically down-regulated by HPV16-E5 in our study, it is quite likely that cellular proliferation, growth and apoptosis are altered in response to the expression of E5 in the infected host cells. Indeed, HPV16-E5 down-regulates the tumor suppressors p21 and p27, increasing cell proliferation and the percentage of cells in the S phase45,46 (Tsao, 1996; Venuti, 2011). Thus, our studies confirm the effect of HPV16-E5 in cervical cancer development and further identify its downstream targets, namely miR196a and HOXB8.
Methods
Cervical tissue characteristics
Human cervical tissue and cervical cancer samples were collected under a protocol approved by the institutional review board of the Second Hospital of Tsinghua University. Informed consent was obtained from patients with uterine leiomyoma or squamous cell carcinoma of cervix (FIGO Ib stage) undergoing hysterectomy or radical hysterectomy, respectively. All the patients in this study were 40 to 50 years old and their HPV-infection status were determined using a genoarray test kit (Hybribio Company, Hong Kong, China). Among forty-one patients with pathologically normal cervices, ten HPV-16 positive and ten HPV-negative samples were randomly selected for this study. All ten patients with squamous cell carcinoma of the cervix (FIGO Ib stage) were infected with high-risk HPV. Of these patients, nine samples were positive for HPV16 and one was positive for HPV33. Thus, the specimens were divided to three groups for the miRNA-microarray analysis: Group 1: HPV-negative normal cervix (n = 10); Group 2: HPV16-positive normal cervix (n = 10); Group 3: HPV16-positive squamous cell cervical carcinoma (n = 9). No significant differences in age were noted among these three groups. All experiments were performed in accordance with the approved guidelines.
Cell lines
CaSki, SiHa (HPV-16-positive), HeLa (HPV-18-positive) and C-33A (HPV-negative) cervical cancer cell lines were purchased from Cell Resource Center, IBMS, CAMS/PUMC (Beijing, China) and were cultured as previously described47. Briefly, all four cell lines were grown in Dulbecco's Modified Eagle's Medium (Gibco, China) supplemented with 10% Fetal Bovine Serum and 1% penicillin/streptomycin (Gibco, China) at 37°C and 5% CO2.
MiRNA microarray analysis
Tissue specimens were immediately snap-frozen in liquid nitrogen until all the specimens in this study were collected. Total RNA was extracted using the TRIZOL RNA isolation kit (Invitrogen, USA) following the manufacturer's instructions. miRNAs were isolated from total RNA using the MirVana miRNA isolation kit (Ambion, USA). miRNAs were labeled using T4 RNA ligase. Microarray probes were oligonucleotides with sequences complementary to 435 human miRNAs prepared on a Smart Array TM (CapitalBio Corp., Beijing, China). The differences in miRNA gene expression were quantified using Significance Analysis of Microarrays (SAM, version 2.1). Ten HPV-negative normal cervix samples (normal control) were divided into two pools of five samples to eliminate individual differences in miRNA expression. Microarray hybridization was performed using miRNA samples from two HPV-negative normal pools, ten HPV16-positive normal cervices and nine HPV16-positive cervical cancers.
miRNA and mRNA real-time quantitative RT-PCR analysis
Total RNA was extracted from the cell lines used in this study using the RNeasy Kit (Qiagen, USA) following the manufacturer's instructions. To measure miR196a levels, 2 μg total RNA was subjected to analysis using Bulge-Loop™ miRNA qRT-PCR detection kit and small nucleolar U6 was used as the housekeeping small RNA reference gene (RiboBio, Guangzhou, China). In total, 500 ng total RNA was reversed transcribed using the iScript Reverse Transcription Supermix and measured with Ssofast Eva Green Supermix (Bio-Rad, USA) using a Bio-Rad CFX96 Real Time system (Bio-Rad, USA) according to the manufacturer's instructions. GAPDH was used as a housekeeping gene control for normalization of the expression level of all mRNAs. All primer sequences are listed in Supplementary Table 1.
Modulating miR-196a expression in cervical cancer cell lines
miR196a levels were modulated by miR196a inhibitor or mimics (RiboBio, Guangzhou, China). miRNA mimics are chemically synthesized double-stranded RNAs and miRNA inhibitors are chemically synthesized single-stranded RNAs. These agents can be transfected into a variety of cell types. The application of these two RNAs has been reported in numerous research articles48,49. miRNA mimics were transfected into SiHa cells at a concentration of 50 nM, whereas 150 nM of inhibitor was transfected into HeLa cells using Lipofectamine 2000™ (Invitrogen, USA) following the manufacturer's instructions. The corresponding controls for the miR-196a inhibitor and mimic were provided in the same kit. After transfection, miR-196a expression was quantitated by qRT-PCR as described above.
Modulating HPV16 E5, E6 and E7 expression in cervical cancer cell lines
HPV16-E2, -E5, -E6 and -E7 overexpression was achieved by transfecting vectors carrying the corresponding genes. First, the three genes were amplified from SiHa cell genomic DNA and cloned into the lentiviral vector p2K7neo as previously described50. The expression of all three genes was driven by the human EF1α promoter. The primers used to PCR amplify these genes are listed in Supplementary Table 1. Lentiviral supernatant was collected to infect HeLa cells as described previously50.
To silence HPV16 E5, E6 and E7 expression, shRNAs were constructed using the Block-iT inducible H1 lentiviral system (Invitrogen, USA) according to the manufacturer's protocols. shRNAs were first inserted into pENTR/H1/TO vectors and LR was recombined into pLenti4/BLOCK-iT™-DEST. Lentiviral supernatant was collected to infect SiHa Cells as described previously50. Cells were collected 48 hours after lentiviral infections and subjected to quantitatively real time RT-PCR.
Cells proliferation assay
Cell proliferation assays were performed using the DNA Proliferation in vitro Detection Kit following the manufacturer's instructions (RiboBio, Guangzhou, China). Briefly, cells were sequentially incubated with the thymidine analog5-ethynyl-2′-deoxyuridine (EdU) for 3 hours, Apollo567 for 30 min and Hoechst 33342 for 30 min. Cell images were captured using a fluorescence microscope (Nikon Eclipse-Ti, Japan). Cell proliferation assays were performed using DNA Proliferation in vitro Detection Kit following manufacturer's instructions (RiboBio, Guangzhou, China) and previously published protocol51. Briefly, cells were sequentially incubated with thymidine analogue, 5-ethynyl-2′-deoxyuridine (EdU), for 3 hours; followed by fixation of PFA, staining of Apollo567 for 30 min and staining of Hoechst 33342 for 30 min. EdU incorporates into actively proliferating cells, so EdU stainings of these cells can be detected with fluorescent microscope and expected to be in the nuclei. Cell images were captured using fluorescence microscope (Nikon Eclipse-Ti, Japan). Serial optical sections were captured using NIS-Elements software and projected to provide two-dimensional maximum brightness images. More than 50 cells in three different areas of three independent cultured wells (n = 9) for each sample were counted for EdU-positive staining followed by statistical analysis.
Cell growth assay
Cell growth curves were plotted using the Cell Counting Kit-8 assay (KeyGEN Biotech, Nanking, China) according to the manufacturer's instructions. Cells were seeded in 96-well plates. After incubation with the CCK-8 reagent (2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5- (2,4-disulfophenyl)-2H-tetrazolium monosodium salt) for 2.5 hours, absorbance at 450 nm was measured using Infinite M200 (Tecan, Switzerland). Each sample was assessed in triplicate at each time point.
Annexin V-FITC/PI apoptotic assay
Apoptosis assays were conducted using the Annexin V-FITC/PI Apoptosis Kit following the manufacturer's instructions (KeyGEN Biotech, Nanking, China). Briefly, >10,000 cells were resuspended in 500 μl binding buffer and incubated with Annexin V-FITC and propidium iodide for 15 min. The cell suspensions were then subjected to flow cytometry analysis (FACSCalibur, BD, USA). Each experiment was repeated in triplicate.
Caspase 3/7 assay
Caspase 3/7 assays were performed using Apo-ONE® Homogeneous Caspase-3/7 Assay following the manufacturer's protocol (Promega, USA). Caspase 3/7 activity in cell lysates was determined by measuring the absorbance at 499 nm using Infinite M200 (Tecan, Switzerland). Each experiment was repeated in triplicate.
Statistical analysis
SPSS software (Version 13.0, SPSS Inc., USA) was used to perform statistical analysis. All averages of measurements were obtained by experimental repeats unless otherwise indicated. All experimental data are expressed as means ± standard deviation. The statistical significance of differences between two groups was performed using the two-sided t-test. Repeated measures data from the CCK8 assay were assessed using Mauchly's Test of Sphericity and repeated measures general linear model. P-values less than 0.05 were considered to be statistically significant.
References
Jemal, A. et al. Global cancer statistics. CA Cancer J Clin 61, 69–90, 10.3322/caac.20107 (2011).
DiSaia, P. J. & Creasman, W. J. [Preinvasive disease of the Cervix.]. Clinical Gynecologic Oncology [1–34] (Elsevier, Philadelphia, 2002).
Schiffman, M., Castle, P. E., Jeronimo, J., Rodriguez, A. C. & Wacholder, S. Human papillomavirus and cervical cancer. Lancet 370, 890–907, 10.1016/S0140-6736(07)61416-0 (2007).
Sargent, A. et al. Prevalence of type-specific HPV infection by age and grade of cervical cytology: data from the ARTISTIC trial. Br J Cancer 98, 1704–1709, 10.1038/sj.bjc.6604324 (2008).
Bouvard, V., Storey, A., Pim, D. & Banks, L. Characterization of the human papillomavirus E2 protein: evidence of trans-activation and trans-repression in cervical keratinocytes. EMBO J 13, 5451–5459 (1994).
Hughes, F. J. & Romanos, M. A. E1 protein of human papillomavirus is a DNA helicase/ATPase. Nucleic Acids Res 21, 5817–5823 (1993).
Scheffner, M., Werness, B. A., Huibregtse, J. M., Levine, A. J. & Howley, P. M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 63, 1129–1136 (1990).
Menges, C. W., Baglia, L. A., Lapoint, R. & McCance, D. J. Human papillomavirus type 16 E7 up-regulates AKT activity through the retinoblastoma protein. Cancer Res 66, 5555–5559, 10.1158/0008-5472.can-06-0499 (2006).
Huibregtse, J. M., Scheffner, M. & Howley, P. M. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J 10, 4129–4135 (1991).
Lechner, M. S. et al. Human papillomavirus E6 proteins bind p53 in vivo and abrogate p53-mediated repression of transcription. EMBO J 11, 3045–3052 (1992).
Band, V., Dalal, S., Delmolino, L. & Androphy, E. J. Enhanced degradation of p53 protein in HPV-6 and BPV-1 E6-immortalized human mammary epithelial cells. EMBO J 12, 1847–1852 (1993).
Munger, K. & Howley, P. M. Human papillomavirus immortalization and transformation functions. Virus Res 89, 213–228 (2002).
Liao, S. et al. HPV16/18 E5, a promising candidate for cervical cancer vaccines, affects SCPs, cell proliferation and cell cycle and forms a potential network with E6 and E7. Int J Mol Med 31, 120–128, 10.3892/ijmm.2012.1168 (2013).
DiMaio, D. & Petti, L. M. The E5 proteins. Virology 445, 99–114, 10.1016/j.virol.2013.05.006 (2013).
Maufort, J. P., Shai, A., Pitot, H. C. & Lambert, P. F. A role for HPV16 E5 in cervical carcinogenesis. Cancer Res 70, 2924–2931, 10.1158/0008-5472.CAN-09-3436 (2010).
Cui, J., Eldredge, J. B., Xu, Y. & Puett, D. MicroRNA expression and regulation in human ovarian carcinoma cells by luteinizing hormone. PLoS One 6, e21730, 10.1371/journal.pone.0021730 (2011).
Lee, J. W. et al. Altered MicroRNA expression in cervical carcinomas. Clin Cancer Res 14, 2535–2542, 10.1158/1078-0432.ccr-07-1231 (2008).
Khella, H. W. et al. Exploring the role of miRNAs in renal cell carcinoma progression and metastasis through bioinformatic and experimental analyses. Tumour Biol, 10.1007/s13277-011-0255-5 (2011).
Fanini, F., Vannini, I., Amadori, D. & Fabbri, M. Clinical implications of microRNAs in lung cancer. Semin Oncol 38, 776–780, 10.1053/j.seminoncol.2011.08.004 (2011).
Schetter, A. J. & Harris, C. C. Alterations of microRNAs contribute to colon carcinogenesis. Semin Oncol 38, 734–742, 10.1053/j.seminoncol.2011.08.009 (2011).
Calin, G. A. & Croce, C. M. MicroRNA signatures in human cancers. Nat Rev Cancer 6, 857–866, 10.1038/nrc1997 (2006).
Johnson, C. D. et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res 67, 7713–7722, 10.1158/0008-5472.can-07-1083 (2007).
Miska, E. A. How microRNAs control cell division, differentiation and death. Curr Opin Genet Dev 15, 563–568, 10.1016/j.gde.2005.08.005 (2005).
Rusca, N. & Monticelli, S. MiR-146a in Immunity and Disease. Mol Biol Int 2011, 437301, 10.4061/2011/437301 (2011).
Esteller, M. Non-coding RNAs in human disease. Nat Rev Genet 12, 861–874, 10.1038/nrg3074 (2011).
Hu, X. et al. A microRNA expression signature for cervical cancer prognosis. Cancer Res 70, 1441–1448, 10.1158/0008-5472.can-09-3289 (2010).
Lu, J. et al. MicroRNA expression profiles classify human cancers. Nature 435, 834–838, 10.1038/nature03702 (2005).
Yanaihara, N. et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 9, 189–198, 10.1016/j.ccr.2006.01.025 (2006).
Lui, W. O., Pourmand, N., Patterson, B. K. & Fire, A. Patterns of known and novel small RNAs in human cervical cancer. Cancer Res 67, 6031–6043, 10.1158/0008-5472.CAN-06-0561 (2007).
Martinez, I. et al. Human papillomavirus type 16 reduces the expression of microRNA-218 in cervical carcinoma cells. Oncogene 27, 2575–2582, 10.1038/sj.onc.1210919 (2008).
Greco, D. et al. Human papillomavirus 16 E5 modulates the expression of host microRNAs. PLoS One 6, e21646, 10.1371/journal.pone.0021646 (2011).
Hou, T. et al. MicroRNA-196a promotes cervical cancer proliferation through the regulation of FOXO1 and p27Kip1. Br J Cancer 110, 1260–1268, 10.1038/bjc.2013.829 (2014).
Villegas-Ruiz, V. et al. Heterogeneity of microRNAs expression in cervical cancer cells: over-expression of miR-196a. Int J Clin Exp Pathol 7, 1389–1401 (2014).
Tusher, V. G., Tibshirani, R. & Chu, G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 98, 5116–5121, 10.1073/pnas.091062498 (2001).
Schwarz, E. et al. Structure and transcription of human papillomavirus sequences in cervical carcinoma cells. Nature 314, 111–114 (1985).
Chen, C., Zhang, Y., Zhang, L., Weakley, S. M. & Yao, Q. MicroRNA-196: critical roles and clinical applications in development and cancer. J Cell Mol Med 15, 14–23, 10.1111/j.1582-4934.2010.01219.x (2011).
Yekta, S., Shih, I. H. & Bartel, D. P. MicroRNA-directed cleavage of HOXB8 mRNA. Science 304, 594–596, 10.1126/science.1097434 (2004).
Davidson, B. et al. Gene expression signatures differentiate ovarian/peritoneal serous carcinoma from breast carcinoma in effusions. J Cell Mol Med 15, 535–544, 10.1111/j.1582-4934.2010.01023.x (2011).
Vider, B. Z. et al. Human colorectal carcinogenesis is associated with deregulation of homeobox gene expression. Biochem Biophys Res Commun 232, 742–748, 10.1006/bbrc.1997.6364 (1997).
Schotte, D. et al. Identification of new microRNA genes and aberrant microRNA profiles in childhood acute lymphoblastic leukemia. Leukemia 23, 313–322, 10.1038/leu.2008.286 (2009).
Zhang, Y. et al. Profiling of 95 microRNAs in pancreatic cancer cell lines and surgical specimens by real-time PCR analysis. World J Surg 33, 698–709, 10.1007/s00268-008-9833-0 (2009).
Braig, S., Mueller, D. W., Rothhammer, T. & Bosserhoff, A. K. MicroRNA miR-196a is a central regulator of HOX-B7 and BMP4 expression in malignant melanoma. Cell Mol Life Sci: CMLS 67, 3535–3548, 10.1007/s00018-010-0394-7 (2010).
Li, Y. et al. Ratio of miR-196s to HOXC8 messenger RNA correlates with breast cancer cell migration and metastasis. Cancer Res 70, 7894–7904, 10.1158/0008-5472.CAN-10-1675 (2010).
Mueller, D. W. & Bosserhoff, A.-K. MicroRNA miR-196a controls melanoma-associated genes by regulating HOX-C8 expression. Int J Cancer 129, 1064–1074, 10.1002/ijc.25768 (2011).
Venuti, A. et al. Papillomavirus E5: the smallest oncoprotein with many functions. Mol Cancer 10, 140, 10.1186/1476-4598-10-140 (2011).
Tsao, Y. P., Li, L. Y., Tsai, T. C. & Chen, S. L. Human papillomavirus type 11 and 16 E5 represses p21(WafI/SdiI/CipI) gene expression in fibroblasts and keratinocytes. J Virol 70, 7535–7539 (1996).
Meissner, J. D. Nucleotide sequences and further characterization of human papillomavirus DNA present in the CaSki, SiHa and HeLa cervical carcinoma cell lines. J Gen Virol 80, 1725–1733 (1999).
Wu, J. et al. MicroRNA-34a inhibits migration and invasion of colon cancer cells via targeting to Fra-1. Carcinogenesis 33, 519–528, 10.1093/carcin/bgr304 (2012).
Zheng, B. et al. MicroRNA-409 suppresses tumour cell invasion and metastasis by directly targeting radixin in gastric cancers. Oncogene 31, 4509–4516, 10.1038/onc.2011.581 (2012).
Kee, K., Angeles, V. T., Flores, M., Nguyen, H. N. & Reijo Pera, R. A. Human DAZL, DAZ and BOULE genes modulate primordial germ-cell and haploid gamete formation. Nature 462, 222–225, 10.1038/nature08562 (2009).
Feng, S. et al. MicroRNA-192 targeting retinoblastoma 1 inhibits cell proliferation and induces cell apoptosis in lung cancer cells. Nucleic Acids Res 39, 6669–6678, 10.1093/nar/gkr232 (2011).
Acknowledgements
This study was supported by the Natural Science Foundation of China (30872938), the Ministry of Science and Technology of China (2012CB966702), the Natural Science Foundation of Guangxi (2013jjBA40189) and Tsinghua-Yue-Yuen Medical Science Foundation (20200574).
Author information
Authors and Affiliations
Contributions
L.C. and J.L. performed the majority of the experiments. L.L., Y.Z. and W.C. participated in experiments and sample collection. Z.C. and H.Z. provided facility and sample procurement support. K.K., C.C. and C.L. designed the experiments and wrote the entire manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Liu, C., Lin, J., Li, L. et al. HPV16 early gene E5 specifically reduces miRNA-196a in cervical cancer cells. Sci Rep 5, 7653 (2015). https://doi.org/10.1038/srep07653
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep07653
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.