Abstract
VAR2CSA stands today as the leading vaccine candidate aiming to protect future pregnant women living in malaria endemic areas against the severe clinical outcomes of pregnancy associated malaria (PAM). The rational design of an efficient VAR2CSA-based vaccine relies on a profound understanding of the molecular interactions associated with P. falciparum infected erythrocyte sequestration in the placenta. Following immunization of a llama with the full-length VAR2CSA recombinant protein, we have expressed and characterized a panel of 19 nanobodies able to recognize the recombinant VAR2CSA as well as the surface of erythrocytes infected with parasites originating from different parts of the world. Domain mapping revealed that a large majority of nanobodies targeted DBL1X whereas a few of them were directed towards DBL4ε, DBL5ε and DBL6ε. One nanobody targeting the DBL1X was able to recognize the native VAR2CSA protein of the three parasite lines tested. Furthermore, four nanobodies targeting DBL1X reproducibly inhibited CSA adhesion of erythrocytes infected with the homologous NF54-CSA parasite strain, providing evidences that DBL1X domain is part or close to the CSA binding site. These nanobodies could serve as useful tools to identify conserved epitopes shared between different variants and to characterize the interactions between VAR2CSA and CSA.
Similar content being viewed by others
Introduction
Malaria remains one of the most prevalent infectious diseases in the world, affecting 207 million people per year and causing an estimated 627,000 subsequent deaths, mostly among children below five years of age and pregnant women1. Among the 5 plasmodia species commonly infecting Humans, Plasmodium falciparum is responsible of the most severe malaria cases and deaths. The pathogenicity of P. falciparum has been linked to the ability of P. falciparum infected erythrocytes (IEs) to adhere to the host endothelium or to the syncytio-trophoblastic layer of the placenta2. By mediating cytoadhesion of IEs to host receptors, P. falciparum has developed an immune evasion strategy that prevents the transit of IEs through the spleen's red pulp and their subsequent retention and clearance by the red pulp macrophages3,4. IEs cytoadhesion is mediated by members of the P. falciparum erythrocyte membrane protein 1 (PfEMP1), a parasite protein expressed at the surface of IEs and encoded by the highly diverse var gene family5,6,7.
In malaria endemic areas, individuals progressively acquire protective clinical immunity during childhood and adults are generally protected against the severe clinical outcomes of the disease8. However, first-time pregnant women become once again susceptible to malaria. Pregnancy associated malaria (PAM) is linked to the massive accumulation of IEs and monocytes in the placental intervillous spaces9, leading to maternal anemia and hypertension as well as stillbirth, preterm delivery, intra-uterine growth retardation and low birth-weight10,11. Overall, PAM considerably increases the morbidity and mortality of both mother and child. Indeed, reports indicate that as many as 363,000 neonates and at least 10,000 maternal deaths may be attributable to PAM every year11.
IEs isolated from placentas of women suffering from PAM (IEs-PAM) present a unique adhesive phenotype contrasting with IEs isolated from other tissues. Indeed IEs-PAM bind to the sulfated glycosaminoglycan chondroitin-4-sulfate (CSA) and not to endothelial receptors such as CD36 and ICAM-112,13. Low-sulfated chondroitin sulfate-proteoglycans (CSPGs) have been detected in the placental intervillous space by the end of the third month of gestation14, thus offering a potential anchor point for the IEs-PAM. However, a recent study provides evidences that women in the first trimester of pregnancy could already be infected with parasites expressing VAR2CSA, suggesting that the placental tropism of P. falciparum is already established during the first 12 weeks of pregnancy15. Following successive pregnancies, women become resistant to PAM as they develop antibodies able to recognize IEs-PAM from different part of the world and block their binding to placental CSA16. This would suggest that the P. falciparum CSA-ligand expressed by placental parasites possess conserved antigenic determinants. The identification of such epitopes would therefore be a crucial advance for the design of an effective vaccine against PAM.
The PfEMP1 variant VAR2CSA has been identified as the parasite ligand to placental CSA17,18,19,20. VAR2CSA is a high molecular weight protein, with a 300 kDa extracellular region organized in 6 Duffy-binding like (DBL) domains and a distinctive cysteine-rich interdomain region (CIDRPAM)21. Although VAR2CSA stands today as the main candidate for a PAM-vaccine, the substantial antigenic polymorphism and the lack of comprehensive structural information regarding the CSA-binding site of the protein limit our understanding of placental sequestration mechanisms and represent therefore significant challenges for any vaccine and therapeutic development against PAM. Recent studies have shown that the presence of a single CSA-binding site is formed by a higher-order domain organization involving multiple domains22,23. The N-terminal region of VAR2CSA plays a major role in CSA adhesion and antibodies targeting that region are able to prevent the adhesion of IEs-PAM to CSA24,25,26,27,28. The identification of conformational regions, conserved between the different polymorphic forms of VAR2CSA as well as the characterization of critical epitopes connected to the high-affinity CSA-binding site would open new avenues in the rational design of a vaccine against PAM.
Camelidae have a unique immune system able to produce functional immunoglobulins G devoid of light chains called heavy-chain only antibodies (HCAbs). As a result, the variable heavy chain of heavy-chain-only antibody (VHH, also renamed Nanobody®) folds autonomously and is the smallest recombinant antigen-binding domain that can be produced (≈ 15 kDa). Nanobodies are organized in four conserved regions, called framework regions (FRs) and three complementary determining regions (CDRs) responsible for antigen recognition29. The use of nanobodies presents several advantages due to their intrinsic characteristics. Their small size permits good tissue penetration in vivo. Moreover, they are very specific and have high affinity for only one cognate target. Nanobodies are easily expressed with high yields in bacteria or yeasts and have an excellent solubility and thermal stability compared to most mice and Human derived antibody fragments29. Due to their advantageous biochemical and economic properties, they have been used for several assays such as protein purification30 and to help protein crystallization in structural studies31,32,33. Importantly, nanobodies have been or are currently being developed as diagnostic tools and novel therapeutic agents for numerous infectious diseases and cancers34.
To further characterize the interactions between VAR2CSA and its receptor, we immunized a llama with the full-length VAR2CSA and generated nanobodies that were subsequently screened for their capacity to recognize the recombinant as well as the native VAR2CSA expressed on the IEs-PAM surface and to inhibit CSA adhesion of CSA-binding erythrocytes infected with the NF54 parasite strain. Using this strategy, we obtained high-affinity nanobodies against VAR2CSA targeting different DBL domains. Of particular interest, one nanobody targeting the N-terminal part of VAR2CSA (DBL1X) was able to recognize the native VAR2CSA protein of the three parasite lines tested. Furthermore, four nanobodies targeting DBL1X reproducibly inhibited the interaction of the homologous NF54-CSA IEs with CSA, providing new evidences that the DBL1X domain is associated with the CSA-binding site. These nanobodies could have important applications in the design of VAR2CSA-based vaccines and in the development of new therapeutics.
Results
Library screening and expression of VAR2CSA-specific nanobodies
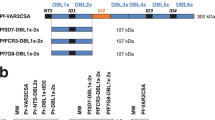
A llama was immunized with the recombinant full-length 3D7-VAR2CSA protein. Following mRNA extraction from the camelid's lymphocytes, we have generated a phage display library comprising 109 different sequences. Clones expressing VAR2CSA-specific nanobodies were enriched by consecutive rounds of panning and screened for binding to the full-length 3D7-VAR2CSA. Following the panning and screening processes, 53 single domain antibody clones that specifically recognized VAR2CSA were selected. Multiple sequence alignments revealed that out of these 53 clones, 40 displayed different amino-acid sequences. Out of these 40 nanobodies, 30 displayed sequence differences in the CDR3 domain involved in antigen recognition. One clone presenting the sequence hallmarks of a VH35 was excluded from our analysis. Twenty nine nanobodies were successfully expressed and purified in sufficient amount (Supplementary Fig. S1). The overall production yields for the different nanobodies were very variable as they ranged from 0.04 to 18 mg per liter of starting culture. The reactivity of the 29 remaining nanobodies was then tested using the full-length VAR2CSA recombinant protein as target antigen (Fig. 1). Twenty one out of the 29 nanobodies recognized to different extents the full-length VAR2CSA protein (3D7-DBL1X-6ε) whereas 8 of them led to a weak or not detectable signal and were therefore not considered any further in this study. In this assay, an irrelevant nanobody (VHHi) targeting Human glycophorin A (GPA)36 was used as a negative control.
Recognition of full-length VAR2CSA recombinant protein by the different nanobodies.
ELISA plates were coated with 1 µg/ml of full-length recombinant VAR2CSA protein (3D7-DBL1X-6ε) and incubated with the 29 nanobodies (0.3 µg/ml). Nanobody binding was detected with an HRP-conjugated goat anti-llama IgG and revealed with TMB substrate. Optical density was measured at 655nm.
Mapping of the VAR2CSA domains targeted by the nanobodies
In order to determine the domain specificity of our nanobodies, we performed ELISA using as target antigens i) the VAR2CSA multi-domains 3D7-DBL1X-6ε, 3D7-DBL1X-3X, 3D7-DBL4ε-6ε and FCR3-DBL3X-4ε ii) the VAR2CSA single-domains 3D7-DBL1X, 3D7-DBL2X, FCR3-DBL3X, 3D7-DBL5ε, 3D7-DBL6ε and FCR3-DBL6ε. Out of these 21 nanobodies, 15 recognized the N-terminal region of the protein (3D7-DBL1X-3X) and 6 recognized the C-terminal part (3D7-DBL4ε-6ε) (Fig. 2a). One nanobody (861) recognized the 3D7-DBL4ε-6ε recombinant protein as well as the FCR3-DBL3X-4ε, indicating that this nanobody most likely targets a common epitope shared between both variants within the DBL4ε domain. The 15 nanobodies targeting 3D7-DBL1X-3X were then screened against the single N-terminal domains (3D7-DBL1X, 3D7-DBL2X and FCR3-DBL3X) (Fig. 2b). Interestingly, they were all targeting the 3D7-DBL1X domain. Out of the 6 nanobodies recognizing the VAR2CSA C-terminal region, two nanobodies (862 and 893) recognized the 3D7-DBL5ε domain and three nanobodies (863, 864 and 881) recognized the 3D7-DBL6ε domain. Notably, these three nanobodies targeting 3D7-DBL6ε did not cross-react with FCR3-DBL6ε (Fig. 2c). The nanobody 861 did not recognize any of the 3D7-DBL5ε, 3D7-DBL6ε and FCR3-DBL6ε domains tested here (Fig. 2c). This is in line with our previous result showing that the VHH861 targeted epitope lies within the DBL4ε domain. None of the 21 nanobodies targeted the DBL2X, CIDRPAM nor the DBL3X domains (Fig. 2d). Out of the 21 nanobodies, VHH878 and VHH881, targeting respectively the DBL1X and DBL6ε, were expressed at too low levels to be further analysed in our work. A flow chart summarizing the nanobody selection and screening results as well as a multiple alignment of the 19 nanobody amino-acid sequences are displayed in Fig. 3a and 3b.
Mapping of the VAR2CSA domains targeted by the nanobodies.
ELISA plates were coated at 1 µg/ml with (a) recombinant VAR2CSA multi-domains 3D7-DBL1X-6ε, 3D7-DBL1X-3X, 3D7-DBL4ε-6ε and FCR3-DBL3X-4ε, (b) recombinant VAR2CSA single-domains 3D7-DBL1X, 3D7-DBL2X, FCR3-DBL3X and (c) recombinant VAR2CSA single-domains 3D7-DBL5ε, 3D7-DBL6ε and FCR3-DBL6ε and incubated with the 29 nanobodies (0.3 µg/ml). Nanobody binding was detected with an HRP-conjugated goat anti-llama IgG and revealed with TMB substrate. Optical density was measured at 655nm. (d) Representation of the different nanobody specificities on VAR2CSA.
Nanobodies recognize mostly conformational epitopes
To determine whether the epitopes targeted by the 19 nanobodies were linear or conformational, recognition of the full-length VAR2CSA recombinant protein under reduced and non-reduced conditions was performed by western blotting. Eleven out of 14 nanobodies against DBL1X were not able to recognize the reduced or the non-reduced protein (Fig. 4a), suggesting that these anti-DBL1X nanobodies recognized highly conformation-dependent epitopes. However, anti-DBL1X VHH889 recognized the reduced and non-reduced recombinant full-length VAR2CSA indicating that this nanobody recognized a linear epitope. All the other nanobodies recognized to different extents the non-reduced protein but showed no or weak reactivity when reducing conditions were used. These nanobodies are therefore targeting conformational epitopes dependent of disulfide bonds formation. Altogether, these results indicate that the epitopes recognized by our nanobodies are mostly conformational.
Nanobodies recognize mostly conformational epitopes.
Reduced and non-reduced recombinant protein VAR2CSA (3D7-DBL1X-6ε) was transferred onto membranes by Western blotting and probed with the nanobodies (0.5 µg/ml). Nanobody binding was detected with an HRP-conjugated goat anti-llama IgG. (a) Western blot with anti-DBL1X nanobodies. (b) Western blot with anti-DBL4ε nanobody. (c) Western blot with anti-DBL5ε nanobodies. (d) Western blot with anti-DBL6ε nanobodies.
Kinetic binding analysis of nanobody-VAR2CSA interaction
Nanobodies were then tested by surface plasmon resonance (SPR) to determine their affinity constants (KD) to the full-length VAR2CSA recombinant protein. One example of SPR sensorgram is shown in Supplementary Fig. S2. The fitted kinetic data derived from the sensorgrams are shown in Table 1. The kinetic analysis reveals that 9 of the nanobodies had a high affinity for recombinant VAR2CSA with a KD in the nanomolar range, while the other 9 nanobodies had a KD in the micromolar range. All the nanobodies against the DBL4ε, DBL5ε and DBL6ε domains possessed a nanomolar affinity constant while only 4 out of 13 nanobodies against DBL1X had a nanomolar affinity constant. KD value of VHH905 could not be determined. The fitted kinetic data showed that VHH862 against the 3D7-DBL5ε had the best kon, koff and KD values (i.e., fastest antigen association, longest retention time on antigen and highest affinity, respectively) (Fig. 5). On the other hand, VHH871 and VHH895 had very low kon and high koff values, respectively.
Nanobodies raised against full-length VAR2CSA recognize placental-type infected erythrocytes
To assess the nanobodies reactivity with the native VAR2CSA protein, erythrocytes infected with the homologous parasite strain NF54 (the in vitro adapted mother clone of 3D7) as well as 2 others strains (FCR3 and 7G8) selected for CSA-binding, were incubated with the purified nanobodies. Throughout the paper, erythrocytes selected for CSA-binding and infected by a select parasite strains will be referred as NF54-CSA, FCR3-CSA and 7G8-CSA. As VAR2CSA is not expressed at the surface of CD36-binding IEs, FCR3 IEs selected for CD36-binding (FCR3-CD36) were used as a control.
Out of the 19 nanobodies, 18 reacted with NF54-CSA (homologous parasite strain) as reflected by a geometric mean ratio above 1.2 (representing a 20% increase in reactivity) (Fig. 6). However, VHH863 targeting the DBL6ε did not react with any of the CSA selected IEs suggesting that the recognized epitope is not accessible in the context of the infected erythrocytes. Interestingly, all the 14 nanobodies targeting DBL1X reacted with NF54-CSA and FCR3-CSA IEs, whereas one of them (VHH871) cross-reacted with all three CSA-binding lines tested. None of the 5 nanobodies targeting the VAR2CSA C-terminal region reacted with FCR3-CSA. The cross-reactive VHH861 recognizing the 3D7- and FCR3-DBL4ε by ELISA only reacted with NF54-CSA IEs and not with FCR3-CSA IEs. However, VHH862, recognizing an epitope within the DBL5ε domain and VHH864, targeting the DBL6ε domain, cross-reacted with 7G8-CSA IEs. Importantly, none of the tested nanobodies reacted with FCR3-CD36 IEs.
Nanobodies raised against VAR2CSA recognize placental-type infected erythrocytes.
Nanobody recognition by flow cytometry of native VAR2CSA expressed at the surface of infected erythrocytes with parasite strains NF54 (homologous parasite strain), FCR3 and 7G8 selected for CSA-binding and FCR3 selected for CD36-binding. Nanobody binding was detected with a mouse anti-His IgG and a PE-conjugated goat anti-mouse IgG. The Geometric Mean PE ratio was calculated using the negative control (without nanobody). The different DBLs targeted by the nanobodies are depicted in the figure. Error bars represent the standard deviation of data obtained from three independent experiments (NF54-CSA) and two independent experiments (7G8-CSA).
Nanobodies against DBL1X block the adhesion of IEs-PAM to CSA
We evaluated the capacity of the 19 nanobodies to inhibit the adhesion of VAR2CSA-expressing infected erythrocytes to CSA. Using a static binding inhibition assay where CSA was immobilized on plastic, we have identified 4 VHHs (879, 883, 884, 889) able to partially block, in a reproducible manner, the interaction of homologous NF54-CSA IEs with CSA (Fig. 7), the mean adhesion inhibition ranging from 50 to 60%. Referring to our target-domain mapping experiments (Fig. 2) we observed that all the blocking nanobodies were directed against the DBL1X domain of VAR2CSA.
Four nanobodies against DBL1X reproducibly inhibit CSA adhesion of the homologous NF54-CSA line.
Adhesion assays on CSA-coated plastic spots were performed using purified erythrocytes infected with the NF54 parasite line and selected for the CSA-binding phenotype. Nineteen different nanobodies were tested for their ability to inhibit the IEs binding to CSA. The percentage of binding for a given condition was calculated using the number of adherent cells obtained with IEs pre-incubated with PBS alone as a reference (100% binding). The different DBLs targeted by the nanobodies are depicted in the figure. Error bars represent the standard deviation of three independent experiments.
Discussion
Due to the high polymorphism of VAR2CSA as well as its complex structure, the identification of cross-reactive epitopes implicated in CSA-binding represents a key element to develop a VAR2CSA-based vaccine aiming to protect future pregnant women from PAM. In order to identify such epitopes, we immunized a llama with the full-length 3D7-VAR2CSA recombinant protein (3D7-DBL1X-6ε) and selected 21 nanobodies that specifically recognized the immunized protein. Fifteen nanobodies recognized the N-terminal part of VAR2CSA (DBL1X-3X), all of them targeting the DBL1X domain. Out of the 5 nanobodies recognizing the C-terminal part of VAR2CSA (DBL4ε-6ε), 1 nanobody targeted the DBL4ε domain, 2 nanobodies the DBL5ε and 3 nanobodies the DBL6ε domain.
Small Angle X-rays Scattering (SAXS) studies have demonstrated that the full-length extracellular region of VAR2CSA presents a compact globular organization22,23, most likely maintained by inter-domain interactions. It was also shown that the full-length VAR2CSA was able to bind to CSA with high-affinity and high specificity compared to single domain constructs and that the CSA-binding site was restricted to the N-terminal part of the protein, within the DBL1X-3X domains24,37. Taken together these results suggest the existence of a conformational CSA-binding pocket within the DBL1X-3X.
According to our results, the DBL1X domain seems to be accessible and/or highly immunogenic in the context of the full-length 3D7-DBL1X-6ε as we managed to isolate 15 different nanobodies targeting this domain. This suggests that the DBL1X domain might contain immuno-dominant epitopes driving the humoral immune response to a bias towards the N-terminal boundary of the protein. The absence of nanobodies selected against the domains DBL2X, CIDRPAM and DBL3X falls in line with the fact that a putative CSA-binding pocket might exist with a restricted accessibility to cryptic epitopes and/or that a region of poor immunogenicity lies within this part of the VAR2CSA protein. Our results have to be compared with a recent similar study where the authors have used the full-length FCR3-VAR2CSA to generate nanobodies. Out of the 17 nanobodies obtained in their study, twelve were against the C-terminal domains (DBL4ε, DBL5ε and DBL6ε) whereas the five others were against the ID1-ID2a encompassing DBL2X. Curiously the authors did not obtained any nanobody against the DBL1X domain38, which could indicate that the antibody response towards different VAR2CSA variants may be different.
By comparison to the Ditlev et al. study38, where 12 out 17 of the nanobodies were recognizing linear epitopes, 18 out of 19 of our nanobodies recognized conformational epitopes. The recent evidence that VAR2CSA has a globular organization22,23 provide important implications in our way to consider its interactions with CSA. Indeed, independent low affinity CSA-binding sites located on different VAR2CSA domains and forming multivalent interactions were previously thought to cause placental sequestration by avidity effects39,40. Experimental designs based on the use of single domain constructs were thus disregarding the presence of potentially important conformational epitopes, resulting from the overall folding of the full-length VAR2CSA protein or of the CSA-binding multi-domain DBL1X-3X. Nanobodies recognizing conformational epitopes are most likely to present similar binding features to that of naturally acquired anti-VAR2CSA antibodies. None of our nanobodies are recognizing a conformational epitope composed of residues from different domains as all of them are reacting with single domains.
The production of high-affinity nanobodies is a crutial aspect for their use either as investigation tools or as therapeutic agents. Indeed, in vivo, the affinity maturation that results from the repeated exposure to the same antigen likely results in higher affinity antibodies. Naturally acquired high-avidity antibodies towards VAR2CSA have been shown to play an important role in protection41 against the severe outcomes resulting from PAM. Since the overall strength of an antibody-antigen complex (avidity) depends, at least in part, of the affinity of each antibody for its own epitope, high-affinity antibodies with low koff rate are, most likely, more capable to play an effective and protective function rather that low affinity ones.
The kinetic analysis revealed that 9 of the nanobodies have a high affinity to the full-length VAR2CSA with a KD in the nanomolar range, while 9 nanobodies have a KD in the micromolar range. Notably, all nanobodies against the DBL4ε, DBL5ε and DBL6ε domains possess a nanomolar affinity constant, while only 4 out of 13 nanobodies against DBL1X have a nanomolar KD. Among all the nanobodies, VHH862 against the 3D7-DBL5ε has the best kon, koff and KD values.
A very large majority (18/19) of nanobodies recognizing the recombinant full-length VAR2CSA protein were able to recognize the native VAR2CSA protein expressed on erythrocytes infected with the autologous strain NF54 and selected for the CSA-binding phenotype. The nanobodies targeting DBL1X (15/15) were also able to cross-react with FCR3-CSA whereas the nanobodies directed against the C-terminal part of the protein (DBL4ε, DBL5ε, DBL6ε) did not. Interestingly, one nanobody (871) directed towards the DBL1X domain, cross-reacted with all the parasite lines tested (NF54-CSA, FCR3-CSA, 7G8-CSA) but not with CD36-selected infected erythrocytes showing a specific recognition of placental type IE-PAM.
Polymorphism analysis of VAR2CSA sequences originated from 106 different parasite strains from different parts of the world has revealed a differential diversity among the DBL domain sequences21. Furthermore, individual DBL domains present distinct patterns of diversifying selection. This indicates that limited and differing portions of each DBL domain are targeted by the host antibody response21. The percentage of sequence identity within the DBL1X domains ranges between 69.5% and 89.2%21. Our results suggest that at least a conserved conformational epitope exists within the DBL1X domain of the 3 VAR2CSA expressing strains. The precise identification of these residues forming the recognition site as well as its accessibility by conventional antibodies still needs to be determined.
Interestingly, we have identified 4 nanobodies directed against the immuno-dominant DBL1X domain that could partially block the binding of VAR2CSA-expressing infected erythrocytes to CSA. In our work, none of the nanobodies directed against the DBL4ε, DBL5ε and DBL6ε of VAR2CSA showed inhibitory capabilities. These results are in line with previously published data demonstrating that the core CSA-binding site of VAR2CSA is located in the N-terminal part of the protein, the binding pocket most probably comprising the DBL2X-CIDRPAM and a flanking region within the DBL1X domain22,37. During our selection process, we did not obtain any nanobodies targeting the DBL2X nor DBL3X. Nevertheless, another study performed by Ditlev et al. identified 5 nanobodies directed against the ID1-ID2a multi-domain. Other reports showed that antibodies directed against other domains such as DBL4ε could possess some inhibitory capabilities42. Antibodies can block the interaction between VAR2CSA and its ligand by either i) directly bind to key residues involved in the interaction interface with CSA, ii) bind to proximal epitopes and block the accessibility of CSA to these latter residues by steric hindrance or iii) modify the VAR2CSA conformation upon binding to the antigen. Possibly because of their relative small size compared to conventional immunoglobulins, our nanobodies targeting the DBL4ε, DBL5ε or DBL6ε were not able to interfere with the VAR2CSA/CSA interaction by steric hindrance or by destabilizing the protein structure to a sufficient extend.
We have demonstrated here the feasibility of developing nanobodies against the leading PAM vaccine candidate VAR2CSA. Nanobodies able to recognize different DBL domains could be used as tools for epitope mapping and VAR2CSA structural studies as they may assist as chaperones in protein crystal production. VAR2CSA structure solving would provide novel insights into the function of this complex variant protein31. Furthermore, cross-reactive and inhibitory nanobodies able to block the interaction between IEs-PAM and the placenta could prove to be valuable reagents for the development of novel immunotherapeutic approaches34.
Methods
Ethics statement
All animal vaccination experiments were executed in strict accordance with good animal practices, following the EU animal welfare legislation and after approval of the local ethical committee (Committee for the Use of Laborary Animals at the Vrije Universiteit Brussel, VUB). Every effort was made to minimize suffering.
Expression and purification of recombinant proteins
VAR2CSA recombinant proteins were produced either in the permanent line established from primary embryonic human kidney (HEK293F) purchased from Life technologies or in Escherichia coli. The multi-domain constructs 3D7-DBL1X-6ε, 3D7-DBL1X-3X and 3D7-DBL4ε-6ε were expressed in HEK293F cells, as soluble, secreted recombinant proteins22,24. Proteins were then purified on a HisTrap FastFlow Ni-affinity column (GE Healthcare), followed by an ion exchange chromatography on a SP Sepharose column (GE Healthcare) and a gel filtration chromatography on a Superdex 200 16/60 column (GE Healthcare), as previously described22,24.
The single-domain constructs 3D7-DBL1X, 3D7-DBL2X, FCR3-DBL3X, 3D7-DBL5ε, 3D7-DBL6ε, FCR3-DBL6ε and the multi-domain FCR3-DBL3X-4ε were expressed either in the Rosetta-Gami (Novagen), Origami B (Novagen) or SHuffle® (NEB Biolabs) strains of E. coli as cytoplasmic soluble proteins22,24. Proteins were purified using a metal affinity column (TALON, Clontech), followed by a heparin affinity column (GE Healthcare) when needed and by a gel filtration chromatography (Superdex 75 16/60, GE Healthcare), as previously described22,24.
Llama immunization, nanobody phage library construction and screening
A llama was immunized with the recombinant full-length VAR2CSA protein (3D7-DBL1X-6ε) produced in HEK293F cells. Llamas were manipulated by authorized staff, preferentially an experienced veterinarian. We allow animals to acclimatize to new housing conditions for one week before immunization starts. Immunization and nanobody library construction was carried out according to established methods43,44,45. Briefly, llama lymphocytes mRNA was extracted, amplified by RT-PCR with specific primers and a cDNA library size of 109 nanobodies with 75% correct sized nanobody insert was generated. The library was cloned in a pMESy4 phagemid vector45 (Genbank KF415192), amplified in a permissive E. coli strain (TG1), subsequently infected with M13K07 helper phage for recombinant phage expression. Phages expressing the nanobodies on their tip were enriched by panning on the recombinant full-length VAR2CSA protein. After enrichment, individual clones were screened for production of VAR2CSA-specific nanobodies by ELISA.
Expression and purification of nanobodies
Nanobodies were expressed in the periplasm of either BL21 (Novagen) or SHuffle® (NEB Biolabs) strains of E. coli. Expression in the periplasmic space of BL21 was done using TB (Terrific Broth) medium and fermentation left for 20h at 28°C after 0.5 mM IPTG (isopropyl thiogalactoside) induction. For SHuffle®, expression was done using LB (Luria Bertani) medium at 30°C and fermentation left for 24 h at 20°C after 0.1 mM IPTG induction. Periplasmic extract was prepared by osmotic shock: cell pellets were resuspended first using 1X TES buffer (0.2 M TRIS, 0.5 mM EDTA, 0.5 M sucrose, pH 8), incubated at 4°C on an orbital shaker ice for 1 h and diluted with a double volume of ¼X TES buffer and incubated at 4°C on an orbital shaker for 1 h. Nanobodies were purified from periplasmic extracts by IMAC (immobilized metal affinity chromatography) using a Ni-NTA Superflow resin (Qiagen).
Domain mapping of nanobodies
ELISA plates (Nunc) were coated with 100 µl per well of each VAR2CSA recombinant protein diluted in PBS at 1 µg/ml (multi-domains: 3D7-DBL1X-6ε, 3D7-DBL1X-3X, 3D7-DBL4ε-6ε and FCR3-DBL3X-4ε; single-domains: 3D7-DBL1X, 3D7-DBL2X, FCR3-DBL3X, 3D7-DBL5ε, 3D7-DBL6ε and FCR3-DBL6ε) and incubated at 4°C overnight. BSA (1% in PBS) was coated for background measurement. After coating, the wells were blocked with PBS 1% BSA 0.05% Tween20 (PBST-BSA), 150 µl per well, at 37°C for 1 h. After removing the blocking solution, nanobodies were added at 0.3 µg/ml diluted in PBST-BSA and incubated at 37°C for 1 h. The wells were then washed 3 times with 150 µl of PBS 0.05% Tween20 (PBST). Nanobody binding was detected with a horseradish peroxidase-conjugated (HRP) goat anti-llama IgG (Agrisera, AS10 1420), diluted 1:800 in PBST-BSA and incubated at 37°C for 1 h. After washing with PBST, the plates were developed with 100 µl per well of TMB (3,3′,5,5′-tetramethylbenzidine) substrate (Biorad) and absorbance was measured at 655 nm.
Western blot assays
Recombinant full-length VAR2CSA protein was loaded (1 µg per lane) on a 4–12% Bis-Tris SDS-PAGE gel (Invitrogen) under reducing and non-reducing conditions. Proteins from the gel were transferred onto a polyvinylidene difluoride (PVDF) membrane. The membrane was then blocked with a casein-containing buffer (Qiagen, Cat.no.34660) for 1 h at RT. The nanobodies (0.5 µg/ml) were added to the blocking buffer and incubated for 1 h at RT. The membrane was washed twice with TBS 0.5% Tween20 (TBST) for 10 min. HRP-conjugated goat anti-llama IgG was added (1:2000, diluted in blocking buffer) to the membrane and incubated for 1 h at RT. The membrane was washed twice with TBST and once with TBS for 10 min before detection with SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific).
Surface Plasmon Resonance
Interactions between the nanobodies and VAR2CSA recombinant protein were studied by surface plasmon resonance (SPR), using a Biacore X100 instrument (GE Healthcare). All experiments were performed in HBS-EP buffer (GE Healthcare) at 25°C. Recombinant full-length VAR2CSA protein was immobilized on the analysis Fc2 channel of a CM5 chip (GE Healthcare) by amine coupling to a total loading of 800 RU. Reference channel Fc1 was blocked with 1 M ethanolamine-HCl pH 8.5 using the same chemistry. Nanobodies were injected at 30 µlmin−1 in dilution series over the VAR2CSA-coated chip. The highest concentration of nanobody was 1 µM and ten twofold serial dilutions were also injected. Between the injections, the chip surface was regenerated with 2 injections of 15 µl of 10 mM HCl pH2. The specific binding response to VAR2CSA was obtained by subtracting the response given by the analytes on Fc2 by the response on Fc1. The kinetic sensorgrams were fitted to a global 1:1 interaction Langmuir model using the manufacturer's software.
Parasite culture
Three P. falciparum laboratory adapted parasite lines were used in this study: NF54, FCR3 and 7G8. Erythrocytes infected by a select parasite strains and selected for CSA-binding phenotype are referred as NF54-CSA, FCR3-CSA and 7G8-CSA. One control parasite (FCR3) selected for CD36-binding phenotype is referred as FCR3-CD36. Parasites were grown in O+ human erythrocytes in RPMI 1640 medium containing Hepes (25 mM) and L-glutamine (2 mM) (Gibco) supplemented with 5% Albumax, 5% human serum, 0.1 mM hypoxanthine and 20 µg/ml gentamicin46. Prior to each experiment, the CSA or CD36 -binding phenotypes of the parasitized red blood cells were verified on receptors immobilized on plastic Petri dishes as previously described47. Parasites were routinely genotyped by PCR using MSP1/MSP2 primers48 and tested for potential mycoplasma contamination (LookOut Mycoplasma PCR Detection Kit by SIGMA). Synchronized parasite cultures (3–7% parasitemia) at mid/late trophozoite stages were purified using VarioMACS and CS columns (Miltenyi Biotec) as previously described49.
Flow cytometry
The capacity of nanobodies to recognize the native VAR2CSA expressed at the surface of infected erythrocytes was tested by flow cytometry. Parasite culture at mid/late trophozoite stages was purified by VarioMACS and washed two times with PBS 0.2% BSA. For each assay, 1 million of IEs were incubated with 50 µl of purified nanobody (100 µg/ml diluted in PBS 0.2% BSA), for 1 h at 4°C. The IEs were washed twice with PBS 0.2% BSA and incubated for 30 min at 4°C with a mouse anti-PentaHis IgG (Qiagen, Cat.no.34660) at 5 µg/ml diluted in PBS 0.2% BSA. After 30 min, the IEs were washed twice with PBS 0.2% BSA and incubated for 30 min at 4°C with a PE-conjugated goat F(ab’)2 anti-mouse IgG (Beckman Coulter, IM0855, diluted 1:100 in PBS 0.2% BSA). Paraformaldehyde (PFA) 4% in PBS was added after a last wash and the IEs were resuspended in PBS after two additional washes. Data was acquired using a BD FACScanto II flow cytometer (Becton-Dickinson, San Jose, CA) and analysis was performed using the FLOWJO 8.1 software (Tree Star Inc.). Cellular debris and free parasites were excluded from the analysis by appropriated gating using the forward and side scatters. The reactivity of the tested nanobodies to surface-expressed VAR2CSA was reflected by an increase geometric mean in fluorescence intensity in the PE channel (Geometric Mean PE). The results were expressed as the fold ratio between the Geometric Mean PE of a given nanobody and the reference Geometric Mean PE representing the background reactivity of infected erythrocytes incubated only with secondary (anti-His) and tertiary antibodies (anti-mouse-PE).
Inhibition of IEs adhesion
Inhibition assays were performed using plastic Petri dishes (Falcon 1058) with 0.5 cm diameter spots. Spots were coated overnight at 4°C with 10 µl of 1 mg/ml CSA sodium salt from bovine trachea (SIGMA) diluted in PBS. After coating, spots were washed 3 times with 20 µl PBS and blocked with PBS 1% BSA, 20 µl per spot, for 1 h at RT. Parasite cultures at mid/late trophozoite stage were purified by VarioMACS. Binding assays were performed using 10 µl of 5.106 IEs/ml pre-incubated (for 30 min at 37°C) with 10 µl of VHH (200 μg/ml) per spot. After 1 h incubation at RT, unbound IEs were washed away by adding 50 ml of PBS 4 times. Bound IEs were then fixed with 2% glutaraldehyde in PBS for 2 h at RT. The average number of adherent IEs was determined in three independent experiments.
References
World Malaria Report 2013. World Health Organization, Geneva, Switzerland. ISBN: 9 789241 56469 4. (2013).
Miller, L. H., Baruch, D. I., Marsh, K. & Doumbo, O. K. The pathogenic basis of malaria. Nature 415, 673–679, 10.1038/415673a415673a (2002).
Barnwell, J. W., Howard, R. J. & Miller, L. H. Influence of the spleen on the expression of surface antigens on parasitized erythrocytes. Ciba Found Symp 94, 117–136 (1983).
Buffet, P. A. et al. The pathogenesis of Plasmodium falciparum malaria in humans: insights from splenic physiology. Blood 117, 381–392, 10.1182/blood-2010-04-202911blood-2010-04-202911 (2011).
Smith, J. D. et al. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell 82, 101–110, 0092-8674(95)90056-X (1995).
Baruch, D. I. et al. Cloning the P. falciparum gene encoding PfEMP1, a malarial variant antigen and adherence receptor on the surface of parasitized human erythrocytes. Cell 82, 77–87, 0092-8674(95)90054-3 (1995).
Smith, J. D., Rowe, J. A., Higgins, M. K. & Lavstsen, T. Malaria's deadly grip: cytoadhesion of Plasmodium falciparum-infected erythrocytes. Cell Microbiol 15, 1976–1983, 10.1111/cmi.12183 (2013).
McGregor, I. A. Immunity and malaria in man. Trop Doct 4, 104–109 (1974).
Walter, P. R., Garin, Y. & Blot, P. Placental pathologic changes in malaria. A histologic and ultrastructural study. Am J Pathol 109, 330–342 (1982).
Umbers, A. J., Aitken, E. H. & Rogerson, S. J. Malaria in pregnancy: small babies, big problem. Trends Parasitol 27, 168–175, 10.1016/j.pt.2011.01.007S1471-4922(11)00020-1 (2011).
Desai, M. et al. Epidemiology and burden of malaria in pregnancy. Lancet Infect Dis 7, 93–104, S1473-3099(07)70021-X (2007).
Beeson, J. G. et al. Plasmodium falciparum isolates from infected pregnant women and children are associated with distinct adhesive and antigenic properties. J Infect Dis 180, 464–472, JID981531 (1999).
Fried, M. & Duffy, P. E. Adherence of Plasmodium falciparum to chondroitin sulfate A in the human placenta. Science 272, 1502–1504 (1996).
Agbor-Enoh, S. T. et al. Chondroitin sulfate proteoglycan expression and binding of Plasmodium falciparum-infected erythrocytes in the human placenta during pregnancy. Infect Immun 71, 2455–2461 (2003).
Doritchamou, J. et al. First-trimester Plasmodium falciparum infections display a typical "placental" phenotype. J Infect Dis 206, 1911–1919, 10.1093/infdis/jis629jis629 (2012).
Fried, M., Nosten, F., Brockman, A., Brabin, B. J. & Duffy, P. E. Maternal antibodies block malaria. Nature 395, 851–852, 10.1038/27570 (1998).
Viebig, N. K. et al. Disruption of var2csa gene impairs placental malaria associated adhesion phenotype. PLoS One 2, e910, 10.1371/journal.pone.0000910 (2007).
Viebig, N. K. et al. A single member of the Plasmodium falciparum var multigene family determines cytoadhesion to the placental receptor chondroitin sulphate A. EMBO Rep. 6, 775–781, 7400466 (2005).
Salanti, A. et al. Evidence for the involvement of VAR2CSA in pregnancy-associated malaria. J Exp Med 200, 1197–1203, jem.20041579 (2004).
Salanti, A. et al. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol Microbiol 49, 179–191, 3570 (2003).
Bockhorst, J. et al. Structural polymorphism and diversifying selection on the pregnancy malaria vaccine candidate VAR2CSA. Mol Biochem Parasitol 155, 103–112, S0166-6851(07)00163-6 (2007).
Srivastava, A. et al. Full-length extracellular region of the var2CSA variant of PfEMP1 is required for specific, high-affinity binding to CSA. Proc Natl Acad Sci U S A 107, 4884–4889, 10.1073/pnas.10009511071000951107 (2010).
Khunrae, P. et al. Full-length recombinant Plasmodium falciparum VAR2CSA binds specifically to CSPG and induces potent parasite adhesion-blocking antibodies. J Mol Biol 397, 826–834, 10.1016/j.jmb.2010.01.040S0022-2836(10)00086-0 (2010).
Srivastava, A. et al. Var2CSA minimal CSA binding region is located within the N-terminal region. PLoS One 6, e20270, 10.1371/journal.pone.0020270PONE-D-11-03862 (2011).
Clausen, T. M. et al. Structural and functional insight into how the Plasmodium falciparum VAR2CSA protein mediates binding to chondroitin sulfate A in placental malaria. J Biol Chem 287, 23332–23345, 10.1074/jbc.M112.348839M112.348839 (2012).
Bigey, P. et al. The NTS-DBL2X region of VAR2CSA induces cross-reactive antibodies that inhibit adhesion of several Plasmodium falciparum isolates to chondroitin sulfate A. J Infect Dis 204, 1125–1133, 10.1093/infdis/jir499jir499 (2011).
Bordbar, B. et al. Identification of Id1-DBL2X of VAR2CSA as a key domain inducing highly inhibitory and cross-reactive antibodies. Vaccine 30, 1343–1348, 10.1016/j.vaccine.2011.12.065S0264-410X(11)02003-2 (2012).
Doritchamou, J. et al. Differential adhesion-inhibitory patterns of antibodies raised against two major variants of the NTS-DBL2X region of VAR2CSA. Vaccine 31, 4516–4522, 10.1016/j.vaccine.2013.07.072S0264-410X(13)01058-X (2013).
Muyldermans, S. Nanobodies: natural single-domain antibodies. Annu Rev Biochem 82, 775–797, 10.1146/annurev-biochem-063011-092449 (2013).
Adams, H. et al. Specific immuno capturing of the staphylococcal superantigen toxic-shock syndrome toxin-1 in plasma. Biotechnol Bioeng 104, 143–151, 10.1002/bit.22365 (2009).
Tereshko, V. et al. Toward chaperone-assisted crystallography: protein engineering enhancement of crystal packing and X-ray phasing capabilities of a camelid single-domain antibody (VHH) scaffold. Protein Sci 17, 1175–1187, 10.1110/ps.034892.108ps.034892.108 (2008).
Park, Y. J., Pardon, E., Wu, M., Steyaert, J. & Hol, W. G. Crystal structure of a heterodimer of editosome interaction proteins in complex with two copies of a cross-reacting nanobody. Nucleic Acids Res 40, 1828–1840, 10.1093/nar/gkr867gkr867 (2012).
Rasmussen, S. G. et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature 477, 549–555, 10.1038/nature10361nature10361 (2011).
Rahbarizadeh, F., Ahmadvand, D. & Sharifzadeh, Z. Nanobody; an old concept and new vehicle for immunotargeting. Immunol Invest 40, 299–338, 10.3109/08820139.2010.542228 (2011).
Muyldermans, S. Single domain camel antibodies: current status. J Biotechnol 74, 277–302, S1389-0352(01)00021-6 (2001).
Habib, I. et al. V(H)H (nanobody) directed against human glycophorin A: a tool for autologous red cell agglutination assays. Anal Biochem 438, 82–89, 10.1016/j.ab.2013.03.020S0003-2697(13)00140-1 (2013).
Dahlback, M. et al. The chondroitin sulfate A-binding site of the VAR2CSA protein involves multiple N-terminal domains. J Biol Chem 286, 15908–15917, 10.1074/jbc.M110.191510M110.191510 (2011).
Ditlev, S. B. et al. Utilizing nanobody technology to target non-immunodominant domains of VAR2CSA. PLoS One 9, e84981, 10.1371/journal.pone.0084981PONE-D-13-40830 (2014).
Gamain, B. et al. Identification of multiple chondroitin sulfate A (CSA)-binding domains in the var2CSA gene transcribed in CSA-binding parasites. J Infect Dis 191, 1010–1013, JID33609 (2005).
Rasti, N. et al. Nonimmune immunoglobulin binding and multiple adhesion characterize Plasmodium falciparum-infected erythrocytes of placental origin. Proc Natl Acad Sci U S A 103, 13795–13800, 0601519103 (2006).
Tutterrow, Y. L. et al. High avidity antibodies to full-length VAR2CSA correlate with absence of placental malaria. PLoS One 7, e40049, 10.1371/journal.pone.0040049PONE-D-12-09930 (2012).
Magistrado, P. A. et al. High efficacy of anti DBL4varepsilon-VAR2CSA antibodies in inhibition of CSA-binding Plasmodium falciparum-infected erythrocytes from pregnant women. Vaccine 29, 437–443, 10.1016/j.vaccine.2010.10.080S0264-410X(10)01593-8 (2011).
De Genst, E. et al. Molecular basis for the preferential cleft recognition by dromedary heavy-chain antibodies. Proc Natl Acad Sci U S A 103, 4586–4591, 0505379103 (2006).
Hassanzadeh-Ghassabeh, G., Saerens, D. & Muyldermans, S. Generation of anti-infectome/anti-proteome nanobodies. Methods Mol Biol 790, 239–259, 10.1007/978-1-61779-319-6_19 (2011).
Pardon, E. et al. A general protocol for the generation of Nanobodies for structural biology. Nat Protoc 9, 674–693, 10.1038/nprot.2014.039nprot.2014.039 (2014).
Trager, W. & Jensen, J. B. Human malaria parasites in continuous culture. Science 193, 673–675 (1976).
Viebig, N. K., Nunes, M. C., Scherf, A. & Gamain, B. The human placental derived BeWo cell line: a useful model for selecting Plasmodium falciparum CSA-binding parasites. Exp Parasitol 112, 121–125, S0014-4894(05)00233-X (2006).
Snounou, G. et al. Biased distribution of msp1 and msp2 allelic variants in Plasmodium falciparum populations in Thailand. Trans R Soc Trop Med Hyg 93, 369–374 (1999).
Staalsoe, T., Giha, H. A., Dodoo, D., Theander, T. G. & Hviid, L. Detection of antibodies to variant antigens on Plasmodium falciparum-infected erythrocytes by flow cytometry. Cytometry 35, 329–336, 10.1002/(SICI)1097-0320(19990401)35:4<329::AID-CYTO5>3.0.CO;2-Y (1999).
Acknowledgements
This work was supported by an ATIP-AVENIR grant from the Institut National de la Santé et de la Recherche Médicale (B.G.) and by the “Investissements d'avenir” funded Laboratory of excellence GR-Ex. Sofia Nunes-Silva was supported by a labex GR-Ex fellowship. The labex GR-Ex, reference ANR-11-LABX-0051, is funded by the program “Investissements d'avenir” of the French National Research Agency, reference ANR-11-IDEX-0005-02. A.C. was supported by a grant from the Fondation pour la Recherche Médicale [SPF20110421450]. A.V. is the recipient of a PhD fellowship from FWO-Vlaanderen. E.P. is supported by the grant 7/40 of the Interuniversity Attraction Poles (IAP) Program of the Belgian Science Policy Office.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: S.N., S.G., E.P., S.R., A.C. and B.G. Performed the experiments: S.N., S.G., M.V., A.V. and S.D. Analysed the data: S.N., S.G., S.D., E.P., J.S., S.R., A.C. and B.G. Contributed reagents/materials/analysis tools: S.C. Wrote the paper: S.N., S.G., E.P., A.C. and B.G. All authors reviewed the final version of manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Info
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Nunes-Silva, S., Gangnard, S., Vidal, M. et al. Llama immunization with full-length VAR2CSA generates cross-reactive and inhibitory single-domain antibodies against the DBL1X domain. Sci Rep 4, 7373 (2014). https://doi.org/10.1038/srep07373
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep07373
This article is cited by
-
A novel epitope tagging system to visualize and monitor antigens in live cells with chromobodies
Scientific Reports (2020)
-
Peptides in headlock – a novel high-affinity and versatile peptide-binding nanobody for proteomics and microscopy
Scientific Reports (2016)
-
Structure of the DBL3X-DBL4ε region of the VAR2CSA placental malaria vaccine candidate: insight into DBL domain interactions
Scientific Reports (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.