Abstract
The influenza virus PB1 protein is the core subunit of the heterotrimeric polymerase complex (PA, PB1 and PB2) in which PB1 is responsible for catalyzing RNA polymerization and binding to the viral RNA promoter. Among the three subunits, PB1 is the least known subunit so far in terms of its structural information. In this work, by integrating template-based structural modeling approach with all known sequence and functional information about the PB1 protein, we constructed a modeled structure of PB1. Based on this model, we performed mutagenesis analysis for the key residues that constitute the RNA template binding and catalytic (TBC) channel in an RNP reconstitution system. The results correlated well with the model and further identified new residues of PB1 that are critical for RNA synthesis. Moreover, we derived 5 peptides from the sequence of PB1 that form the TBC channel and 4 of them can inhibit the viral RNA polymerase activity. Interestingly, we found that one of them named PB1(491–515) can inhibit influenza virus replication by disrupting viral RNA promoter binding activity of polymerase. Therefore, this study has not only deepened our understanding of structure-function relationship of PB1, but also promoted the development of novel therapeutics against influenza virus.
Similar content being viewed by others
Introduction
Influenza A viruses are segmented, negative RNA viruses, which are dependent on a heterogenic trimeric polymerase complex consisting of PA (polymerase acidic), PB1 (polymerase basic 1) and PB2 (polymerase basic 2) subunits to transcribe and replicate their RNA genomes1. An elucidation of the structure-function relationship of the influenza replication machinery is indispensable not only for a profound mechanistic understanding of the viral infection, activity and pathogenesis, but also for designing more effective therapeutics against the recurring pathogens that have caused and will continue to cause significant morbidity and mortality in human population2,3,4. Therefore, many efforts have been devoted to understand the molecular mechanisms of the viral replication machinery5,6,7,8,9,10. Within the influenza heterotrimeric polymerase complex, PB1 is the RNA-dependent RNA polymerase (RdRP) subunit6,11,12,13,14,15,16, which carries out viral mRNA and genomic RNA synthesis in reactions with the cap-binding subunit PB2 and the endonuclease subunit PA9,10. As the central functional subunit for catalysis of influenza viral RNA synthesis, the molecular mechanism of PB1 has been extensively explored, including the identification of several critical sites in the four conserved RdRP motifs that are essential for the RNA polymerase activity17,18,19, mapping of the epitopes on PB120, determination of its nuclear location signal21,22, characterization of its interactions with PA, PB27,8,14,23 and RNA promoter24,25 and more. Since PB1 plays a central role in synthesis of influenza virus RNA genome, it has also become a promising target for developing new anti-influenza drugs12,26,27,28,29,30.
Despite the extensive attention to PB1 which has led to many discoveries of PB1 functionality, the progress from its structural analysis is rather limited. So far, there is no crystal structure available for PB1 protein except its N terminal 25 amino acids and C terminal about 80 amino acids7,8. This is mainly due to its low solubility when expressed alone in bacteria or the low productivity of the active heterotrimeric polymerase complex when expressed in insect cells and mammalian cells5. Therefore, the structural basis for PB1 functionality is still unclear. Moreover, the lack of the structural information of PB1 hinders a deep understanding of the influenza viral replication, infection and pathogenesis.
Fortunately, structures of several RdRPs of other positive-strand RNA viruses have been determined and reported to have similar architecture despite some structural divergence31,32,33. Moreover, based on sequence and secondary structure analysis, at least four motifs have been characterized as the “polymerase module”, which are generally conserved in negative-strand, positive-strand and double-strand RNA viruses17. Biswas et al further verified the involvement of these conserved motifs in PB1 polymerase activity by mutational analysis18. Therefore, it is reasonable to assume that the known structures of RdRPs can be used as structural template to model the structure of PB1. Recently, much progress has been achieved in modeling of protein structures from known structural templates, an approach called template-based modeling (TBM)34. For example, we have developed a TBM method, called FR-t5, which can improve structural modeling by considering the local structure preference35. TBM method is widely used, before the structures are solved, to decipher the structure and function relationship of proteins like human APOBEC3G36, GBV-C RdRP37, L protein of Lassa virus38,39, KERP140 and so on.
In this study, by integrating known functional information of PB1 into FR-t535, we have constructed a structure model of PB1. The modeled PB1 structure correlates well with its known functional characterizations, offering structural insights into its functionality. Moreover, based on the model, we identified the residues forming a RNA template binding and catalytic (TBC) channel. Many new residues surrounding the TBC channel are verified critical for influenza virus RNA synthesis by using RNP reconstitution system. Then, based on the modeled structure, we have derived 5 peptides from PB1 to inhibit viral polymerase activity. More importantly, we found that one peptide PB1(491–515) from template binding and catalytic channel of PB1 can inhibit influenza virus replication by disrupting the vRNA promoter binding activity of influenza virus polymerase. Our work has not only deepened understanding of the structure-function relationship of PB1, but also offered new lights into the design of anti-viral therapeutics by targeting the essential protein of influenza virus.
Results
Structure modeling of influenza A virus PB1 protein
Usually, TBM can reliably build the structure for a protein based on the structure of its homologous protein if they have a high sequence similarity (over 30%). For PB1, its sequence similarity to the viral RdRPs with known structures is close to or below 30% (~20–32%) (Supplementary Table 1). Traditional TBM methods can not be used to construct reliable structure of PB1. To model the PB1 structure with high accuracy, we designed a computational framework by integrating the known functional information of PB1 into the general TBM method FR-t5 that consists of three steps35 (Fig. 1A). First, target sequence was threaded on every potential template within structure database and their alignment was scored. Then, the optimal template was selected based on the rank of their normalized alignment scores. Finally, the structural model was constructed according to the alignment between target sequence and its optimal template. Two pieces of knowledge about PB1 RNA polymerase activity were considered in the modeling process using FR-t5. One is the region from 110 to 637 as the PB1 RdRP domain to be modeled according to Asano et al's work11 (Fig. 1B). The other is the four conserved sites D305, G406, D445 and K481 located on four conserved polymerase motifs A, B, C and D respectively to guide the sequence-structure alignment in the structure modeling process (Fig. 1B and Methods) since they have been shown to be required for activity of all RdRPs including that of PB117,18. Indeed, the functionally related structure templates for PB1 were more effectively identified in the function-guided FR-t5 than those without using site constrains (Fig. 1C, D). The RdRP of Norwalk Virus (NV) (PDB ID: 2B43, 1208-1695AA) was identified as the top-scored template and thus was used to construct the structural model for PB1. Fig. 1E and Supplementary Table 2 shows that the alignment of RdRP of NV (PDB ID: 2B43, 1208-1695AA) with the PB1 sequence (71-612AA) presents good match especially for five motifs which were conserved in all RdRPs. And more importantly, the good match of secondary structure elements between PB1 and NV RdRP indicates the reliability of the alignment (Fig. S1).
Structural modeling of influenza A virus PB1 protein.
(A) The modeling flowchart. (B) Selection of the PB1 core region 110–637 as initial seed segment to search its potential structural templates. Four conserved residues (D305, G406, D445 and K481) on the RdRP motifs (motif A to D) are highlighted in red. The binding regions of PA and PB2 on PB1's N and C terminuses are labeled in gray. NLS, Nuclear Localization Signal; POL, Polymerase. (C–D) The comparison of the threading results by using (C) or not using (D) functional site constraints. The percentages of RdRP and Non-RdRP for top 5, 10 and 20 templates were presented in black and gray. (E) The sequence alignment between influenza A PB1 segment 71–612 and the selected template, Norwalk Virus (NV-) RdRP. Five conserved motifs (preA, A, B, C and D) were highlighted. Amino acids are colored according to its polarity: basic amino acids in blue, acidic in red.
A brief description of the modeled structure for PB1
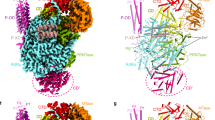
Based on the alignment results, the structural model of PB1 segment 71-612AA was modeled with the optimal template (NV-RdRP: 2B43D) and refined with SWISS-MODEL. Previous studies showed that the “closed right-hand” conformation is a characteristic of all viral RdRPs, which is composed of three domains, namely the palm, fingers and thumb domains. In our modeled PB1 structure (Fig. 2A), the two regions 267-313AA and 406-501AA form the palm domain, 71-266AA and 314-405AA form the fingers domain and the thumb domain includes a continuous segment 502-600AA. In addition to the overall “closed right-hand” topology, five structural motifs have also been identified among all viral RdRPs including one (pre-motif A) at fingers domain and four (motif A, B, C and D) located at palm domain (Fig. 2A), which are implicated in metal binding, template recognition and catalytic activity17,18,31. Functionally, the most important feature of all viral RdRPs is the formation of a viral RNA template binding and catalytic (TBC) channel that is responsible for binding template, substrates and cofactors, as well as catalyzing the nucleotidyl transferase reaction31,41,42. We can see that PB1 contains TBC channel, formed by palm, fingers and thumb domains, in the central of the structure (Fig. 2A). PB1-RNA complex was modeled according to the complex structure of NV RdRP and RNA (PDB ID: 3BSN) (Fig. 2B). Based on the conservation analysis of amino acids on PB1 (see Methods), the inner surface residues that line the TBC channel contains more conserved residues than on the outer surface (Fig. 2B). This not only indicates the functional importance of the inner region for RNA synthesis but also suggests the reliability of our modeled structure. Next, we analyzed the electrostatic surface properties of PB1. Interestingly, we found there are three positive charge patches (PCP 1, 2, 3) on the model (Fig. 2C, D). Moreover, we have found that the NTP tunnel of PB1 is located on PCP1 (Fig. 2C). It is in line with the fact that NTP tunnel is responsible for diffusing NTPs in positive stranded RNA virus RdRP.
Characteristics of modeled PB1 structure.
(A) The overall view of the model. Three structural domains, “Fingers”, “Palm” and “Thumb” domain were separated with dash lines approximately. Five conserved motifs (preA, A, B, C and D) were shown as flat ribbons colored with purple, blue, drab, green and orange respectively. TBC (template binding and catalytic) channel is indicated in the central of the model. 40 newly identified residues constituted the TBC channel were mapped on the PB1 model, presented side-chains with red. (B) The distribution of non-conserved sites (highlighted by green mesh) on the PB1 model. The structure is rotated around Y axis by 180° refer to (A). The template binding and catalytic (TBC) channel and NTP tunnel are indicated by two curved arrows. The docking of RNA duplex composed of template RNA (blue) and synthesized RNA chains (orange) was based on the RdRP-RNA complex structure of the Norwalk virus (PDB ID: 3BSN). The sites highlighted are the same to that in (A). (C–D) Electrostatic variation from negative to positive charges presented with the color differences from red to blue gradually. Three regions are highly positively charged, designated as positive charge patch (PCP).
Previous studies showed that the “Flap” like structure was used as a structure-marker to classify whether the RdRP can use dsRNA as template or not when initiating RNA synthesis31,32,43,44. Interestingly, we found the deletions of the “flap” loop (Fig. 3A) for PB1 and NV RdRP correlated with their abilities to bind to double-strand template-primer RNA (dsRNA) since the “flap” loop was only present in RdRPs that binds single-strand RNA (ssRNA)31,43,44. We also have collected the previously identified important residues on PB1 as shown in Supplementary Table 3. Remarkably, most of the previously reported functionally important sites were mainly located at the five conserved motifs which are around the modeled TBC channel (Fig. 3B), while the nuclear localization signals (187–195 and 203–216AA)21,22 and antigenic epitope (586–599AA)20 are located on the surface of the modeled structure (Fig. 3B). Thus our modeled structure has provided potential structural basis for the previously identified functional regions, enabling a deep understanding of PB1 structure-function relationship.
The functional features of PB1.
(A) Local sequence and structural alignments between PB1 and RdRP templates. Norwalk Virus (NV-RdRP) and Dengue Virus (DV-RdRP) were colored with orange and gray separately. The “flap” insertion segment on DV relative to PB1 and NV was marked with rectangle. As the 2nd structure (2ND-STR) elements alignment, H means helix, C means coil. (B) The mapping of the 17 reported functional sites and 3 functional regions on the PB1 model. Reported 17 functional important sites (Supplementary Table 3), two nuclear localization signal (NLS) regions (187–195 and 203–216) and antigenic epitope (586–599) were colored red, purple and blue separately. Epitope was highlighted with surface mesh.
Identification of new residues of PB1 critical for RNA synthesis
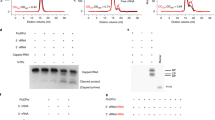
Besides providing a comprehensive view of PB1 structure-function relationship, our structural model also can allow us to systematically identify 40 new essential residues that constitute the TBC channel, with accessible surface area > 10 Å2, the catalytic center of the protein (Supplementary Table 4). To verify their critical roles in the process of RNA synthesis, 9 sites (M179, K229, K278, K279, K308, E491, F492, F495 and F496) around the TBC channel from Supplementary Table 4 were randomly picked and their effects on polymerase activity and RNA synthesis activity were analyzed by mutational analysis (see Methods). The functions of these selected residues are largely unknown. For controls we also randomly selected 6 sites (K288, F381, K391, K433, F551 and K586) outside these regions (Fig. 4A). In order to observe the mutational effect clearly, we did reverse charge mutations (such as lysine to glutamic acid or glutamic acid to arginine) or changed the physiochemical property by substituting methionine or phenylalanine to serine. Previous study has discovered that the D445H substitution can disrupt the influenza virus polymerase activity while V451A can not18. Therefore, we used these two mutants as controls in our experimental system (Fig. 4B, C). The expression amounts of PB1 were on the similar level (data not shown). Then the effects of mutations on polymerase activity were analyzed by RNP reconstitution assay using firefly luciferase as reporter gene. Fig. 4B shows, among the mutations located at TBC channel, except for the one mutation at site 495 has mild effects on the polymerase activity using 1.0% as the cutoff, the remaining nine mutations at site 179, 229, 278, 279, 308, 445, 491, 492 and 496 can destroy the enzyme activity remarkably. While for the six sites located outside the TBC channel, only two mutations at sites 288 and 381 can significantly decrease polymerase activity and the other four mutations have little or mild effects. By conducting primer extension assay based on a minireplicon system of influenza A virus using CAT as RNA reporter gene (details see Methods), we further analyzed the effects of these mutants on viral mRNA, vRNA and cRNA synthesis. The results showed that, for the sites that constitute the TBC channel, except mutation at 495, other mutations at site 179, 229, 278, 279, 308, 445, 491, 492 and 496 can also disrupt the RNA synthesis activity of the enzyme remarkably. Moreover, for the six sites located outside the TBC channel, four mutations have little or mild effects to the polymerase activity or RNA synthesis (Fig. 4B, C). The polymerase activity of PB1 K288E is very closed to the cut off (Fig. 4B) and its RNA synthesis activity is not totally disrupted as shown in Fig. 4C. The region containing F381 may evolve in interacting with other subunits or host factors indicating the importance of the site F381. These results are in agreement with those from the polymerase activity assay (comparing Fig. 4B with Fig. 4C). Then, we have rescued viruses within WSN33 background with 15 selected mutations (K433E, V451A, not done), respectively and analyzed their effects on virus production. As shown in Fig. S2, only the viruses containing K391E, F495S and K586E can be rescued with high titers and M179S and F551S with relatively lower titers, while the others can not be rescued. The mRNA level of PB1 M179S is detectable as shown in Fig. 4C, which explained that the virus containing M179S can be rescued with low titer. These results are consistent with those from primer extension assay and polymerase activity assay.
Effects of mutations in the PB1 subunit on polymerase activity and RNA synthesis.
(A) The location of all selected sites on the PB1 model. Sites colored red if its polymerase activity is less than 1% of that of wild type PB1 as shown in (B), while others blue. Circled sites indicate that the locations of these sites are around the template binding and catalytic channel. (B) The polymerase activity of influenza A in the presence of the indicated PB1 mutants. The activity detected with samples contained wild type PB1 (WT) was set to 100% and a transfection mixture with the omission of PB1 (-PB1) used as negative control. (C) In vivo RNA synthesis mediated by PB1 mutants. In vivo primer extension of vRNA, mRNA and cRNA isolated from 293T cells, as indicated above the line, transfected either with pPolI-CAT-RT and pcDNA3.1 (indicated by “C”), with pPolI-CAT-RT, pcDNA-PB2, pcDNA-PA (-PB1), or with pPolI-CAT-RT, pcDNA-PB2, pcDNA-PA and wild type (WT) or mutant pcDNA-PB1. Signals of vRNA, mRNA, cRNA and 5 s RNA, used as control, are indicated.
Moreover, in order to verify our model more extensively, we have further mutated other 11 residues on surface of PB1 and analyzed their effects on viral RNA synthesis. It showed that only one (R560E) out of 11 mutants can disrupt the influenza polymerase activity and RNA synthesis activity completely, while the other 10 remained significant polymerase activities (Fig. S3). These results further supported our structural model since most of the mutations made on the surface of PB1 are not critical for the polymerase activity and RNA synthesis activity. And also, we have analyzed the conservation of all the mutated sites. Fig. S4 shows that most of the selected residues are conserved. It is difficult to find novel residues that are critical for virus RNA synthesis if only based on sequence conservation. The mutational analyses provide additional insights to support and elaborate the modeled structure of PB1, enabling us to identify the novel critical regions or residues that constitute the TBC channel.
Model-based derivation of peptides to inhibit influenza polymerase activity and virus replication
As demonstrated above, our structural modeling has revealed the critical sites and regions that constitute the TBC channel of PB1, suggesting the crucial roles of these sites and regions in influenza virus replication. We further asked whether these regions can provide effective anti-viral therapeutics for control of influenza virus infection by disrupting the activity of influenza polymerase. We followed 3 rules to select potential inhibiting peptides. First, they all have stable secondary structures in our model. Second, they are all located around the template binding and catalytic channel. Third, the functions of two selected regions are well explored such as 176–195AA contains NLS21,22 and 231–255AA is responsible for binding RNA24. We therefore derived 5 short peptides which were composed of about 25 amino acids around the modeled TBC channel, namely peptide 176–195, 231–255, 271–295, 481–515 and 516–540. Also, as references, we selected 5 short peptides far from the channel according to our model noted as peptide 296–330, 376–405, 406–435, 436–455 and 541–565. All peptide-GFP fusion proteins were over expressed in HEK293T cells with plasmids of minireplicon system of influenza A virus and a plasmid constitutively express Renilla luciferase which was used for normalizing variation in transfection efficiency. The expression level of every fragment-GFP fusion protein was semi-quantified with GFP antibody by Western blot. The amounts of Flag-GFP were higher than or equal to other fusion proteins (Fig. 5A). Results indicated that four of five peptides around the TBC channel (176–195, 231–255, 481–515 and 516–540) can inhibit RdRP's function efficiently except peptide 271–295 (Fig. 5A). While for other 5 peptides far from the TBC channel, only one peptide 516–540 can inhibit RdRP's function efficiently (Fig. 5B). These results further confirmed our hypothesis that regions surrounding the TBC channel are very important to the RNA synthesis. Among the four peptides that have the inhibitory effects on PB1 polymerase activity, we also found the peptide 481–515 was able to inhibit influenza A/WSN/33 and A/Quail/G1/HK/97 virus replication (Fig. 5C–D) while others need further investigation. Moreover, the IAV-Luc virus45, a replicable influenza virus expressing secreted Gaussia luciferase, was used to determine the inhibitory effects of the peptide 481–515. Fig. 5E shows that the peptide 481–515 can inhibit IAV-Luc replication effectively at 12 h post infection (p.i.), while not so effective at 24 h p.i. And also, as shown in Fig. 5F, like PB11-25-GFP, PB1481-515-GFP can inhibit WSN33 replication at 12 h p.i. since the NP expression was inhibited.
The PB1 structure model derived peptides inhibit influenza virus polymerase activity and virus replication.
(A) Polymerase inhibitory activity of 10 model-derived peptide-GFP fusion proteins in influenza A virus minireplicon systems. The activity detected with samples containing Flag-GFP was set to 100%. PB11-25-GFP (1–25) was used as positive control. A transfection mixture without PB2 used as negative control.* indicates p < 0.05 (Student t-test). Error bars mean standard deviation (n = 3). (B) Location of these 10 peptides on the PB1 model. Five peptides with remarkable inhibitory effects were marked and colored with red while other five peptides with green. The N- and C- terminus of each peptide was marked with scissors. (C–D) The inhibitory effects of PB1481-515-GFP to H1N1 (A/WSN/33) (C) and H9N2 (A/Quail/HK/G1/97) (D) viruses replication in A549 cells. The activity detected with samples containing Flag-GFP (Flag-GFP) was set to 100% after subtracting background of uninfected samples (Mock). PB11-25 was used as positive control. (E). PB1481-515 can inhibit influenza virus IAV-Luc replication. 293T cells were transfected with PX/PB11-25/PB1481-515-GFP expressing plasmids, respectively. 24 h post transfection, cells were challenged with influenza virus (IAV-Luc) carrying Gaussia luciferase reporter gene at MOI = 0.25. 12 or 24 h p.i., activity of Gaussia luciferase from supernatant was detected using microplate reader. The activity from samples containing PX-GFP was set to 100%. (F). PB1481-515 can inhibit influenza virus (A/WSN/33) replication. 293T cells were transfected with PX/PB11-25/PB1481-515-GFP expressing plasmids. 24 h post transfection, cells were challenged with influenza virus (A/WSN/33) at MOI = 0.02. 12 h p.i., Western blot of whole cells extracts were conducted to determine the expression level of actin, GFP fusion proteins and influenza virus NP.
PB1491-515 can disrupt influenza polymerase activity in vitro by inhibiting its RNA promoter binding activity
We thus ask whether the peptide in isolation can compete with the TBC channel for viral RNA binding and interfere with influenza RdRP activity since it is around the modeled TBC channel. As to the peptide PB1481-515, we found that it also can inhibit the polymerase activity of influenza virus after deletion of the N terminal 10 amino acids (named as PB1491-515) compared to PB11-25-GFP (Fig. 6A). Furthermore, in order to see whether this peptide alone can inhibit influenza virus replication in vitro, the peptide PB1491-515-TAT with PB1491-515 fused to a membrane permeable sequence from HIV TAT46,47 was synthesized and added to the cell culture of A549 cells to test its inhibitory effect on influenza virus replication. A peptide of 25 amino acids correlated to 69–93AA of BDV-P (denoted as PX) was used as negative control since it showed no inhibition to influenza virus replication in previous studies27,48. Then, the peptides were tested to see whether it can inhibit virus replication using a PolI driven plasmid expressing Gaussia luciferase as reporter gene for detecting viral infection49. A549 cells were treated by peptides PB1491-515-TAT or PX-TAT as control before and after infected with virus (A/Quail/HK/G1/97). As we expected, PB1491-515-TAT can inhibit virus replication efficiently at 10 μM while had no side effects to cells growth compared to PX-TAT (Fig. 6B).
The peptide PB1491-515 can inhibit the activity of influenza virus RNA dependent RNA polymerase in vivo and in vitro effectively.
(A) The inhibitory effects of PB1491-515-GFP to activity of influenza virus RdRP in influenza A virus minireplicon systems. (B) The inhibitory effects of synthesized peptide PB1491-515-TAT to virus (A/Quail/HK/G1/97) infection. A549 cells were treated with indicated synthesized peptides before and after virus infection. The activity detected with samples containing PX-TAT was set to 100% after subtracting background of uninfected samples. (C) In vitro ApG primed transcription activity of PB2-TAP purified influenza A virus polymerase with synthesized peptides PX-TAT or PB1491-515-TAT at indicated concentrations (μM). TP means transcription products. (D) 3' vRNA promoter binding activity of influenza virus RdRP after adding indicated peptides. The position of RNA cross-linked polymerase is indicated. Sample without polymerase (no protein) is used as negative control.
Next, we tested its inhibitory activity to influenza virus polymerase in vitro transcription activity. Firstly, influenza virus polymerase complex were purified using TAP pull down assay as described earlier15. In vitro ApG primed transcription assay was conducted by incubation of polymerase complex and peptide at 30°C for 30 min firstly and then the ApG and RNA promoter were added into the reaction mixture23. As Fig. 6C shown, the peptide PB1491-515-TAT can disrupt the transcription activity of polymerase at 5 μM while the PX-TAT can not inhibit the polymerase activity even at higher concentration. Since promoter binding is the first procedure when polymerase conducted its transcription activity, we asked whether the peptide can inhibit polymerase activity by inhibiting its binding with RNA promoter. As shown in Fig. 6D, PB1491-515-TAT can inhibit influenza virus polymerase binding to RNA promoter efficiently in a dose dependent manner. Therefore we argue that the inhibitory effects of PB1491-515 on influenza polymerase activity and virus replication could be due to its competition with TBC channel for viral RNA binding, suggesting that inhibition of the binding between PB1 TBC channel and viral RNA templates offer potential novel anti-flu therapeutics.
Discussion
Up to now, structural knowledge about influenza virus PB1 protein has been very little except for its short N-terminal and C-terminal7,8 and there have not been any FDA approved therapeutics against PB1 polymerase to inhibit viral replication. By integrating computational modeling and functional assays, we described the structure-function relationship of the PB1 subunit of influenza A virus for the first time as we known. This work also provided new insights into designing novel therapeutics to influenza virus.
In our study, in order to avoid the negative effect for alignment from the low homological sequence segment, we chosen a core segment (111–640) to find the optimal template. Then we used conserved sites to restrain sequence-structure alignment and to make sure four conserved sites in all viral polymerase (D305, G406, D445 and K481) to be aligned correctly. The final template was selected based on the functional clusters analysis and motifs conservation. These conserved functional information improved the results remarkably (Fig. 1C–D). Therefore, the TBM method of integrating target-specific functional constraints could be developed into a more general threading method for protein structure modeling. Though low sequence identity (11%) and insertions or deletions would affect the accuracy of the modeling result, good motif conservations and secondary structure elements identity (Fig. S1) indicated the reliability of the structure topology. More importantly, the modeled structure can well explain the functions of the previously identified functional regions. For example, the conserved RdRP motifs were modeled to constitute the TBC channel whereas NLS and antigenic epitopes of PB1 were located on the surface of our model, correlating with their functions.
Although all known structural data confirm the proposal that all polynucleotide polymerases share similar structures and mechanisms of catalysis31,32,33, up to the present, there is still no crystal structure of negative-strand RdRP since it is difficult to express or to crystallize. The solved structures of RNA polymerase can be divided into two groups depending on whether it can bind dsRNA or not. The structure difference between the two groups is that whether or not it contains “Flap” like structure which can prevent RdRP binding to dsRNA31,43,44. Hass et al modeled structure of Lassa virus RdRP domain, which is from negative stranded RNA virus, using HCV RdRP as template39. The limitation is obvious that HCV's NS5B can not bind to dsRNA because there is a flap in thumb domain while Lassa virus's L protein can bind dsRNA43,44. The same to PB1 as L and NV RdRP, they both can use dsRNA as template23,50 although PB1 can also use ApG as primer to synthesis mRNA.
The modeled structure of PB1 also provides structural insights into the mechanism of transcription and replication especially for vRNA promoter binding. The vRNA promoter of influenza A virus is composed by 13 and 12 nt of the influenza virus 5' and 3' ends respectively23. Earlier researches found that the vRNA promoter binding site mainly located on the PB1 subunit24. The N1 region (231–249AA) was involved in binding to the 5' end of the vRNA promoter while 5A region contained R571 and R572 was responsible for the 3' vRNA binding24,25. In our structure model of PB1, the N1 region located on the inner surface of template binding and catalytic channel, it is consistent with the results reported by Jung et al24. The 5A region was on opposite direction to N1 region in our model. The reason may be that the template we used was at its inactive form and when it binds to the substrate, the thumb region may have a movement towards the finger region like HIV RT when it matured or other positive stranded RNA virus RdRPs51. This structure shift may be initiated by binding to the 5' vRNA promoter. Based on our model, we also found a “hinge” region between palm and thumb region of PB1 which may facilitate the movement of thumb domain.
Targeting PB1 opens new avenues to develop efficient anti-flu therapeutics. Recently, pioneer works by Ortigoza et al through screening of chemical compounds, have identified small molecule inhibitors ASN2 can target PB152. Su et al also have found molecule 367 can inhibit influenza virus replication by binding to PB129. Until to now, there was no FDA approved drug targeting PB1 to inhibit virus replication. Based our model, we also found five new peptide inhibitors to influenza virus polymerase activity (Fig. 5). Four out of five peptides that can inhibit polymerase activity were surrounding the TBC channel. These peptides could inhibit the polymerase activity probably by disrupting the conformation changes of PB1, the interactions between PB1 and substrates or template and so on, which needs further investigation. However, only one peptide (541–560) that far from the TBC channel can inhibit influenza virus polymerase activity. It may stabilize the conformation change initiated by binding to RNA substrate like pep A and pep B in HIV RT to inhibit replication of HIV53. Moreover, the synthetic peptide PB1491-515 which derived from the interface between the thumb region and the template RNA, can inhibit the viral replication when introduced into the cells. The reason for the other four peptides that can inhibit influenza virus polymerase activity in the RNP reconstitution system, but can not inhibit virus replication is unknown. We speculate that the presence of other viral proteins might preclude the inhibition effects of the four peptides in infected cells. Further we found the peptide disrupted the binding between the polymerase and RNA promoter, confirming the reliability of our modeling of TBC channel as structural basis for influenza RNA synthesis, but also open new avenues for structure-based design of novel therapeutics targeting TBC channel. However, the mechanism underlying the peptide can inhibit the binding between the polymerase and RNA promoter needs further investigation.
Although PB1 and its template RdRP of Norwalk Virus have considerable conservation in the five conserved motifs, it should be noted that the overall sequence similarity (31.7%) between them is not high. Therefore, the low similarity may affect the modeling accuracy of the regions with long insertion or deletion. However, given the high conservation of the polymerase motifs and of the polymerase catalytic and RNA binding channel, together with our supporting functional analysis, it is likely that the model, at least for the core part of the enzyme is correct. Surely, this model will await the real crystal structure of PB1 or the polymerase complex to correct it. However, at this moment, this PB1 model offers a potential structural view of the functional domains of the PB1 that may provide a novel target for developing anti-influenza virus therapeutics.
Methods
Cells and plasmids
HEK293T, human type II alveolar epithelial (A549) and MDCK cells were maintained in Dulbecco's modified Eagle's medium containing 10% Fetal Bovine Serum (FBS; Gibicol) and penicillin/streptomycin (100 units/ml and 50 μg/ml, respectively) for MDCK. All cells were incubated at 37°C and 5% CO2. Transfection assay was conducted according to the manual. pPolI-NP-luc, cDNA of H5N1(A/goose/Guangdong/1/96) and IAV-Luc virus were generously provided by professors Martin Schwemmle (University of Freiburg, German), Yingfang Liu (Chinese Academy of Sciences, China) and Ling Chen (Chinese Academy of Sciences, China) respectively.
Structure Modeling
A function-guided fold recognition framework (Fig. 1A) by integrating specific conserved information into the general threading method FR-t535 was developed to obtain the optimal structure template for PB1. Firstly, PB1 segment 110–637 was selected as initial seed domain and modeled since it is the polymerase domain as previously described (Fig. 1B)14,17,18,54. Secondly, four conserved sites (D305, G406, D445 and K481) for the activity of all RdRPs17,18 were used to guide the sequence-structure alignment in our developed threading method FR-t535. Namely, these four sites were forced to be conserved in the dynamic programming algorithm. Thirdly, the optimal template was selected according to the sequence/secondary structure similarities and their biological functions. Finally, considering the integrity of sequence alignment, the model of PB1 segment 71–612 was constructed with the optimal template (NV-RdRP: 2B43D) and refined with SWISS-MODEL55.
Modeling Materials
Influenza A virus PB1 sequence (A/goose/Guangdong/1/96) came from the Lab of Prof. Yingfang Liu. Influenza B and C virus PB1 sequences came from NCBI with accession numbers as NP_056657.a and Q6I7C3.1 separately. Secondary structure prediction of influenza A virus PB1 was done with PsiPred56 while secondary structures of the Top3 templates (2B43D, 2J7WA and 1U09A) were calculated with DSSP57. The threading template library composed of 90334 single-chain structures constructed from PDB database.
Construct PB1-RNA complex model
To get the PB1-RNA elongation complex model, we used the Norwalk virus polymerase and primer-template RNA complex (PDB ID: 3BSN) as template58. Structure matching between PB1 model and Norwalk virus polymerase and PB1-RNA complex modeling were all carried on with software Chimera1.5.3rc59.
Evolutional analyses for Influenza A virus PB1 protein
5580 full-length, de redundancy influenza A virus PB1 sequences were retrieved from the NCBI influenza database60. All sequences were aligned using MUSCLE61. The Shannon Entropy (SE) of each site i was calculated. If SE(I) < −0.1, site i was defined as non-conserved sites else conserved sites.
Mini replicon and reverse genetics systems for influenza A virus
293T cells were transfected using PEI reagent (Sigma) in 6-well plates. Each well contained plasmids expressing A/WSN/33 PB2, PB1, PA, NP and a luciferase reporter plasmid pPolI-NP-luc. The mixture also contained 100 ng plasmid constitutively expressing Renilla luciferase, which used for normalizing variation in transfection efficiency. Cells were lysised 24 h post transfection and 20 μl were used to detect the signal of firefly luciferase and renilla luciferase using Dual Luciferase Assay system (Promega) by a microplate reader (Tecan, GENiosPlus). The activity detected with samples containing Flag-GFP was set to 100%. Recombinant WSN33 viruses were rescued using 293T cells as previously described30,45. The rescued viruses were titrated on MDCK cells.
Primer Extension Assay
As previously reported23, pPolI-CAT was used as reporter in primer extension assay. 40 h post transfection of pcDNA-PB2/PB1/PA/NP and pPolI-CAT, total RNA was extracted with trizol (BBI, shenggong). CAT primers (5' CGCAAGGCGACAAGGTGCTGA 3', 5' ATGTTCTTTACGATGCGATTGGG 3') were used for detecting vRNA and m/cRNA respectively. 5S RNA was detected by the primer (5' ACCCTGCTTAGCTTCCGAGA 3'), the length of the product was 62 nt RNA.
Viral Inhibition Assay
Recombinant influenza A/WSN/33 virus was prepared as reported previously23 and influenza viruses A/Quail/HK/G1/97 (H9N2, G1) and IAV-Luc virus were amplified in MDCK cells. A549 cells in 24-well plates were transfected with 1 μg of plasmids encoding Peptide-GFP fusion proteins and 0.4 μg of a Gaussia luciferase reporter plasmid pPolI-Gluc to detect viral polymerase activity49. At 12 h post transfection, cells were infected with either A/WSN/33 or G1 virus at multiplicity of infection (MOI) of 0.015 for 1.5 h. 12 h p.i., the supernatant were collected and tested using Gluc Assay Kit (New England Biolabs). The activity detected with samples containing Flag-GFP was set to 100% after subtracting background. 293T cells in 12-well plates were transfected with 1 μg of plasmids encoding Peptide-GFP fusion proteins. 24 h post transfection, cells were challenged with IAV-Luc or WSN33 virus. Then 12 h or 24 h p.i., the Gaussia luciferase activity from supernatant or NP level from cell lysates were detected, respectively. The Gaussia luciferase activity from samples containing PX-GFP was set to 100%.
Virus protection assay in vitro
All peptides were purchased from Shanghai ChinaPeptides Co., Ltd. A549 cells in 24-well plates were transfected with 1 μg of a Gaussia luciferase reporter plasmid pPolI-Gluc to detect virus infection as described previously49. 10 h post transfection, peptides were added into the medium and were incubated at 37°C for 1.5 h. Then, cells were washed with PBS for twice and virus was added at a MOI of 0.015 for 1 h. After that, cells were washed again with PBS and incubated with medium containing peptides. 24 h p.i., activity of Gaussia luciferase in supernatant was tested using Gluc Assay Kit (New England Biolabs). The activity detected with samples containing PX-TAT was set to 100%.
ApG-primed transcription assay
Reactions were conducted with partially purified TAP-tagged polymerase and 5' end of the vRNA promoter (5'-AGUAGAAACAAGGCC-3') and 3' end of the vRNA (5'-GGCCUGCUUUUGCU-3') (Shanghai GenePharma) as described before15,23. Peptides and polymerase were incubated at 30°C for 30 min before adding the vRNA promoter.
Promoter binding assay
A/WSN/33 virus derived plasmids pcDNA-PB2/PB1/PA were described previously23. RNA binding assay were performed as previously described with synthesized peptides15,23,27. In brief, the purified influenza polymerase complex was incubated with synthesized peptides for 30 min before adding the vRNA promoter. Both 5' and 3' vRNA promoter were used when testing the 3' vRNA promoter binding activity, while only 5' vRNA promoter was added when testing the 5' vRNA promoter binding activity. The polymerase complex was cross linked with the vRNA promoter by UV and then separated on 8% SDS-PAGE.
Western Blot
PB1 was detected by rabbit polyclonal serum (1:1500), alpha-Tubulin was detected by Rabbit antibody (1:2000); Anti-GFP antibody (1:2000, Santa Cruz). Primary antibodies were kept at room temperature for 2 h and then incubated at room temperature for 1 h with HRP-conjugated secondary antibodies (1:10000, Beijing ZSBio).
References
Neumann, G., Brownlee, G. G., Fodor, E. & Kawaoka, Y. Orthomyxovirus replication, transcription and polyadenylation. Curr Top Microbiol Immunol 283, 121–143 (2004).
Boivin, S., Cusack, S., Ruigrok, R. W. & Hart, D. J. Influenza A virus polymerase: structural insights into replication and host adaptation mechanisms. J Biol Chem 285, 28411–28417, 10.1074/jbc.R110.117531 (2010).
Gao, R. et al. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 368, 1888–1897, 10.1056/NEJMoa1304459 (2013).
Fang, L. Q. et al. Mapping spread and risk of avian influenza A (H7N9) in China. Sci Rep 3, 2722, 10.1038/srep02722 (2013).
Tarendeau, F. et al. Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit. Nat Struct Mol Biol 14, 229–233, 10.1038/nsmb1212 (2007).
Torreira, E. et al. Three-dimensional model for the isolated recombinant influenza virus polymerase heterotrimer. Nucleic Acids Res 35, 3774–3783, 10.1093/nar/gkm336 (2007).
He, X. et al. Crystal structure of the polymerase PA(C)-PB1(N) complex from an avian influenza H5N1 virus. Nature 454, 1123–1126, 10.1038/nature07120 (2008).
Sugiyama, K. et al. Structural insight into the essential PB1-PB2 subunit contact of the influenza virus RNA polymerase. EMBO J 28, 1803–1811, 10.1038/emboj.2009.138 (2009).
Yuan, P. et al. Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site. Nature 458, 909–913, 10.1038/nature07720 (2009).
Guilligay, D. et al. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nat Struct Mol Biol 15, 500–506, 10.1038/nsmb.1421 (2008).
Asano, Y., Mizumoto, K., Maruyama, T. & Ishihama, A. Photoaffinity labeling of influenza virus RNA polymerase PB1 subunit with 8-azido GTP. J Biochem 117, 677–682 (1995).
Ruigrok, R. W., Crepin, T., Hart, D. J. & Cusack, S. Towards an atomic resolution understanding of the influenza virus replication machinery. Curr Opin Struct Biol 20, 104–113, 10.1016/j.sbi.2009.12.007 (2010).
Resa-Infante, P., Jorba, N., Coloma, R. & Ortin, J. The influenza virus RNA synthesis machine: advances in its structure and function. RNA Biol 8, 207–215 (2011).
Ohtsu, Y., Honda, Y., Sakata, Y., Kato, H. & Toyoda, T. Fine mapping of the subunit binding sites of influenza virus RNA polymerase. Microbiol Immunol 46, 167–175 (2002).
Deng, T., Sharps, J., Fodor, E. & Brownlee, G. G. In vitro assembly of PB2 with a PB1-PA dimer supports a new model of assembly of influenza A virus polymerase subunits into a functional trimeric complex. J Virol 79, 8669–8674, 10.1128/JVI.79.13.8669-8674.2005 (2005).
Chu, C. et al. Functional analysis of conserved motifs in influenza virus PB1 protein. PLoS One 7, e36113, 10.1371/journal.pone.0036113 (2012).
Poch, O., Sauvaget, I., Delarue, M. & Tordo, N. Identification of four conserved motifs among the RNA-dependent polymerase encoding elements. EMBO J 8, 3867–3874 (1989).
Biswas, S. K. & Nayak, D. P. Mutational analysis of the conserved motifs of influenza A virus polymerase basic protein 1. J Virol 68, 1819–1826 (1994).
Muller, R., Poch, O., Delarue, M., Bishop, D. H. & Bouloy, M. Rift Valley fever virus L segment: correction of the sequence and possible functional role of newly identified regions conserved in RNA-dependent polymerases. J Gen Virol 75 (Pt 6), 1345–1352 (1994).
Khurana, S. et al. Antigenic fingerprinting of H5N1 avian influenza using convalescent sera and monoclonal antibodies reveals potential vaccine and diagnostic targets. PLoS Med 6, e1000049, 10.1371/journal.pmed.1000049 (2009).
Hutchinson, E., Orr, O., Man Liu, S., Engelhardt, O. & Fodor, E. Characterisation of the interaction between the influenza A virus polymerase subunit PB1 and the host nuclear import factor Ran Binding Protein 5. J Gen Virol 92, 1859–1869, 10.1099/vir.0.032813-0 (2011).
Nath, S. T. & Nayak, D. P. Function of two discrete regions is required for nuclear localization of polymerase basic protein 1 of A/WSN/33 influenza virus (H1 N1). Mol Cell Biol 10, 4139–4145 (1990).
Fodor, E. et al. A single amino acid mutation in the PA subunit of the influenza virus RNA polymerase inhibits endonucleolytic cleavage of capped RNAs. J Virol 76, 8989–9001 (2002).
Jung, T. E. & Brownlee, G. G. A new promoter-binding site in the PB1 subunit of the influenza A virus polymerase. J Gen Virol 87, 679–688, 10.1099/vir.0.81453-0 (2006).
Li, M. L., Ramirez, B. C. & Krug, R. M. RNA-dependent activation of primer RNA production by influenza virus polymerase: different regions of the same protein subunit constitute the two required RNA-binding sites. EMBO J 17, 5844–5852, 10.1093/emboj/17.19.5844 (1998).
Boltz, D. A., Aldridge, J. R., Jr, Webster, R. G. & Govorkova, E. A. Drugs in development for influenza. Drugs 70, 1349–1362, 10.2165/11537960-000000000-00000 (2010).
Ghanem, A. et al. Peptide-mediated interference with influenza A virus polymerase. J Virol 81, 7801–7804, 10.1128/JVI.00724-07 (2007).
Gong, J. et al. Potential targets and their relevant inhibitors in anti-influenza fields. Curr Med Chem 16, 3716–3739 (2009).
Su, C. Y. et al. High-throughput identification of compounds targeting influenza RNA-dependent RNA polymerase activity. Proc Natl Acad Sci U S A 107, 19151–19156, 10.1073/pnas.1013592107 (2010).
Li, C. et al. A peptide derived from C-terminus of PB1 inhibits influenza virus replication by interfering with viral polymerase assembly. FEBS J 280, 1139–1149, 10.1111/febs.12107 (2012).
Ferrer-Orta, C., Arias, A., Escarmis, C. & Verdaguer, N. A comparison of viral RNA-dependent RNA polymerases. Curr Opin Struct Biol 16, 27–34, 10.1016/j.sbi.2005.12.002 (2006).
Steitz, T. A. A mechanism for all polymerases. Nature 391, 231–232, 10.1038/34542 (1998).
van Dijk, A. A., Makeyev, E. V. & Bamford, D. H. Initiation of viral RNA-dependent RNA polymerization. J Gen Virol 85, 1077–1093 (2004).
Fiser, A. Template-based protein structure modeling. Methods Mol Biol 673, 73–94, 10.1007/978-1-60761-842-3_6 (2010).
Hu, Y. et al. Incorporation of local structural preference potential improves fold recognition. PLoS One 6, e17215, 10.1371/journal.pone.0017215 (2011).
Zhang, K. L. et al. Model structure of human APOBEC3G. PLoS One 2, e378, 10.1371/journal.pone.0000378 (2007).
Ferron, F., Bussetta, C., Dutartre, H. & Canard, B. The modeled structure of the RNA dependent RNA polymerase of GBV-C virus suggests a role for motif E in Flaviviridae RNA polymerases. BMC Bioinformatics 6, 255, 10.1186/1471-2105-6-255 (2005).
Vieth, S., Torda, A. E., Asper, M., Schmitz, H. & Gunther, S. Sequence analysis of L RNA of Lassa virus. Virology 318, 153–168, 10.1016/j.virol.2003.09.009 (2004).
Hass, M., Lelke, M., Busch, C., Becker-Ziaja, B. & Gunther, S. Mutational evidence for a structural model of the Lassa virus RNA polymerase domain and identification of two residues, Gly1394 and Asp1395, that are critical for transcription but not replication of the genome. J Virol 82, 10207–10217, 10.1128/JVI.00220-08 (2008).
Perdomo, D. et al. The alpha-helical regions of KERP1 are important in Entamoeba histolytica adherence to human cells. Sci Rep 3, 1171, 10.1038/srep01171 (2013).
Gong, P. & Peersen, O. B. Structural basis for active site closure by the poliovirus RNA-dependent RNA polymerase. Proc Natl Acad Sci U S A 107, 22505–22510, 10.1073/pnas.1007626107 (2010).
Choi, K. H. et al. The structure of the RNA-dependent RNA polymerase from bovine viral diarrhea virus establishes the role of GTP in de novo initiation. Proc Natl Acad Sci U S A 101, 4425–4430, 10.1073/pnas.0400660101 (2004).
Zhong, W. et al. Template/primer requirements and single nucleotide incorporation by hepatitis C virus nonstructural protein 5B polymerase. J Virol 74, 9134–9143 (2000).
Hong, Z. et al. A novel mechanism to ensure terminal initiation by hepatitis C virus NS5B polymerase. Virology 285, 6–11, 10.1006/viro.2001.0948 (2001).
Pan, W. et al. Visualizing influenza virus infection in living mice. Nat Commun 4, 2369, 10.1038/ncomms3369 (2013).
Fawell, S. et al. Tat-mediated delivery of heterologous proteins into cells. Proc Natl Acad Sci U S A 91, 664–668 (1994).
Vives, E., Richard, J. P., Rispal, C. & Lebleu, B. TAT peptide internalization: seeking the mechanism of entry. Curr Protein Pept Sci 4, 125–132 (2003).
Wunderlich, K. et al. Identification of a PA-binding peptide with inhibitory activity against influenza A and B virus replication. PLoS One 4, e7517, 10.1371/journal.pone.0007517 (2009).
Zhu, W. et al. A reporter system for assaying influenza virus RNP functionality based on secreted Gaussia luciferase activity. Virol J 8, 29, 10.1186/1743-422X-8-29 (2011).
Fodor, E., Seong, B. L. & Brownlee, G. G. Photochemical cross-linking of influenza A polymerase to its virion RNA promoter defines a polymerase binding site at residues 9 to 12 of the promoter. J Gen Virol 74 (Pt 7), 1327–1333 (1993).
Agopian, A., Depollier, J., Lionne, C. & Divita, G. p66 Trp24 and Phe61 are essential for accurate association of HIV-1 reverse transcriptase with primer/template. J Mol Biol 373, 127–140, 10.1016/j.jmb.2007.07.044 (2007).
Ortigoza, M. B. et al. A novel small molecule inhibitor of influenza A viruses that targets polymerase function and indirectly induces interferon. PLoS Pathog 8, e1002668, 10.1371/journal.ppat.1002668 (2012).
Morris, M. C. et al. A new potent HIV-1 reverse transcriptase inhibitor. A synthetic peptide derived from the interface subunit domains. J Biol Chem 274, 24941–24946 (1999).
Kolpashchikov, D. M., Honda, A. & Ishihama, A. Structure-function relationship of the influenza virus RNA polymerase: primer-binding site on the PB1 subunit. Biochemistry 43, 5882–5887, 10.1021/bi036139e (2004).
Arnold, K., Bordoli, L., Kopp, J. & Schwede, T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201, 10.1093/bioinformatics/bti770 (2006).
Buchan, D. W. et al. Protein annotation and modelling servers at University College London. Nucleic Acids Res 38, W563–568, 10.1093/nar/gkq427 (2010).
Kabsch, W. & Sander, C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637, 10.1002/bip.360221211 (1983).
Zamyatkin, D. F. et al. Structural insights into mechanisms of catalysis and inhibition in Norwalk virus polymerase. J Biol Chem 283, 7705–7712, 10.1074/jbc.M709563200 (2008).
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612, 10.1002/jcc.20084 (2004).
Bao, Y. et al. The influenza virus resource at the National Center for Biotechnology Information. J Virol 82, 596–601, 10.1128/JVI.02005-07 (2008).
Edgar, R. C. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5, 113, 10.1186/1471-2105-5-113 (2004).
Acknowledgements
This study was supported by National Natural Science Foundation of China (31371338), National Foundation of Talent Youth (31125016) and the Major National Earmark Project for Infectious Diseases (2014ZX10004002-001) to T.J., NIH R01 AI078389, AI069120 to G.C., National Natural Science Foundation of China (31100950) to A.W. and National Natural Science Foundation of China (31300632) to H.Z. Thanks professor Martin Schwemmle (University of Freiburg, German) and Yingfang Liu (Chinese Academy of Sciences, China) for providing pPolI-NP-luc and cDNA of H5N1(A/goose/Guangdong/1/96), respectively. Thanks for the computational supporting from the DAWN computational cluster of the Institute of Biophysics, Chinese Academy of Sciences.
Author information
Authors and Affiliations
Contributions
C.L., J.W., Y.G., Z.C., H.Z., Y.W., J.D. and L.W. performed experiments. A.W. and Y.P. performed the computation. C.L., A.W., G.C., T.D. and T.J. wrote the paper. T.J., T.D., G.C. and X.Q. conceived and directed the project.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Li, C., Wu, A., Peng, Y. et al. Integrating computational modeling and functional assays to decipher the structure-function relationship of influenza virus PB1 protein. Sci Rep 4, 7192 (2014). https://doi.org/10.1038/srep07192
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep07192
This article is cited by
-
The ubiquitination landscape of the influenza A virus polymerase
Nature Communications (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.