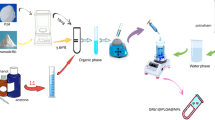
Abstract
In this work, synergism and antagonism among active ingredients of traditional Chinese medicine (TCM) were studied at system-level by using molecular imprinting technology. Reduning Injection (RDNI), a TCM injection, was widely used to relieve fever caused by viral infection diseases in China. Molecularly imprinted polymers (MIPs) synthesized by sol-gel method were used to separate caffeic acid (CA) and analogues from RDNI without affecting other compounds. It can realize the preparative scale separation. The inhibitory effects of separated samples of RDNI and sample combinations in prostaglandin E2 biosynthesis in lipopolysaccharide-induced RAW264.7 cells were studied. The combination index was calculated to evaluate the synergism and antagonism. We found that components which had different scaffolds can produce synergistic anti-inflammatory effect inside and outside the RDNI. Components which had similar scaffolds exhibited the antagonistic effect and the antagonistic effects among components could be reduced to some extent in RDNI system. The results indicated MIPs with the characteristics of specific adsorption ability and large scale preparation can be an effective approach to study the interaction mechanism among active ingredients of complex system such as TCM at system-level. And this work would provide a new idea to study the interactions among active ingredients of TCM.
Similar content being viewed by others
Introduction
The mechanism of traditional Chinese medicine (TCM) was mainly caused by the interactions between complex TCM system and biological system1. TCM has been used for anti-inflammatory for thousands of years and accumulated lots of clinical experience2. TCM is a complex system which contains lots of components with diversities in chemical structures, biological activities and interactions among compounds and the content for each component varies greatly3,4,5. Typically, an herbal formula which consists of several herbs would comprise hundreds of compounds and can affect the biological systems through interactions with multiple cellular targets6,7,8,9,10,11. TCM is so complicated that it's almost impossible to explore the molecular mechanism thoroughly. It's time-consuming and laborious to explore the efficacy of each compound. Moreover, compounds in TCM exert therapeutic effects in combination rather than as individuals8,12,13. The results of individual studies do not necessarily get the overall effect of TCM because of abundant synergistic and antagonistic effects8,14,15,16,17. To understand biology and chemistry at system level, we must identify the compounds of the systems and gain insights into emergent properties through interactions among compounds in the TCM systems.
Molecularly imprinted polymer (MIP) is tailor-made adsorption material used to separate template and analogues from complex matrix18,19,20,21,22,23. The binding sites of MIP have high affinity for the template by interacting with its complementary functional groups or structural elements24,25,26,27. It can be grafted to the surface of silica beads by sol-gel process to realize semi-preparative scale, even preparative scale separation and preparation and can retain the specific adsorption performance at the same time28,29,30. In recent years, several researches were carried out for caffeic acid-MIPs (CA-MIPs) and the CA-MIPs were used to determine and extract CA from complex media31,32,33. Therefore, a method by using MIP to selectively remove the template or a group of analogues from TCM can be a promising approach to study the emergent properties in the TCM systems.
Reduning Injection (RDNI) is a TCM injection which was prepared by Artemisia annua L., Lonicera japonica Thunb. and Gardenia jasminoides E.. It is widely used in clinical to relieve fever caused by viral infection diseases, such as upper respiratory tract infection34 and hand-foot-mouth disease35. The main constituents and contents in RDNI had been determined in previous work36. It mainly included caffeoylquinic acid compounds (CACs), coumarins, iridoids, flavonoids37. In this work, MIP was used to separate CA and CACs from RDNI. Lipopolysaccharide (LPS)-induced prostaglandin E2 (PGE2) release in RAW264.7 cells based on phenotypic assays maintained reasonable experimental efficiency and related to inflammatory diseases. So it was carried out to study the anti-inflammatory effects of separated samples and sample combinations. Combination index (CI), which was proposed by Chou and Talalay38, was used to evaluate the interaction between the components.
Results
Synthesis and Characterization of MIPs
In this section two kinds of polymers were synthesized by the sol-gel method. 8 polymers were prepared to optimize MIP1 (Table 1). The results of chromatographic evaluations showed that MIP1e had the highest imprinted factor (9.07) and good capacity factor (15.96) for CA (Table 2). The preparation condition of MIP2 (Table 1), which was used to separate all CACs in one step, was optimized from the composition of MIP1.
The chromatographic conditions for separating CA by using MIP1 as solid phase were investigated. When the mobile phase was CH3OH-HOAc (500:1, v/v), the retention time of acetone and CA were about 2 min and 19 min, respectively. But the chlorogenic acid (CGA) was not cleared out within 50 min (Fig. 1A). When CH3OH-HOAc (9:1, v/v) was used as the mobile phase, the retention time of acetone, CA and CGA were about 1 min, 2 min and 11 min, respectively (Fig. 1B). The possible reason of this phenomenon was that the molecular structures of CGA, Isochlorogenic acid B (IsoB) and other CACs contain caffeic acid groups, so those compounds can partly embedded in the three-dimensional cavities of MIP. The molecular structures of CA and other 6 CACs of RDNI were shown in Fig. 2. The another reason was that the numbers of hydrogen bond donors and acceptors of CGA, IsoB and other CACs were more than those of CA, so the binding affinities for those compounds with MIP were stronger than that of CA. Finally, gradient elution was used to separate CA and other CACs. The column was firstly washed with CH3OH-HOAc (500:1, v/v) to elute CA. Then it was washed with CH3OH-HOAc (9:1, v/v) to elute other CACs.
Chromatogram of MIP1 column for acetone, CA and CGA by using different mobile phase.
The mobile phase of (A) was CH3OH-HOAc (500:1, v/v). The mobile phase of (B) was CH3OH-HOAc (9:1, v/v). (1): Acetone; (2): CA; (3): CGA. Detection wavelength and velocity of mobile phase were 324 nm and 0.6 mL min−1, respectively.
Adsorption Isotherms and Adsorption Kinetics
The calibration curve of CA solution was y = 17.745x - 0.0491 (R2 = 0.9992), where y was concentration of the solution and x was the absorbance at 324 nm. The concentration range was from 0.0050 to 0.20 mM.
Adsorption isotherms of MIP1 and NIP1 were shown in Fig. 3A. The saturated adsorption equilibrium of MIP1 and NIP1 for CA were achieved at 8.0 and 6.0 mM, respectively. The maximum adsorption capacities of MIP1 and NIP1 were about 38.3 and 17.9 mg g−1, respectively. The data revealed high imprinting performance of the proposed approach. Adsorption kinetic curves of MIP1 and NIP1 were shown in Fig. 3B. The saturated adsorption of MIP1 and NIP1 were reached in about 600 and 400 min, respectively.
Separation and HPLC Determinations of Samples from RDNI
Fig. 4A was the chromatogram of MIP1 column for the separation of CA from RDNI. Peak 1 was comprised of the other compounds of RDNI except CACs. Peak 2 represented CA of RDNI. Peak 3 contained the CACs except CA of RDNI. To separate the CACs from RDNI, MIP2 were packed into the glass chromatographic column (13 mm I.D. × 200 mm). First, the separation properties of MIP2 column for CA and CGA were tested. Fig. 4C and Fig. 4D were the chromatograms of CA and CGA on MIP2 column by using CH3OH-HOAc-HCOOH (100:1:3, v/v/v) as mobile phase. The peak of about 20 min was the matrix. The retention time of CA and CGA on MIP2 column was about 100 min and 125 min, respectively. When RDNI was loaded (Fig. 4B), there were two peaks in the chromatogram. Peak 4, which was the same with Peak 1, contained the other compounds of RDNI except CACs. Peak 5 was comprised of the CACs of RDNI. Finally, the effluent liquid of Peak 1 and Peak 3 were collected and freeze dried as sample of RDNI which lacked of CA (S1). The effluent liquid of Peak 2 was collected and freeze dried as CA of RDNI. Peak 4, Peak 5 was collected and freeze dried as sample of RDNI which lacked of CACs (S2) and CACs of RDNI (S3), respectively. SN1 and SN2 were the freeze-dried powder of effluent liquid of RDNI by NIP1 and NIP2 column, respectively.
Separating chromatograms of MIP1 column and MIP2 column.
(A) was the chromatogram of the separation of CA from RDNI by MIP1 column. The chromatographic conditions were as follows: 0 ~ 50 min: CH3OH-HOAc (500:1, v/v), after 50 min: CH3OH-HOAc (9:1, v/v); the flow rate of mobile phase was 2.4 mL min−1; the detective wavelength was 324 nm. (B) was the chromatogram of the separation of CACs from RDNI by MIP2 column. (C) and (D) were chromatograms of CA solution and CGA solution by MIP2. The mobile phase was CH3OH-HOAc-HCOOH (100:1:3, v/v/v); flow rate of mobile phase which controlled by gravity was about 1.5 mL min−1; the detective wavelength which subject to the conditions of the instrument was 340 nm.
To evaluate the separation effect of MIP1 and MIP2, HPLC fingerprints and component contents of samples were determined. The linear equation of CA, CGA, IsoB and gardenoside (GAR) were y = 52.155x - 88.519 (R2 = 0.9999), y = 30.118x - 178.39 (R2 = 0.9999), y = 33.051x - 102.16 (R2 = 0.9999) and y = 14.369x - 67.05 (R2 = 0.9999), respectively. Y was the concentration of each compound in the solution, x was the absorbance (CA, CGA and IsoB were at 324 nm, GAR was at 237 nm). The linear ranges for CA, CGA, IsoB and GAR were 5.5 ~ 218.0 μg mL−1, 30.0 ~ 838.0 μg mL−1, 9.8 ~ 390.0 μg mL−1 and 25.4 ~ 1016.0 μg mL−1, respectively. The sample amount and component content of RDNI and the separated samples were shown in Table 3. After extraction and separation by MIP and NIP column, contents of four compounds of samples were basically the same with those of RDNI freeze-dried powder. Fig. 5A, Fig. 5B and Fig. 5C were the fingerprint chromatograms of RDNI, SN1 and SN2 under 324 and 237 nm. The results indicated that the non-specific adsorptions of MIP1 and MIP2 had no significant impact on the composition of RDNI.
In Fig. 5D (fingerprint chromatogram of S1), there was no chromatographic peak of CA in about 26 min, but the chromatographic peak of other compounds were not changed. 2.1 mg CA were collected from 10 mL RDNI. 500 μg of separated CA were dissolved into 10 mL methanol to detect and calculate the purity. The fingerprint chromatogram and mass spectrogram of CA which was separated from RDNI was shown in Fig. 6A and Fig. 6B. The purity of CA separated from RDNI was calculated through the linear equation and it was 90.5%. The ion peak m/z179.0 was almost the only peak in Fig. 6B. The main absorption peak of Fig. 5E (fingerprint chromatogram of S2) was GAR in about 27 min under 237 nm and the absorption peak in about 13, 22 and 36 min under 237 nm were other non-caffeoylquinic acid compounds of RDNI. Absorption peak of seven caffeoylquinic acid compounds (CA, CGA, NCGA, CQA, IsoA, IsoB and IsoC) were shown in Fig. 5F (fingerprint chromatogram of S3). The results indicated that MIP1 and MIP2 were suitable to separate CA and CACs from RDNI, respectively and MIPs synthesized by sol-gel method can realize preparative scale separation.
Inhibitory Effects of Single Compound and Collected Sample
The compounds or samples were added and cells of each group were incubated for another 18 h. The status of cells were observed and the results showed that cells cultured with samples at the concentration of 500 µg mL−1 or compounds at the concentration of 500 µM did not change the cell viability.
The EC50 of SN1 and SN2 (104.2 and 105.1 μg mL−1, Fig. 7D) were almost the same with that of freeze-dried powder of RDNI (RDN, 104.0 μg mL−1, Fig. 7D), indicated that the non-specific adsorptions of MIP1 and MIP2 had no significant influence on the activity of RDNI.
CA, IsoB and scopoletin (SCO) showed the inhibitory effects against LPS induced PGE2 release. The dose-effect curves and EC50 values of CA, IsoB and SCO were shown in Fig. 7A, B and C. When CA was removed from RDNI, the activity of S1 (EC50 was 119.3 μg mL−1, Fig. 7D) against the release of PGE2 was decreased compared with RDNI. It indicated that CA was an important anti-inflammatory compound in RDNI system. EC50 of S2 (54.7 μg mL−1, Fig. 7D) was lower than that of RDNI. But the EC50 of S3 (119.8 μg mL−1, Fig. 7D) was larger than that of RDNI. The possible reason was that compounds which had the similar skeletons could competitively bind with the same target protein in the pathway, so the activity of sample S3 (CACs from RDNI) was weaker than that of RDNI. And when the S3 was separated from RDNI, the activity of S2 was enhanced.
Inhibitory Effects of Drug Combinations
Cell experiments of combinations of compounds and samples were studied. The combination index (CI) value of combinations of CA and S2, CA and RDNI, IsoB and S2, IsoB and RDNI were all less than 1.0, so these four combinations showed the synergistic effects (Table 4). The CI value of CA and S2, IsoB and S2 were smaller than that of CA and RDNI, IsoB and RDNI with the increase of concentration of CA and IsoB. So the synergisms of CA and S2, IsoB and S2 were stronger than that of CA and RDNI, IsoB and RDNI, respectively. Combination of SCO and S2 produced strong antagonistic effects (CI values of 5 concentrations were all larger than 1.0). But combinations of SCO and S3, SCO and RDNI both showed synergistic effects (Table 4). The possible reason for the results could be that compounds with different skeletons can produce synergistic effect and meanwhile the antagonism of similar skeleton compounds can be reduced to some extent in the RDNI system.
To study the mechanism of the synergistic and antagonistic effects and verify the above inference, cell experiments of combinations among CA, IsoB and SCO were carried out. We found that combinations of CA and SCO, IsoB (4.5 ~ 35.6 μM) and SCO (6.0 ~ 47.9 μM) were both exhibited synergism to decrease the release of PGE2 with the increasing of concentration (Table 5). However, when IsoB was 71.3 μM and SCO was 95.7 μM, the combination showed antagonism and the inhibition rate of this concentration level was basically the same with the concentration level of 35.6 μM and 47.9 μM. Since CA and SCO, IsoB and SCO (each structure was shown in Fig. 2) had the different skeletons, they could interact with different target proteins in the pathway of LPS induced PGE2 release process, so that to produce synergism39,40. But when the concentration reaches a certain level, the inhibition rate will not increase with the increasing of the concentration, so it became antagonism. Combination of CA and IsoB showed antagonism in all cases (Table 5). Because both CA and IsoB were CACs with similar skeleton, they would competitively bind to the same active sites. Meanwhile, due to large molecular structure of IsoB, it may hinder the approaching of CA to the binding sites. So the reason for the reducing of activity of S3 was the increasing of competitive inhibition of compounds with similar skeleton and the decreasing of synergism of compounds with different skeletons. The reason for the increasing of activity of S2 was that after CACs were separated, the concentrations of other active compounds were relatively increased (e.g. Gardenoside and SCO) and the disappearance of antagonism of S3.
After experiments of combinations of single compound with samples, series of combinations of equimolar CA and SCO with different concentrations of RDN were performed. The results (Table 6) indicated that combinations of CA and SCO produced synergism not only in the compound combination but also in the RDNI system. Compared with combination of CA and SCO, the synergism of combination of CA, SCO and RDNI was weakened. The possible reason was that the antagonism between CA and CACs, SCO and S2 both existed in the RDNI system. On the other hand, the results also indicated the antagonism can be reduced to some extent in the RDNI system.
Discussion
We have studied the synergistic and antagonistic anti-inflammatory effects of active compounds in a traditional Chinese medicine (RDNI) at system-level by using molecular imprinting technology for the first time. The core-shell MIPs were used as stationary phase of liquid chromatography to separate CA and CACs from RDNI without affecting other compounds.
The in vitro experiment results showed that both synergism and antagonism among compounds and separated samples existed inside and outside the RDNI (TCM) system. Compounds with different skeletons can produce synergistic effect through binding with different target proteins in the pathway. Compounds with similar skeletons can produce antagonistic effect due to competitive binding with the same target protein in the pathway.
It also indicated that TCM as a system could regulate the synergism and antagonism by interactions among compounds. And molecular imprinting technology could be an effective approach to study the interaction mechanism of TCM. This work would provide a novel approach for studying the interaction mechanism of TCM at system-level and provide new insights for the further study of TCM to enhance effectiveness by improving synergism and reducing antagonism.
Methods
Reagents and materials
RDNI was provided by Jiangsu Kanion Pharmaceutical Co., Ltd. (Lianyungang, PR China). Caffeic acid (CA, 98%) was purchased from Nanjing Zelang Medical Technology Co., Ltd. (Nanjing, PR China). Chlorogenic acid (CGA, 98%), Gardenoside (GAR, 98%) were purchased from National Institute for Food and Drug Control (Beijing, PR China). Isochlorogenic acid A (IsoA, 98%), Isochlorogenic acid B (IsoB, 98%), Isochlorogenic acid C (IsoC, 98%), Neochlorogenic acid (NCGA, 98%) and 4-Caffeoylquinic acid (CQA, 98%) were purchased from Tianjin Yifang Science and Technology Co., Ltd. (Tianjin, PR China). Scopoletin (SCO, 98%) was purchased from Shanghai PureOne Bio technology Co., Ltd. (Shanghai, PR China). Silica beads (19 ~ 37 μm, 33 ~ 63 μm and 62 ~ 105 μm), 3-aminopropyltriethoxysilane (APTS, 98%) and tetraethoxysilane (TEOS, 98%) were purchased from Alfa Aesar Co. Ltd. (Tianjin, PR China). Acetone was obtained from Beijing Chemical Works (Beijing, PR China). Hydrochloric acid and tetrahydrofuran (THF) were purchased from Guangdong Xilong Chemical Co., Ltd. (Guangdong, PR China). Methanol was purchased from J&K Scientific Led. (Beijing, PR China). Glacial acetic acid (HOAc) was purchased from Tianjin Kemiou Chemical Reagent Co., Ltd. (Tianjin, PR China). Methanol and HOAc were of HPLC grade. All other chemicals used were of analytical grade. Water used in the experiment was deionized water.
The sieve (500-mesh) was purchased from Fengxing Manufacturing Co., Ltd. (Shanghai, PR China). The Luna C18 (2) analytical column (4.6 mm I.D. × 250 mm, 5 μm) was purchased from Phenomenex Inc. (USA). The glass chromatographic column (13 mm I.D. × 200 mm) was purchased from Beijing Synthware Glass Co., Ltd (Beijing, PR China). Empty stainless steel column (4.6 mm I.D. × 50 mm) and syringe filter (0.45 μm) were purchased from Dikma Technology Inc. (Beijing, PR China). Empty stainless steel column (10.0 mm I.D. × 100 mm) was purchased from Dalian Elite Analytical Instruments Co., Ltd (Dalian, PR China).
Murine macrophage cell line RAW264.7 was purchased from Cell Culture Center (CCC) of the Chinese Academy of Medical Sciences (Beijing, PR China). Dulbecco's Modified Eagle's medium (DMEM) supplemented with penicillin (100 U mL−1) and streptomycin (100 U mL−1) was purchased from Gibco (Carlsbad, CA). Fetal bovine serum (FBS) was purchased from Hangzhou Sijiqing Biological Engineering Materials Co., Ltd. (Hangzhou, PR China). PGE2 EIA kit was purchased from Enzo Life Sciences (Farmingdale, USA). LPS was purchased from Nanjing Baikang Biological Technology Co., Ltd. (Nanjing PR China).
Equipment
Separation and collection of CA were performed on a Gilson analytical and semi-preparative high performance liquid chromatography (HPLC) which comprised a 322 Pump and a 152 UV/Vis detector (Gilson Science & Technology, Beijing, PR China). Separation and collection of caffeoylquinic acid compounds (CACs) were performed on a HD 21C-B UV detector and a HDC-01 signal acquisition unit from Shanghai Jihui Scientific Analytical Instruments Co., Ltd (Shanghai, PR China). The HPLC experiments were conducted on a HP 1100 series HPLC (Agilent Technologies, USA). A Shimadzu UV-2550 Ultraviolet and visible spectrophotometer was used for the batch adsorption experiments (Kyoto, Japan). High-resolution mass spectra (HRMS) were detected by a Bruker Apex IV FTMS mass spectrometer (ESI) (Bruker Daltonics, Germany).
The optical density of cell experiment was measured by a SpectraMax M2e Microplate Reader (Molecular Devices, Menlo Park, USA).
Synthesis of Molecular Imprinting Polymers
The MIPs were synthesized by using CA, silica beads, APTS, TEOS and THF as template, supporting matrix, functional monomer, cross-linker and solvent, respectively. Before use, silica beads (100 g) were suspended in aqueous solution of hydrochloric acid (2.0 mol L−1, 600 mL) at 60°C for 6 hours to activate the surface hydroxyl groups. Then the silica beads were washed with deionized water and anhydrous ethanol and dried at 120°C41.
The solution of CA and APTS in THF (7 mL) was magnetic stirred for 2 hours at room temperature. Then activated silica beads (3.5 g) were added into the solution and kept stirring for 2 hours. Then added TEOS and kept stirring for 1 hour. At last aqueous solution of glacial acetic acid (1.0 mol L−1, 1 mL) was added into the suspension and kept stirring for 18 hours to finish the polymerization. The resultant MIPs were filtrated and washed with methanol for 3 times and dried at 70°C. Then the polymers were sieved through a 500-mesh sieve. To remove the template, the obtained MIPs were washed by CH3OH-HOAc (9:1, v/v). Finally, the polymers were washed with methanol and dried at 70°C. The non-imprinted polymers (NIPs) were prepared using the same protocol without addition of template.
HPLC Evaluation of MIPs
MIP or NIP was fully packed into an empty stainless steel column (4.6 mm I.D. × 50 mm). Then the column was connected to HPLC system and washed with CH3OH-HOAc (9:1, v/v). After no template was detected, the column was washed to baseline by methanol. Chromatographic experiment was carried out under 324 nm by using CH3OH-HOAc (500:1, v/v) as mobile phase. The flow rate of mobile phase was 0.6 mL min−1. The injection volume of sample solutions and acetone were 20 μL and 5 μL, respectively. Capacity factor (k) and imprinted factor (IF) were used to evaluate the retention behavior of the polymers. K and IF were calculated by Eq. (1) and Eq. (2), respectively.


where tanalyte and tacetone were the retention times of analyte and acetone, respectively; tacetone was regarded as dead time; kMIP and kNIP were the capacity factors of MIP and NIP, respectively.
Adsorption Experiments
Adsorption experiments were studied by ultraviolet and visible spectrophotometer. To investigate the adsorption equilibrium, 100 mg of polymer particles (MIP or NIP) were mixed with 5 mL methanol or CA solution in a 10 mL tube, respectively. The concentrations of CA solution were 1.0, 2.0, 3.0, 4.0, 5.0, 6.0, 7.0, 8.0, 9.0 and 10.0 mM. The methanol was used as the control to eliminate the influence of solvent. The tubes were slightly shaken for 24 h at room temperature. After sedimentation the supernatants were filtered by syringe filter (0.45 μm). The filtrate was diluted by methanol before detection. The detection wavelength was 324 nm. Equilibrium adsorption capacity (Qe, mg g−1) was calculated according to Eq. (3).
To investigate the adsorption kinetics of MIP and NIP, 50 mg of particles were put in two cuvettes, then 3.5 mL of methanol or 0.2 mM CA solutions were injected into the cuvette along the edge slowly, respectively. The methanol was used as the control. The absorbance was detected every 1 minute from the solution was added. The whole test was carried out at room temperature. Adsorption capacity (Q, mg g−1) was calculated by Eq. (4):


where C0, Ct and Ce (mM) were the initial concentration, t time concentration and equilibrium concentration of CA solution, respectively; V (mL) was the volume of solution; M (180.15) was the molecular weight of CA; m (mg) was the weight of MIPs.
Separation of CA and CACs from RDNI
Separation of CA from RDNI was performed on a Gilson analytical and semi-preparative high performance liquid chromatography. First, MIP1 was fully packed into the empty stainless steel column (10.0 mm I.D. × 100 mm). Then the column was connected to Gilson HPLC system and washed with CH3OH-HOAc (9:1, v/v) until no template was detected. The column was washed with mobile phase (CH3OH-HOAc, 500:1, v/v) to baseline before loading samples. RDNI (100 mL) was frozen dry and redissolved in 50 mL of 90% methanol solution. The injection volume was 200 μL.
The separation of CACs from RDNI was carried out on a glass chromatographic column (13 mm I.D. × 200 mm). 20 g of MIP2 was dispersed in 20 mL methanol and then the suspension was poured into the glass column. After natural sedimentation, the column was connected with a UV detector, a signal acquisition unit and a computer to assemble the LC system for separation. The column was washed on-line by CH3OH-HOAc (9:1, v/v) and methanol until no template was detected and a stable baseline was obtained. The injection volume of RDNI was 1.0 mL.
Determinations of HPLC Fingerprints and Component Contents of Samples
The HPLC chromatographic conditions for fingerprints and determinations of compounds were as follows: a Phenomenex Luna C18 (2) chromatographic column (4.6 mm I.D. × 250 mm, 5 μm), mobile phase A was methanol, mobile phase B was 0.1% phosphoric acid solution (v/v). The gradient elution program of mobile phase was that: 0 ~ 20 min, 12% ~ 30% A; 20 ~ 60 min, 30% ~ 50% A. The detection wavelengths were 237 nm and 324 nm. The velocity of mobile phase was 1.0 mL min−142.
CA, CGA, IsoB and GAR were accurate weighed and dissolved with 50% methanol (v/v) to prepare the mixed standard solution. The concentrations of CA, CGA, IsoB and GAR were 218, 838, 390 and 1016 μg mL−1, respectively. Then the mixed solution was diluted 40, 20, 10, 5 and 2.5 times by 50% methanol (v/v), respectively.
A certain amount of RDNI samples which separated by MIP or NIP were accurate weighed. Then the samples were dissolved with 50% methanol (v/v) to 10 mL for the determination of CA and IsoB. The samples were dissolved with 50% methanol (v/v) to 100 mL for the determination of CGA and GAR.
The mixed standard solutions (10 μL) and sample solutions (10 μL) were injected each time to determine the fingerprints and contents of compounds, respectively. The contents of CA, CGA and IsoB were determined under 324 nm and contents of GAR were determined under 237 nm.
Cell Culture and LPS Stimulation
RAW264.7 cells were cultured in plastic dishes containing high-glucose DMEM supplemented with antibiotics (100 U mL−1 streptomycin and penicillin) and 10% (v/v) FBS. The cells were maintained at 37°C in a humidified incubator containing 5% CO2.
RAW264.7 cells were plated on 24-well culture plates at a density of 1 × 105 cells mL−1 in 400 μL at 37°C overnight. 100 μL serum-free DMEM was added as the normal group. 95 μL serum-free DMEM with DMSO (final concentration was 0.1%) was added as DMSO group. 95 μL serum-free DMEM was added as model group and 95 μL serum-free DMEM with samples was added as sample group. Then the cells were pre-treated for 1 h at 37°C in a humidified incubator containing 5% CO2. Finally, 5 μL serum-free DMEM was added to the DMSO group. 5 μL LPS was added to the model group and sample group, respectively. Final volume and final concentration of each group was 500 μL and 1 µg mL−1, respectively. Then cells of each group were incubated for another 18 h. Each group was performed in triplicate43,44,45.
Determination of PGE2 Concentration and Calculation of Inhibition Rate
The culture supernatants were collected and diluted 5 times with FBS-free DMEM and assayed with PGE2 EIA kit according to the manufacturer's protocols. The inhibition rate of each sample for PGE2 release was calculated by Eq. (5):

where A was the average content of PGE2 in the model group; B was the average content of PGE2 in the sample group; C was the average content of PGE2 in the DMSO group.
Calculation of Combination index
For the drug combinations (two or more drugs), there may be three effects: synergism, additive effect and antagonism. If the effect of drug combination was stronger than the addition effect of each drug individual, it was synergism. And if the effect of drug combination was weaker than the addition effect of each drug individual, it was antagonism46,47,48,49. To determine the synergistic or antagonistic effect of sample combinations in this work, Chou and Talalay analysis was used38. Combination index (CI) was calculated by Eq. (6) to quantify the synergism or antagonism for two or more drugs. The calculation was carried out by CompuSyn50.

where CI < 1, = 1 and > 1 indicated synergism, additive effect and antagonism, respectively. (D)i is the concentration of Drug i when the inhibition is x% for “(D)1+(D)2+(D)3+…+(D)i combination”. (Dx)i is the concentration of Drug i when the inhibition is x% for Drug i “alone”.
References
Auffray, C., Chen, Z. & Hood, L. Systems medicine: the future of medical genomics and healthcare. Genome Med. 1, 2 (2009).
Drasar, P. & Moravcova, J. Recent advances in analysis of Chinese medical plants and traditional medicines. J. Chromatogr. B 812, 3–21 (2004).
Wang, H., Zou, H., Ni, J. & Guo, B. Comparative separation of biologically active components in Rhizoma chuanxiong by affinity chromatography with alpha(1)-acid glycoprotein and human serum albumin as stationary phases. Chromatographia 52, 459–464 (2000).
Zhang, Y. et al. A Novel Analgesic Isolated from a Traditional Chinese Medicine. Curr. Biol. 24, 117–123 (2014).
Ma, T. et al. Bridging the gap between traditional Chinese medicine and systems biology: the connection of Cold Syndrome and NEI network. Mol. BioSyst. 6, 613–619 (2010).
Gu, J. Y., Chen, L. R., Yuan, G. & Xu, X. J. A Drug-Target Network-Based Approach to Evaluate the Efficacy of Medicinal Plants for Type II Diabetes Mellitus. Evid. Based Complement. Alt. 2013, 203614 (2013).
Li, S. & Zhang, B. Traditional Chinese medicine network pharmacology: theory, methodology and application. Chin. J. Nat. Med. 11, 110–120 (2013).
Liu, Y. H., Ming, K. J., Sai, C. L. & Khang, G. N. Synergistic effect of traditional Chinese medicine. Asian J. Chem. 19, 867–882 (2007).
Zhao, M., Zhou, Q., Ma, W. & Wei, D. Q. Exploring the ligand-protein networks in traditional chinese medicine: current databases, methods and applications. Evid. Based Complement. Alt. 2013, 806072 (2013).
Shen, M. Y. et al. Drug-likeness analysis of traditional Chinese medicines: 1. property distributions of drug-like compounds, non-drug-like compounds and natural compounds from traditional Chinese medicines. J. Cheminform. 4, 31 (2012).
Xu, X. J. New concepts and approaches for drug discovery based on traditional Chinese medicine. Drug Discov. Today 3, 247–253 (2006).
Li, S., Zhang, B. & Zhang, N. B. Network target for screening synergistic drug combinations with application to traditional Chinese medicine. BMC Syst. Biol. 5, S10 (2011).
Wang, L. et al. Dissection of mechanisms of Chinese medicinal formula Realgar-Indigo naturalis as an effective treatment for promyelocytic leukemia. Proc. Natl. Acad. Sci. U. S. A. 105, 4826–4831 (2008).
Gu, P. Q. & Chen, H. J. Modern bioinformatics meets traditional Chinese medicine. Brief. Bioinform. 10.1093/bib/bbt063 (2013).
Li, J. et al. Synergism and rules from combination of Baicalin, Jasminoidin and Desoxycholic acid in refined Qing Kai Ling for treat ischemic stroke mice model. PLoS One 7, e45811 (2012).
Ma, X. H. et al. Synergistic therapeutic actions of herbal ingredients and their mechanisms from molecular interaction and network perspectives. Drug Discov. Today 14, 579–588 (2009).
Ulrich-Merzenich, G., Panek, D., Zeitler, H., Vetter, H. & Wagner, H. Drug development from natural products: Exploiting synergistic effects. Indian J. Exp. Biol. 48, 208–219 (2010).
Alexander, C. et al. Molecular imprinting science and technology: a survey of the literature for the years up to and including 2003. J. Mol. Recognit. 19, 106–180 (2006).
Pardeshi, S., Dhodapkar, R. & Kumar, A. Molecularly imprinted microspheres and nanoparticles prepared using precipitation polymerisation method for selective extraction of gallic acid from Emblica officinalis. Food Chem. 146, 385–393 (2014).
Gu, J. Y., Zhang, H., Yuan, G., Chen, L. R. & Xu, X. J. Surface-initiated molecularly imprinted polymeric column: In situ synthesis and application for semi-preparative separation by high performance liquid chromatography. J. Chromatogr. A 1218, 8150–8155 (2011).
Ranada, M. L., Akbulut, M., Abad, L. & Guven, O. Molecularly imprinted poly(N-vinyl imidazole) based polymers grafted onto nonwoven fabrics for recognition/removal of phloretic acid. Radiat. Phys. Chem. 94, 93–97 (2014).
Ozcan, A. A. & Demirli, S. Molecular Imprinted Solid-Phase Extraction System for the Selective Separation of Oleuropein from Olive Leaf. Sep. Sci. Technol. 49, 74–80 (2014).
Dai, C. M., Zhang, J., Zhang, Y. L., Zhou, X. F. & Liu, S. G. Application of Molecularly Imprinted Polymers to Selective Removal of Clofibric Acid from Water. PLoS One 8, e78167 (2013).
Bakas, I. et al. Molecular imprinting solid phase extraction for selective detection of methidathion in olive oil. Anal. Chim. Acta 734, 99–105 (2012).
Xu, X. J., Zhu, L. L. & Chen, L. R. Separation and screening of compounds of biological origin using molecularly imprinted polymers. J. Chromatogr. B 804, 61–69 (2004).
Vasapollo, G. et al. Molecularly imprinted polymers: present and future prospective. Int. J. Mol. Sci. 12, 5908–5945 (2011).
Ge, Y. & Turner, A. P. F. Molecularly Imprinted Sorbent Assays: Recent Developments and Applications. Chem-Eur J. 15, 8100–8107 (2009).
Giovannoli, C., Anfossi, L., Biagioli, F., Passini, C. & Baggiani, C. Solid phase extraction of penicillins from milk by using sacrificial silica beads as a support for a molecular imprint. Microchim. Acta 180, 1371–1377 (2013).
Luo, W. et al. Synthesis of surface molecularly imprinted silica micro-particles in aqueous solution and the usage for selective off-line solid-phase extraction of 2,4-dinitrophenol from water matrixes. Anal. Chim. Acta 618, 147–156 (2008).
Chen, T. F. et al. Semi-Preparative Scale Separation of Emodin from Plant Extract by Using Molecularly Imprinted Polymer as Stationary Phase. Chromatographia 77, 893–899 (2014).
Zhu, T., Li, S. N. & Row, K. H. Molecularly Imprinted Monolithic Material for the Extraction of Three Organic Acids from Salicornia herbacea L. J. Appl. Polym. Sci. 121, 1691–1696 (2011).
Michailof, C., Manesiotis, P. & Panayiotou, C. Synthesis of caffeic acid and p-hydroxybenzoic acid molecularly imprinted polymers and their application for the selective extraction of polyphenols from olive mill waste waters. J. Chromatogr. A 1182, 25–33 (2008).
Valero-Navarro, A. et al. Synthesis of caffeic acid molecularly imprinted polymer microspheres and high-performance liquid chromatography evaluation of their sorption properties. J. Chromatogr. A 1218, 7289–7296 (2011).
Huang, X. M., Liu, Y. J., He, Y. Z., Chen, F. F. & Wang, Y. K. Clinical observation of Reduning injection in treatment of patients with acute upper respiratory tract infections. Chin. J. Clin. Pharmacol. Ther. 11, 470–473 (2006).
Zhang, Y. The Clinical Efficacy of Reduning in the Treatment of HFMD. Guide of China Medicine 9, 33–34 (2011).
Li, Y. J. et al. Direct analysis in real time ionization/quadrupole time-of-flight tandem mass spectrometry for rapid identification of iridoid glycosides and caffeoylquinic acids in Re Du Ning Injections. Anal. Methods-UK 5, 7081–7089 (2013).
Li, H. B. Studies on Pharmacodynamic Material Basis of Reduning Injection Ph.D thesis, Nanjing University of Chinese Medicine (2013).
Chou, T. C. & Talalay, P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv. Enzyme Regul. 22, 27–55 (1984).
Zhang, X. Z. et al. Network pharmacology study on the mechanism of traditional Chinese medicine for upper respiratory tract infection. Mol. BioSyst. 10, 2517–2525 (2014).
Zhang, X. Z. et al. Insights into the inhibition and mechanism of compounds against LPS-induced PGE2 production: a pathway network-based approach and molecular dynamics simulations. Integr. Biol. 10.1039/C4IB00141A (2014).
Zhuang, Y., Luo, H. P., Duan, D. L., Chen, L. R. & Xu, X. J. In situ synthesis of molecularly imprinted polymers on glass microspheres in a column. Anal. Bioanal. Chem. 389, 1177–1183 (2007).
Bi, Y. A. et al. The Fingerprint Research and Multi-component Qualitative Analysis of Reduning Injection via HPLC. Mode. Tradit. Chin. Med. Mater. Med 12, 298–303 (2010).
Zhang, X. Z. et al. Bauer ketones 23 and 24 from Echinacea paradoxa var. paradoxa inhibit lipopolysaccharide-induced nitric oxide, prostaglandin E2 and cytokines in RAW264.7 mouse macrophages. Phytochemistry 74, 146–158 (2012).
Tewtrakul, S., Wattanapiromsakul, C. & Mahabusarakam, W. Effects of compounds from Garcinia mangostana on inflammatory mediators in RAW264.7 macrophage cells. J. Ethnopharmacol. 121, 379–382 (2009).
Jin, X. H. et al. Effects of blue honeysuckle (Lonicera caerulea L.) extract on lipopolysaccharide-induced inflammation in vitro and in vivo. Exp. Eye Res. 82, 860–867 (2006).
Wagner, H. & Ulrich-Merzenich, G. Synergy research: Approaching a new generation of phytopharmaceuticals. Phytomedicine 16, 97–110 (2009).
Urich-Merzenich, G., Panek, D., Zeitler, H., Wagner, H. & Vetter, H. New perspectives for synergy research with the "omic"-technologies. Phytomedicine 16, 495–508 (2009).
Hemalswarya, S. & Doble, M. Potential synergism of natural products in the treatment of cancer. Phytother. Res. 20, 239–249 (2006).
Williamson, E. M. Synergy and other interactions in phytomedicines. Phytomedicine 8, 401–409 (2001).
Chou, T. C. Theoretical basis, experimental design and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol. Rev. 58, 621–681 (2006).
Acknowledgements
This study was financed by the “Innovative Drug Development” Key Project of the Ministry of Science and Technology of China (Grant No. 2012ZX09501001-004, 2013ZX09402202 and 2013ZX09402203). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
W.X., X.J.X. and L.R.C. conceived the study. T.F.C. and J.Y.G. designed the experiments. T.F.C., J.Y.G. and Y.M.M. performed the experiments with the help of X.Z.Z., L.C. and Z.Z.W. T.F.C. and J.Y.G. analyzed the data and wrote the manuscript. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Chen, T., Gu, J., Zhang, X. et al. System-level Study on Synergism and Antagonism of Active Ingredients in Traditional Chinese Medicine by Using Molecular Imprinting Technology. Sci Rep 4, 7159 (2014). https://doi.org/10.1038/srep07159
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep07159
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.