Abstract
Autosomal recessive cerebellar ataxias and autosomal recessive hereditary spastic paraplegias (ARHSPs) are clinically and genetically heterogeneous neurological disorders. Herein we describe Japanese siblings with a midlife-onset, slowly progressive type of cerebellar ataxia and spastic paraplegia, without intellectual disability. Using whole exome sequencing, we identified a homozygous missense mutation in DDHD2, whose mutations were recently identified as the cause of early-onset ARHSP with intellectual disability. Brain MRI of the patient showed a thin corpus callosum. Cerebral proton magnetic resonance spectroscopy revealed an abnormal lipid peak in the basal ganglia, which has been reported as the hallmark of DDHD2-related ARHSP (SPG 54). The mutation caused a marked reduction of phospholipase A1 activity, supporting that this mutation is the cause of SPG54. Our cases indicate that the possibility of SPG54 should also be considered when patients show a combination of adult-onset spastic ataxia and a thin corpus callosum. Magnetic resonance spectroscopy may be helpful in the differential diagnosis of patients with spastic ataxia phenotype.
Similar content being viewed by others
Introduction
Autosomal recessive cerebellar ataxias (ARCAs) and autosomal recessive hereditary spastic paraplegias (ARHSPs) are clinically and genetically heterogeneous neurological disorders. Mutations in more than 30 different genes have been identified for ARCAs and those of 38 different genes have been identified for ARHSPs. Genetic diagnosis of ARCAs and ARHSPs has been difficult, because of genetic and clinical heterogeneity. In addition, the accompanying symptoms can vary even within the same disease (e.g. spastic paraplegia 7 (SPG7) can present with both a pure and complex phenotype1), making correct diagnosis more difficult. Recent advances in parallel sequencing technology are making a major contribution, not only to the identification of novel responsible genes2, but also to finding genetic causes even in unrecognized phenotypes3 and to the comprehensive genetic diagnosis of ARCAs and ARHSPs4,5. Herein, we describe Japanese siblings exhibiting a midlife-onset spastic ataxia with a novel homozygous DDHD2 mutation found by exome-sequencing.
Results
Among the children of first-cousin parents, two sisters were affected (Figure 1A). The early developmental milestones of the proband (II-6) were normal. At the age of approximately 45 years old, she developed gait unsteadiness and dysarthria. She was 69 years old at the last examination and could not stand without holding on to something. She had gaze-evoked horizontal nystagmus, dysarthria, extensor plantar reflexes, mild limb ataxia, moderate truncal ataxia, postural tremor in the head and upper extremities, decline of vibratory sense in the lower extremities and urinary incontinence. Patellar tendon reflexes were increased, while Achilles tendon reflexes were absent. Cognitive impairments including callosal apraxia were not observed. Laboratory biochemistry results were normal, including serum liver enzymes, ammonia, thyroid hormones, copper, α-fetoprotein, vitamin E and very long-chain fatty acids. Serum antibody for Human T lymphotropic virus type 1 was negative. Galactocerebrosidase activity in leukocytes was normal. Blood amino-acid analysis and urinary organic acid analysis revealed no apparent deviance. A nerve conduction study disclosed a mild slowing of motor and sensory nerve conduction velocities (between 36.0 and 46.3 m/s) with reduced compound muscle action potentials. Brain magnetic resonance imaging revealed mild atrophy of the cerebellum and a thinness of the splenium of the corpus callosum (Figure 1B). Neither atrophy nor cross sign were observed in the brainstem. The patient was negative for the genetic alterations associated with spinocerebellar ataxia (SCA)1, SCA2, SCA3, SCA6, SCA7, SCA12, SCA17 and dentatorubral pallidoluysian atrophy. Her elder sister (II-2) developed gait unsteadiness at the age of 38 years and by age 55 years, could not walk independently. She showed saccadic eye pursuit, dysarthria, dysphagia, limb muscle weakness, extensor plantar reflexes, limb and truncal ataxia and urinary incontinence. She died at 65, most likely because of hepatic encephalopathy with hyperammonemia (164 μg/dl, normal range 12–66 μg/dl), high serum lactate level (22.9 mg/dl, normal range 4–16 mg/dl) and pyruvate (2.3 mg/dl, normal range 0.3–0.9 mg/dl) and ketonuria. Serum copper and ceruloplasmin were normal. Eventually, a precise cause of hepatic insufficiency could not be detected. Another two siblings (II-4 and II-5) died at early ages (7 and 5 years, respectively) with unknown cause, but it is unlikely that the causes of their deaths were related to spastic ataxia, considering that they died in childhood. Homozygosity mapping and linkage analysis identified 11 candidate regions totalized to ~240 Mb, with the maximum LOD score equaling 1.32 (Table S2). When an adult patient exhibits both cerebellar ataxia and spasticity, the primary diagnostic considerations are autosomal recessive ataxia of Charlevoix-Saguenay, late-onset Friedreich ataxia or SPG76. However, none of the related genes, SACS, FRDA and SPG7, were located in the candidate regions of our patient. As a result of whole exome-sequencing of the proband, approximately 39.8 million paired-reads were mapped to the human reference genome. A coverage analysis revealed that 95.9% of the bases within the target regions were covered by 10 reads or more. In total, 37,553 variations, which were unregistered in dbSNP137 and registered as uncommon SNPs with minor allele frequency <1%, were detected. Among these, 2,986 variations (including 1,148 homozygous variants) were located in exons or splice sites (within 2 bp of the boundaries). Only the eight homozygous missense single nucleotide variations (SNVs) remained in the ~240-Mb candidate regions with the frequency <1% in exome data from 575 “in house” Japanese controls (Table 1). Sanger sequencing confirmed that all of these SNVs were homozygous in the proband and heterozygous in the unaffected sibling. Among the SNVs, the c.658G > T [p.Val220Phe] of DDHD2 (Figure 1C) was of interest, because mutations of DDHD1 and DDHD2, which code for members of the intracellular phospholipase A1 (PLA1), have recently been found to be the causative genes for ARHSPs (SPG28 and SPG54)7,8. Furthermore, only the SNVs of DDHD2 and FAM222A were consistently predicted to be disruptive in protein function when analyzed with multiple tools including Polyphen2, SIFT and Mutation Taster9,10,11, while the predictions for the other six SNVs were benign or inconclusive (Table 1). Considering the allele frequency of FAM222A SNV in Japanese control exome data (4/575), it is unlikely that the SNV is the cause of extremely rare diseases. The potentially compound heterozygous SNVs detected in the proband are listed, indicating that none of the listed genes is likely to be the cause of the disease (Table S3). We further checked whether any other causative variations were present in known ARCA or ARHSP genes, which are listed in Table S1. We confirmed that no pathological homozygous or compound heterozygous SNVs were found in these genes. Patients with DDHD2 mutations have been reported to show very early-onset (before the age of 6 years) spastic paraplegia with intellectual disability (SPG54), occasionally associated with strabismus and/or hypoplasia of the optic nerve (Table 2)8,12,13,14. Brain MRIs of these patients showed a thin corpus callosum with periventricular white-matter hyperintensity8. As a unique finding, proton magnetic resonance spectroscopy (1H-MRS) revealed an abnormal lipid peak in the basal ganglia and thalamus area (Figure 1D). Considering the highly characteristic 1H-MRS findings and the observations that the patient carried a novel homozygous p.Val220Phe of DDHD2 predicted as deleterious15,16 (and not present in 575 Japanese controls by whole exome sequencing or 429 Japanese controls by Sanger sequencing), we thought that the DDHD2 mutation was the causative agent in this patient. Because most causative mutations of SPG54 were protein-truncating (Figure 1C), loss of DDHD2 function is plausible. We first checked intracellular distribution of p.Val220Phe and wild type (WT) in HEK293T cells, but found no difference (Figure S1). The result indicated that p.Val220Phe does not severely affect the conformation or stabilities of DDHD2. We then assessed an impact of the p.Val220Phe mutation by mapping the mutation on a 3D structure. Val220 is predicted to be involved in a hydrophobic core near the candidate catalytic site, suggesting that the p.Val220Phe mutation may impair lipase activity (Figure 2). Although SPG54-linked point mutations were reported (Table 2), whether the mutations affect enzymatic activity was not examined. We thus analyzed the PLA1 activity of the p.Val220Phe mutant as well as p.Trp103Arg and p.Asp660His mutants, both of which were reported to be linked to SPG5414. The results clearly demonstrated that the p.Val220Phe mutant as well as both the p.Trp103Arg and p.Asp660His mutants has a statistically significant reduction in their PLA1 activity (Figure 3A–C, lane 6–8). Notably, the p.Val220Phe mutant, but not other two mutants still retained a marginal PLA1 activity (Figure 3A–C, lane 6). We also co-transfected equal amounts of the WT- and each mutant-expressing plasmids and then measured PLA1 activity. These conditions mimicked the heterozygous states of healthy carriers with both wild-type and mutant alleles. In these conditions, the PLA1 activity was not severely affected (Figure 3A–C, lane 3–5), indicating that all of the mutants did not have a dominant-negative effect on the WT DDHD2. These results strongly indicated that the p.Val220Phe mutation of DDHD2 was indeed a culprit mutation in this patient.
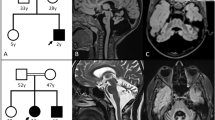
Familial pedigree, brain MRI and proton MRS of a patient with homozygous DDHD2 mutation.
(A): Familial pedigree. * indicates members whose genomic DNA was available for this study (II-3 and II-6). Arrow indicates the proband (II-6). Homozygosity mapping and linkage analysis were performed using DNA from the proband and the unaffected sibling (II-3). (B): Brain MRI of II-6 at 69 years of age. Axial and sagittal sections of fluid-attenuated inversion recovery image are shown. Mild atrophy of the cerebellum and the thinness of the splenium of the corpus callosum (arrow) are observed. (C): Schematic presentation of DDHD2 and mutations. The thick arrow indicates the location of the mutation in the patient. (D): Proton MRS obtained from left thalamus, at a magnetic field of 3 Tesla (echo time 30 ms and 144 ms, respectively). Arrows indicate the pathologic lipid peak at 1.3 ppm. mI: myo-inositol, Cho: choline, Cr: creatine, NAA: N-acetylaspartate.
Structural consideration of an impact of the p.Val220Phe mutation in human DDHD2.
(A–B): A modeled structure of the region around Val220 in human DDHD2 (A) and the crystal structure of the catalytic domain of GPLRP2 chimerized with the C-terminal domain of HPL (PDB id; 1GPL) (B) are shown. The side chains of Val220 in A and the corresponding residue Ile74 in B are shown in red spheres. The catalytic triad in B and a component residue of the triad, Ser351, in A are shown in yellow spheres28. In A, a stretch of the sequence, Gly349-His-Ser-Leu352, which is conserved in most lipases22, is colored green and their side chains were shown as sticks, except for Ser351. Inset: close-up view of the squared region.
PLA1 activity of DDHD2 mutants.
GST and GST-tagged DDHD2 WT and/or DDHD2 mutants were expressed in 293T cells and partially purified. Their PLA1 activities were measured using 32P-labeled DOPA as a substrate, as described in “Methods”. The reaction products were analyzed by TLC. (A): Representative results of TLC analysis. GST (lane 1), GST-DDHD2 WT (lane 2), a mixture of GST-DDHD2 WT and one of GST-DDHD2 mutants (p.Val220Phe (lane 3), p.Trp103Arg (lane 4) and p.Asp660His (lane 5)) and GST-DDHD2 mutants alone (p.Val220Phe (lane 6), p.Trp103Arg (lane 7) and p.Asp660His (lane 8)) were used. Positions of DOPA and LPA are indicated by arrows. The product LPA was clearly detected in the lane with GST-DDHD2 WT but not with GST alone. The amounts of LPA were markedly reduced in the cases of all the three mutants. (B): A representative image of Western blotting analysis. GST and GST fusion proteins used for the above PLA1 assay were analyzed by Western blotting using an anti-GST antibody. One-fifth of amount of each sample used in A was loaded. Lane numbers are the same as in A. (C): Comparison of PLA1 activity. The intensities of spots on a TLC plate were quantified using Multi Gauge V3.0 software (Fujifilm). The PLA1 activity (an amount (nmol) of LPA formed per min) was calculated from the intensities of LPA and DOPA spots. Data are shown as means ± S.D. from 3 or more independent experiments. Numbers in the graph are the same as in A. The asterisks indicate statistically significant differences between the DDHD2 mutants and control DDHD2 WT (asterisks placed above each bracket), or between GST and each condition of DDHD2 (asterisks placed above each bar) (*P < 0.05, **P < 0.001 and ***P < 0.0005, Student's t-test).
Discussion
Compared with the clinical presentations described in previous reports, our patients were quite a bit older at onset, the main phenotype was spastic ataxia and intellectual disability was not observed. Mild polyneuropathy was observed in our case, revealing the phenotypic variability of SPG54. One of our patients showing spastic ataxia phenotype (II-2) developed hepatic encephalopathy with unknown cause. Because we could not detect the evidence of metabolic disease in the proband, whether hepatic encephalopathy was a part of the SPG54 is unknown. It is possible that her sibling (II-2), but not the proband, might have had some metabolic problems other than spastic ataxia, because she did not have apparent evidence of acquired liver diseases (such as viruses or toxic agents) or portacaval shunt. Brain MRIs of our case showed mild white-matter hyperintensity and a thin corpus callosum, sharing these common characteristics with previous cases. In addition, mild cerebellar atrophy was recognized. In diseases presenting with SCA or SPG, abnormal 1H-MRS findings have been reported, especially when the phenotype is the consequence of metabolic disorders. In these cases, some with white matter changes have shown increased myo-inositol/creatine (Cr) and decreased N-acetylaspartate/Cr, reflecting astrogliosis and neuroaxonal loss17,18. Although broad lipid peaks have sometimes been detected in child food adrenoleukodystrophy and peroxisome disorders, emergence of clear, sharp lipid peaks have been reported in very limited diseases such as Sjögren-Larsson syndrome19,20 hypomyelination and congenital cataract21, which show distinct clinical manifestations from SCA and SPGs. No SNVs were detected in the causative gene of Sjögren-Larsson syndrome (ALDH3A2). Because a sharp lipid peak has not been reported in any other SCA or SPGs, the finding could be considered as highly characteristic for SPG548. However, mutations in a group of genes involved in lipid metabolism including CYP7B1 (SPG5), DDHD1 (SPG28), FA2H (SPG35), PNPLA6 (SPG39), GBA2 (SPG46) and CYP2U1 (SPG56) have been found as the causes of ARHSPs. The 1H-MRS findings of the ARHSPs remain to be investigated. DDHD2 is a member of intracellular PLA122, which hydrolyzes an acyl group from phospholipids at the sn-1 position. The tandem SAM-DDHD domain of DDHD2 is essential for binding to phosphatidylinositol phosphate23. Most DDHD2 mutations reported reside in the SAM-DDHD domain (Figure 1C), but it has been never assessed whether the mutations actually impair the PLA1 activity. Previously, only two missense mutations, p.Trp103Arg and p.Asp660His (Table 2), were reported to cause SPG54. Our results showed that three missense mutations including p.Val220Phe significantly reduced PLA1 activity (Figure 3), as predicted from the structural consideration (Figure 2). These data indicated that the loss of PLA1 activity significantly contributes to SPG54 pathogenicity. Furthermore, we demonstrated that the pVal220Phe mutant had marginal PLA1 activity (Figure 3A–C, lane 6). The result may account for the mild phenotype in the present case. The highly characteristic 1H-MRS finding of an abnormal lipid peak, which was also observed in our patient, has been considered to reflect accumulation of phospholipids as a result of abolished PLA1 activities. Although the p.Val220Phe mutation is not located in the SAM-DDHD domain, it is conceivable that the mutation associated with decreased PLA1 activity caused SPG54.
In conclusion, our report indicates that the possibility of SPG54 should also be taken into consideration when patients show a combination of adult-onset spastic ataxia and a thin corpus callosum. Magnetic resonance spectroscopy may be helpful in the differential diagnosis of patients with the spastic ataxia phenotype.
Methods
Patients
Clinical information, radiological images and blood samples were obtained from family members after written informed consent was provided. Experimental protocols were approved by the Independent Review Boards of Yokohama City University and Shinshu University. All experiments were performed in accordance with the institutional guidelines.
Homozygosity mapping and exome sequencing
To identify the disease locus, genome-wide single nucleotide polymorphism (SNP) genotyping of the proband and the unaffected siblings was performed using the Genome-Wide Human SNP Array 6.0 (SNP 6.0 array) (Affymetrix, Inc., Santa Clara, CA). Then, SNP 6.0 array data were subjected to homozygosity mapping using HomozygosityMapper software24. The linkage analysis was performed based on the model of autosomal recessive inheritance with complete penetrance, using the subset of 7860 SNPs with high heterozygosity extracted from the SNP 6.0 array data with the program Linkdatagen, setting the bin size to 0.5 cM25. To find a gene mutation within the loci, whole exome sequencing was performed on the proband. The genomic DNA was processed using the SureSelect Human All Exon Kit v5 (Agilent Technologies, Santa Clara, CA).
Structural consideration of an impact of the p.Val220Phe mutation in human DDHD2
A modeled structure of the region around Val220 in human DDHD2 was constructed from the crystal structure of the catalytic domain of guinea pig pancreatic lipase-related protein 2 (GPLRP2) chimerized with the C-terminal domain of human pancreatic lipase (HPL) (PDB id; 1GPL) using Phyre2 server26.
Phospholipase A1 (PLA1) assay
The full-length DDHD2 cDNA (FXC00119) was obtained from Kazusa DNA Research Institute (Chiba, Japan) and subcloned into pcDNA3.1/V5-His vector (Invitrogen). Site-directed mutagenesis was performed to produce DDHD2 mutants with c.658G > T [p.Val220Phe], c.307T > C [p.Trp103Arg] and c.1978G > C [p.Asp660His] by using a mutagenesis kit (Toyobo, Osaka, Japan). All constructs were verified by Sanger sequencing.
The mammalian expression plasmid pEBG27 was used to express GST-fusion proteins. The WT and DDHD2 mutant cDNAs were inserted into pEBG and transfected into 293T cells using the Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacture's instruction. For a 3.5-cm culture dish, total 2 μg of the plasmids were used. Co-transfection of the WT- and each mutant-expressing constructs was performed in a 1:1 ratio. Their cellular lysates were prepared and mixed with Gluthathione Sepharose 4B beads (GE Healthcare, Piscataway, NJ), as described previously23. The lysates prepared from 293T cells grown on six 3.5-cm culture dishes were mixed with 22.5 μl of beads. After washing, half of the beads were used for PLA1 assay and the one-tenth was used for Western blotting analysis.
Preparation of 32P-labeled 1,2-dioleoyl-sn-phosphatidic acid (DOPA) and PLA1 assay were performed as described previously23. Briefly, reactions were started by adding 100 μl of a reaction buffer [50 mM Tris-HCl, pH 7.5, 100 mM KCl, 100 μM DOPA (Avanti Polar Lipids, Inc. Alabaster, AL) and 4.2 nM 32P-labeled DOPA to beads-containing tubes. The reactions were conducted at 37°C for 15 or 30 min and then stopped by adding 200 μl of 1N HCl and 400 μl of CHCl3/methanol (2:1). The substrate (DOPA) and a product 2-oleoyl-lysophosphatidic acid (LPA) were recovered from an organic phase and separated by thin-layer chromatography (TLC). Radioactivity on a TLC plate was visualized using a Bio-Image Analyzer FLA-9000 (Fujifilm, Tokyo, Japan). For Western blotting analysis, an SDS sample buffer was added to the beads and the mixture was heated at 100°C for 5 min. The eluted proteins were analyzed by Western blotting, followed by staining with a rabbit polyclonal anti-GST antibody (Santa Cruz, CA).
References
van Gassen, K. L. et al. Genotype-phenotype correlations in spastic paraplegia type 7: a study in a large Dutch cohort. Brain 135, 2994–3004, 10.1093/brain/aws224 (2012).
Hammer, M. B. et al. Mutations in GBA2 cause autosomal-recessive cerebellar ataxia with spasticity. Am J Hum Genet 92, 245–251, 10.1016/j.ajhg.2012.12.012 (2013).
Pyle, A. et al. Prominent sensorimotor neuropathy due to SACS mutations revealed by whole-exome sequencing. Arch Neurol 69, 1351–1354, 10.1001/archneurol.2012.1472 (2012).
Doi, H. et al. Identification of a novel homozygous SPG7 mutation in a Japanese patient with spastic ataxia: making an efficient diagnosis using exome sequencing for autosomal recessive cerebellar ataxia and spastic paraplegia. Intern Med 52, 1629–1633 (2013).
Hammer, M. B. et al. Exome sequencing: an efficient diagnostic tool for complex neurodegenerative disorders. Eur J Neurol 20, 486–492, 10.1111/j.1468-1331.2012.03883.x (2013).
de Bot, S. T., Willemsen, M. A., Vermeer, S., Kremer, H. P. & van de Warrenburg, B. P. Reviewing the genetic causes of spastic-ataxias. Neurology 79, 1507–1514, 10.1212/WNL.0b013e31826d5fb0 (2012).
Tesson, C. et al. Alteration of Fatty-Acid-metabolizing enzymes affects mitochondrial form and function in hereditary spastic paraplegia. Am J Hum Genet 91, 1051–1064, 10.1016/j.ajhg.2012.11.001 (2012).
Schuurs-Hoeijmakers, J. H. et al. Mutations in DDHD2, encoding an intracellular phospholipase A(1), cause a recessive form of complex hereditary spastic paraplegia. Am J Hum Genet 91, 1073–1081, 10.1016/j.ajhg.2012.10.017 (2012).
Adzhubei, I. A. et al. A method and server for predicting damaging missense mutations. Nat Methods 7, 248–249, 10.1038/nmeth0410-248 (2010).
Kumar, P., Henikoff, S. & Ng, P. C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4, 1073–1081, 10.1038/nprot.2009.86 (2009).
Schwarz, J. M., Rodelsperger, C., Schuelke, M. & Seelow, D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods 7, 575–576, 10.1038/nmeth0810-575 (2010).
Gonzalez, M. et al. Mutations in phospholipase DDHD2 cause autosomal recessive hereditary spastic paraplegia (SPG54). Eur J Hum Genet 21, 1214–1218, 10.1038/ejhg.2013.29 (2013).
Citterio, A. et al. Mutations in CYP2U1, DDHD2 and GBA2 genes are rare causes of complicated forms of hereditary spastic paraparesis. J Neurol 261, 373–381, 10.1007/s00415-013-7206-6 (2014).
Magariello, A. et al. Further evidence that DDHD2 gene mutations cause autosomal recessive hereditary spastic paraplegia with thin corpus callosum. Eur J Neurol 21, e25–26, 10.1111/ene.12305 (2014).
Adzhubei, I., Jordan, D. M. & Sunyaev, S. R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet/editorial board, Jonathan L. Haines. et al. Chapter 7, Unit7 20, 10.1002/0471142905.hg0720s76 (2013).
Sim, N. L. et al. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res 40, W452–457, 10.1093/nar/gks539 (2012).
Brockmann, K. et al. Proton MRS profile of cerebral metabolic abnormalities in Krabbe disease. Neurology 60, 819–825 (2003).
Jamrozik, Z. et al. Late onset GM2 gangliosidosis mimicking spinal muscular atrophy. Gene 527, 679–682, 10.1016/j.gene.2013.06.030 (2013).
Miyanomae, Y. et al. Cerebral MRI and spectroscopy in Sjogren-Larsson syndrome: case report. Neuroradiology 37, 225–228 (1995).
van Domburg, P. H. et al. Sjogren-Larsson syndrome: clinical and MRI/MRS findings in FALDH-deficient patients. Neurology 52, 1345–1352 (1999).
Rossi, A. et al. Hypomyelination and congenital cataract: neuroimaging features of a novel inherited white matter disorder. AJNR. Am J Neuroradiol 29, 301–305, 10.3174/ajnr.A0792 (2008).
Nakajima, K. et al. A novel phospholipase A1 with sequence homology to a mammalian Sec23p-interacting protein, p125. J Biol Chem 277, 11329–11335, 10.1074/jbc.M111092200 (2002).
Inoue, H. et al. Roles of SAM and DDHD domains in mammalian intracellular phospholipase A1 KIAA0725p. Biochim Biophys Acta 1823, 930–939, 10.1016/j.bbamcr.2012.02.002 (2012).
Seelow, D., Schuelke, M., Hildebrandt, F. & Nurnberg, P. HomozygosityMapper--an interactive approach to homozygosity mapping. Nucleic Acids Res 37, W593–599, 10.1093/nar/gkp369 (2009).
Bahlo, M. & Bromhead, C. J. Generating linkage mapping files from Affymetrix SNP chip data. Bioinformatics 25, 1961–1962, 10.1093/bioinformatics/btp313 (2009).
Kelley, L. A. & Sternberg, M. J. Protein structure prediction on the Web: a case study using the Phyre server. Nature Protoc 4, 363–371, 10.1038/nprot.2009.2 (2009).
Mayer, B. J., Hirai, H. & Sakai, R. Evidence that SH2 domains promote processive phosphorylation by protein-tyrosine kinases. Curr Biol: CB 5, 296–305 (1995).
Withers-Martinez, C., Carriere, F., Verger, R., Bourgeois, D. & Cambillau, C. A pancreatic lipase with a phospholipase A1 activity: crystal structure of a chimeric pancreatic lipase-related protein 2 from guinea pig. Structure 4, 1363–1374 (1996).
Acknowledgements
We would like to thank the patient and her family for their participation in this study.
Author information
Authors and Affiliations
Contributions
H.D. wrote the main manuscript and prepared the figures and tables. S.T., F.T., K.Y. and N. Matsumoto revised the manuscript and gave conceptual advice. M.U., K.Y. and S.I. collected the clinical data and samples. H.D., Y.F.Y. and S.T. conducted linkage analysis. T.B. and K.T. analyzed PLA1 activity of the DDHD2 and prepared Figure 3. M.S. and K.O. analyzed the structure of the DDHD2 mutant and prepared Figure 2. H.D., S.M., M.N., Y.T., N. Miyake and H.S. conducted the analysis of the genetic data.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Doi, H., Ushiyama, M., Baba, T. et al. Late-onset spastic ataxia phenotype in a patient with a homozygous DDHD2 mutation. Sci Rep 4, 7132 (2014). https://doi.org/10.1038/srep07132
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep07132
This article is cited by
-
Autosomal recessive adult onset ataxia
Journal of Neurology (2022)
-
Defining the clinical-genetic and neuroradiological features in SPG54: description of eight additional cases and nine novel DDHD2 variants
Journal of Neurology (2019)
-
Loss of DDHD2, whose mutation causes spastic paraplegia, promotes reactive oxygen species generation and apoptosis
Cell Death & Disease (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.