Abstract
Evolution has produced some remarkable creatures, of which silk gland is a fascinating organ that exists in a variety of insects and almost half of the 34,000 spider species. The impressive ability to secrete huge amount of pure silk protein and to store proteins at an extremely high concentration (up to 25%) make the silk gland of Bombyx mori hold great promise to be a cost-effective platform for production of recombinant proteins. However, the extremely low production yields of the numerous reported expression systems greatly hindered the exploration and application of silk gland bioreactors. Using customized zinc finger nucleases (ZFN), we successfully performed genome editing of Bmfib-H gene, which encodes the largest and most abundant silk protein, in B. mori with efficiency higher than any previously reported. The resulted Bmfib-H knocked-out B. mori showed a smaller and empty silk gland, abnormally developed posterior silk gland cells, an extremely thin cocoon that contain only sericin proteins and a slightly heavier pupae. We also showed that removal of endogenous Bmfib-H protein could significantly increase the expression level of exogenous protein. Furthermore, we demonstrated that the bioreactor is suitable for large scale production of protein-based materials.
Similar content being viewed by others
Introduction
With the developing understanding of disease pathogens and the identification of new molecular targets, biopharmaceutical proteins such as vaccines, hormones and protein-based biomaterials are in increasing demand for both analytical and clinical applications. Bioreactors using genetic modified organisms, which enable production of recombinant proteins in a commercial scale, emerged and underwent decades of development to meet this demand. However, few proteins produced in such bioreactors are now in clinical trials and only one has been approved for marketing1, mainly due to low production yield and costly protein purification process using genetic modified plants or livestock. As the silk producing organ of many insect and spider species, silk gland has been extraordinarily conspicuous and well-studied, owing to the impressive ability to secrete huge amount of silk protein and to store proteins at high concentration without aggregation or denaturalization2. This ability was greatly enhanced in silkworm, Bombyx mori, by thousands of years' domestication. Nowadays, approximately 1,000,000 tons of cocoon and 200,000 tons of silk were produced each year worldwide3. Eating about 20 g mulberry leaves, one commercial B. mori larvae can produce 0.5 g pure silk protein (dry weight, 25% of total worms). Therefore, the silk gland of B. mori holds great promise to be a cost-effective platform for commercial scale production of recombinant proteins.
During the past decade, several transgenic silk gland based expression systems were established and various recombinant proteins including collagen4,5,6, globular protein7, human basic fibroblast growth factor8, human serum albumin9, feline interferon10, human mu-opioid receptor11, mouse monoclonal antibody12 and spider silk13,14,15 were successfully produced in these systems. Meanwhile, many strenuous attempts utilizing different kinds of regulatory elements including promoters16, enhancers17,18, UTRs18,19, mutant strains20 and insulators (unpublished) were performed to increase the expression level of exogenous proteins. However, the actual yields of these proteins were much lower than expected and they can not be dissolved and purified without some hash reagents, in which the proteins may lose their biological activities.
As a giant organ whose main, if not only, mission was synthesis and secretion of silk protein, the extremely extraordinary protein production ability may mainly serve the silk protein production and thus repress the expression of exogenous proteins. Thus we proposed that reducing or removal of the endogenous silk proteins might increase the expression level of exogenous recombinant proteins. The successive emerging of ZFN21,22, transcription activator-like effector nucleases (TALENs)23 and clustered regularly interspersed short palindromic repeats (CRISPR)/Cas924 technologies enable genome editing in various organisms. And more encouragingly, these systems have been established in B. mori, in which targeted mutagenesis had never been achieved before25,26. In the current study, we showed that genetic removal of fibroin heavy chain protein by ZFN re-programmed the silk gland to produce exogenous recombinant proteins at an explosive level.
Results
We first performed targeted mutagenesis of fibroin heavy chain (BmFib-H) gene, which encodes the largest and most abundant silk protein, using custom ZFN. BmFib-H consists of a highly repetitive core flanked by non repetitive 5′ and 3′ ends and encodes a considerably large protein comprising N-terminal and C-terminal hydrophilic domains and 12 highly repetitive Gly-Ala-rich regions27. A pair of ZFN designed to target the 5′-non-repetitive region of BmFib-H (Fig. 1a, supplementary Figs. 1 and 2) was microinjected into 318 embryos of a non-diapausing B. mori strain in mRNA form. Twenty-five broods (65.79%) with mutant worms were obtained from 38 total broods. To confirm the considerable high efficiency, which seemed to be much higher than previously reported ZFN or TALEN induced mutations in B. mori25,26, the same mRNA was also microinjected into Dazao, a diapausing B. mori strain whose genomic sequence was revealed and used as a reference strain28. 189 mutant worms with similar efficiency (73.47%) were obtained from 271 injected Dazao embryos (Fig 1d, supplementary Tables 1 and 2). Three mutant phenotypes were observed in both knock-out experiments. About half of 609 total mutant silkworms could produce extremely thin-layered cocoons and developed normally, while the rest were naked pupae or arrested during the larvae-pupae transition stages (supplementary Fig. 3). A total of 2250 T-clones were generated using PCR fragments amplified from genomic DNA of 450 mutant individuals to indentify the mutant sequences. Similar to ZFN-induced mutations reported previously, various mutation types including deletions, insertions and nucleotide substitutions were found, most of which were micro deletions or deletions companied with small insertions (Fig. 1b,c, supplementary Figs. 5 and 6). No significant differences were found among different phenotypes. However, to our surprise, about one-third of the mutations were in-frame mutations with single nucleotide substitution, deletion of 3n nucleotides, or insertion of 3n nucleotides, resulting point changes of fibroin heavy chain protein (Fig. 1c). And genotypes of all the mutant individuals harboring in-frame mutations were heterozygous with one wild-type allele (data not shown), indicating that the in-frame mutations were dominant. To confirm the dominance/recessive of the mutations, two typical mutations (Fib-H-D3, in which 3 nucleotides were deleted and Fib-H-I1, in which 1 nucleotide was inserted) were subjected to genetic analysis and the results showed that Fib-H-D3 was a dominant mutation and Fib-H-I1 was a recessive mutation (supplementary Tables 4 and 5). Fib-H-I1, in which one nucleotide was inserted at the cleavage site and only a truncated N terminal hydrophilic domain of Fib-H was produced (supplementary Fig. 7), was used in the following studies.
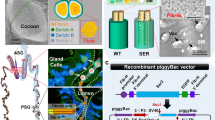
Generation and characterization of BmFib-H knock-out B. mori line.
(a) Schematic representation of the structure of BmFib-H gene depicting the non-repetitive regions (green boxes), repetitive regions (brown boxes) and ZFN target site (red eclair icon). (b) Part of the mutant sequences (deletion) generated by ZFN mediated genome editing. The wild-type sequence is shown at the top. The letter “D” represents deletions and the following numbers represent the size of deletion. The number within the “( )” represents the number of mutations recovered by sequence. The ZFN recognition sites are underlined. Deletions are indicated by dashed lines. (c) Ratios of different mutation types. (d) Microinjections of B. mori embryos and genome editing efficiency. (e–g) Cocoons (e), dissected silk gland (f) and pupae (g) of wild type Dazao and Fib-H-I1. Silk gland were dissected at the wandering stage and imaged under light macro-scope and confocal fluorescent microscope. ASG, MSG, PSG represent the anterior, middle and posterior sections of the silk gland, respectively. Red box represents one silk gland cell. Pupae were imaged and weighted at 4th day post pupation. The number of investigated pupae was 50 for each group. The scale bars represent 1 cm unless specifically labeled within this figure.
Compared with wild-type Dazao, the posterior silk glands (PSG) of Fib-H-I1 were smaller, while the anterior and middle silk glands (ASG and MSG) were normal in size (Fig. 1f and supplementary Fig. 4). We counted the number of cells in MSG and PSG in the 1st and 5th larvae stages and no differences were observed. However, the PSG cells of Fib-H-I1 were smaller and abnormal in shape (Fig. 1f). This phenotype was in accordance with the fact that silk gland is an endoreplicating tissue and its growth solely depends on cell size increase. It is unclear how the cell size increase in PSG is regulated. The results here may provide some important information to reveal this mystery, because we shorted the PSG just by removing its product. More importantly, despite of the size of PSG and PSG cell, the lumen of Fib-H-I1 PSG was hollow inside and MSG was almost empty with a very small amount of sericin proteins secreted by MSG cells (Fig. 1f and supplementary Fig. 4). To confirm this observation, the cocoons of Fib-H-I1 were firstly analyzed using scanning electron microscopy (SEM) and protein electrophoresis. Compared with wild-type Dazao, the cocoons of Fib-H-I1 were much lighter and thinner, while the pupae were little heavier (Fig. 1e, g), indicating a nutrient resorption from cocoon to pupae. SEM photography showed that the silk filaments within the cocoons were much thinner and exhibited a sericin-cocoon like phenotype Fig. 3c). SDS-PAGE and western blotting analysis showed that the cocoons contained only sericins and some small fibroin proteins (supplementary Fig. 7). Although the mechanism of many aforementioned phenotypes is not clear, we were confidential to conclude that we had successfully generated a BmFib-H knock-out B. mori line, which might be an ideal solution for our original hypothesis as its PSGs became empty, while the ability of protein synthesis was not affected.
Production of EGFP fusion proteins in Fib-H-I1.
(a, b) Schematic representation of the transgenic vector. The detailed information of the vector construction was as described as Zhao et al.16 (c) The microinjection results and the fluorescent images of 3xp3-DsRed transgenic marker expression in the nerves system of embryo and compound eyes of adult. (d) Fluorescent images of dissected silk glands of N4-R3, N4/Fib-H-I1-R3 and Fib-H-I1-R3 during the wandering stage. (e) SDS-PAGE of cocoon proteins of N4 (1), N4/Fib-H-I1 (2), Fib-H-I1 (3), N4-R3 (4), N4/Fib-H-I1-R3 (5), Fib-H-I1-R3 (6) and (f) Western blotting analysis of cocoon proteins of N4-R3, N4/Fib-H-I1-R3 and Fib-H-I1-R3 with EGFP polyclonal antibody. Proteins were run at a 6% gel. The scale bars represent 1 cm.
Production of artificial silk protein in BmFib-H knock-out B. mori line.
(a) Schematic representation of the structure of artificial silk protein and fluorescent imaging of different subdivision of transgenic silk gland. (b) Mechanical properties of cocoon sheet of Fib-H-I1 and Fib-H-I1-R3. For all histogram, the test number was 10. (c) SEM observation of WT, Fib-H-I1 and Fib-H-I1-R3 cocoons. (d) Diameters of silk fibers from cocoons of Fib-H-I1 and Fib-H-I1-R3. Diameters were measured from the SEM photograph and 50 samples were used for each test. (e, f) Cocoon weight and cocoon shell rate of Fib-H-I1 and Fib-H-I1-R3. The scale bars represent 1 cm unless specifically labeled within this figure.
To determine whether Fib-H-I1 can be used as a host for recombinant protein expression, we first attempted to express a recombinant enhanced green fluorescent protein (EGFP), which was widely used as a marker for gene expression and had been successfully expressed in the silk gland of B. mori. We previously established a highly efficient expression system, in which the fibroin heavy chain non-repetitive terminal region-EGFP fusion protein content of transgenic B. mori cocoons was up to 15% (w/w)16. One transgenic B. mori line (Fib-H-I1-R3) using the same vector was generated from the mutant line, Fib-H-I1, by piggyBac mediated transformation. Fluorescence could be detected in the PSG cells and lumen of the whole silk gland (Fig. 2d). The results suggested that although PSG cells of Fib-H-I1 were smaller in size and phenotypically abnormal, the ability to express and secret recombinant proteins was as normal as wild type B. mori strains. To further test whether removal of endogenous Bmfib-H gene would increase the expression level of exogenous recombinant proteins, Fib-H-I1-R3 was crossed to wild type N4B. mori strain. Two additional transgenic lines, N4-R3 and N4/Fib-H-I1-R3, which contained the same transgene with Fib-H-I1-R3 but were wild type and heterozygous at the BmFib-H locus, respectively, were generated. To avoid the effect such as copy numbers, the transgene were always maintained as a single copy and heterozygous. The Gel electrophoresis and Western blotting analysis of the cocoons showed a significantly increase in the expression level of recombinant fusion protein (Fig. 2e, f). The estimated content was about 52% and 50% of total N4/Fib-H-I1-R3 and Fib-H-I1-R3 cocoons, respectively. The heterozygotes produced the largest amount of EGFP protein, because the production of normal fibroin heavy chain protein from one wild type allele provide the silkworm a better developed PSG and the presence of normal fibroin heavy chain proteins helped the recombinant EGFP proteins spin out into the cocoons. However, the homozygous are preferred in future applications as the absence of fibroin heavy chain proteins will facilitate the purification process. Anyhow, this high productivity is superior to any of the known transgenic bioreactors (the highest was 15% in the case of EGFP fusion protein production that we reported previously) and cultured cell factories (the highest was 27 g/L in the case of IgG production using an IgG-producing PER.C6 clone29). Therefore, the genome edited silk gland generated here shows great promise as a highly efficient system for large scale production of EGFP fusion protein and perhaps other recombinant proteins.
The most attractive feature of silk gland is its ability to storage protein in an extremely high concentration without aggregation, which is recognized to hold great potential to produce protein-based materials. However, previous attempts using any existing B. mori strains only generate a mixture of a very small amount of target proteins (such as collagen and spider silk) and abundant endogenous silk proteins. Given its “empty” feature and “full” protein production ability, we suspected that Fib-H-I1 silk gland could be an ideal system for expression of protein based materials. For this demonstration, we designed an artificial silk protein (art-Fib-H) by assembling N terminal (contains the 1st repetitive region and 1st amorphous domain) and C terminal (contains the 11th amorphous domain and 12th repetitive region) regions of BmFib-H. For easy detection, an EGFP protein was fused between 1st and 11th amorphous domains (Fig. 3a). One transgenic B. mori line, Fib-H-I1-TR, in which art-Fib-H was expressed under the control of a BmFib-H promoter, was obtained. Fib-H-I1-TR was also crossed to wild type strain (Dazao) to generate WT-TR. Fluorescence detection showed that art-Fib-H could be expressed in PSG cells and secreted into silk gland lumen in both Fib-H-I1-TR and WT-TR. Interestingly, the EGFP fluorescence exhibit distinguished pattern in the lumen of different subdivision: homogeneous in PSG, punctuate distribution in PMSG and ASG and streaming in MMSG and AMSG (Fig. 3a). This might indicate a conformational change of art-Fib-H proteins when moving from PSG to ASG, during which the protein concentration, metal ions content, pH and shear stress were gradually changed to facilitate fiber formation. Compared with Fib-H-I1, the cocoons of Fib-H-I1-TR were thicker and 1.7 fold heavier and the cocoon shell rate was also higher (Fig. 3d, e, f). This result was consistent with the comparisons between Fib-H-I1 and WT (Fig. 1g) and suggested that art-Fib-H was successfully expressed in Fib-H-I1-TR and could form cocoons together with sericin proteins produced in MSG. SEM analysis of the cocoons revealed that silk fibers of Fib-H-I1-TR were thicker than Fib-H-I1and formed typical bifilar fibers in the inner layer of cocoon, which is the typical phenotype of fibroin containing fibers in the inner layer (Fig. 3c, d). The expression of art-Fib-H was also confirmed by gel analysis and western blotting using antibodies against N-terminal of Fib-H, full length Fib-L and P25 (Fig. 4). The mechanical properties of cocoons were also analyzed and the result showed that the cocoons of Fib-H-I1-TR were much stronger than Fib-H-I1 (Fig. 3b), indicating art-Fib-H proteins were correctly assembled into silk fibers. BmFib-H protein and spider dragline silk proteins, which are very similar to each other, were the most famous and typical proteins that used as biomaterials. The successful production of artificial silk designed from BmFib-H protein has showed the possibility to utilize Fib-H-I1 silk gland as a unique high efficient bioreactor for the production of protein-based materials and will surely pave the way for production of other biomaterials.
The detection of protein in the cocoons.
The proteins were analyzed by SDS-PAGE and western blotting. M represents protein markers. The numbers on the left represent the sizes of each band of the protein marker. The red arrows on the right represent the expected size of each protein. Anti Fib-H, Anti Fib-L and Anti P25 represent antibodies against the N terminal Fib-H, the full length Fib-L and P25 respectively. Proteins were run at a 4-20% gel.
Discussion
In summary, we generated several novel B. mori mutants by targeted editing of BmFib-H gene using customized ZFN. The genome editing efficiency was higher than any previously reported25,26,33. In one of the mutant lines, knock-out of BmFib-H gene resulted in a smaller and empty silk gland, abnormally developed posterior silk gland cells, an extremely thin cocoon that contain only sericin proteins and a slightly heavier pupae. Using an EGFP fusion protein as a reporter, we showed that the genome edited silk gland could be used as a highly efficient bioreactor for recombinant protein production, whose productivity is superior to any of the known expression systems. Furthermore, by designing and successful production of an artificial B. mori silk protein, we demonstrated that the bioreactor is suitable for large scale production of protein-based materials.
Genetically modified organisms have been used as bioreactors since 1980s, during which increasing the expression yield and developing strategies for large scale production has been long-lasting challenge. Given the highly protein synthesis activity30, the high protein content (more than 95%)4 and the simplicity of the silk protein components (only 11 known major proteins), the silk gland of B. mori has held promise as a low-cost, high-yield bioreactor. And this promise was proved by successful production of numerous recombinant proteins31. The present silk gland bioreactor generated by genome editing of BmFib-H gene further increase the content of recombinant proteins to about 50% of total cocoons, placing this bioreactor superior to any of the known bioreactors. It is reasonable to assume that other proteins such as collagen, human serum albumin and monoclonal antibodies could be expressed in this superior bioreactor with a similar yield. Furthermore, B. mori has been used for production of silk for thousand of years and are now large-scaled producing in China, Indian, Brazil and some other developing countries every year. It is very easy to achieve industrial scale production of biopharmaceutical proteins, which are in increasing demand for both analytical and clinical applications32. According to Tomita et al.'s calculation3, production of 5 kg of recombinant proteins using our bioreactor only need about 10 kg of cocoon material, which can be achieved in a facility with a floor surface of about 5 m2 and 1 worker. Besides the amazing recombinant production ability, we also showed that the superior bioreactor could be used to produce protein-based materials, which are also in great need for biomedical applications but lack of efficient production system. As silk gland is the only organ that can store proteins at an extremely high concentration without aggregation or denaturalization, we believe that the genome edited silk gland generated here could be a unique high efficient bioreactor for the production of protein-based materials.
The mechanism of silk proteins secretion and transport has long been an interesting but unresolved matter. The H-L subunit was thought to be important for the solubility of fibroin during intracellular transport, secretion and luminal transport through the silk gland33. Based on this knowledge, Fib-L and P25 proteins should be absent in the cocoons of the mutant line Fib-H-I1 and recovered in the line Fib-H-I1-TR as the small version of Fib-H (art-Fib-H) would form a disulfide bond and interact with P25. However, our results (Fig. 4) showed that both Fib-L and P25 proteins existed in the cocoons of Fib-H deficient mutant line, Fib-H-I1 and other lines including WT, WT/Fib-H-I1 and Fib-H-I1-TR. This result indicated that the H-L subunit is not necessary for the secretion of Fib-L. Given the phenotypic effect of Fib-L mutation observed in the Nd-sd mutant, we suspected that the H-L subunit is only necessary for the intracellular transport, secretion and luminal transport of large fibroin proteins such as Fib-H. The small proteins such as Fib-L, P25 and some other silk proteins are able to secrete and transport in an independent manner. However, to completely reveal the mechanism, more investigations such as knock-out or down/up regulating Fib-L and P25 are required.
The fast-changing development of genome editing technologies in recent years and their rapid applications in increasing amount of organisms have greatly reshaped the field of genetic manipulation, especially in those non-model organisms such as B. mori that targeted mutagenesis was not achievable. ZFNs, TALENs CRISPR/Cas9 system emerged subsequently and served as the troika for genome editing tools. Recently, we showed that TALEN and CRISPR/Cas9 could introduce highly efficient targeted mutagenesis, large chromosomal deletion and even sophisticated genomic structure variations in B. mori26,34,35,36. Together with the high efficiency of ZFN for B. mori genome editing proved here, it is feasible for researchers to implement precise and sophisticated manipulation of any chosen B. mori gene. Besides the largest and most abundant fib-H protein, silk gland also secretes another 2 fibroin proteins (Fib-L and P25) and 8 main sericin proteins (Ser1 variants, Ser2 and Ser3). The next step of silk gland genetic engineering would be removing these 10 silk proteins using genome editing tools, which in principle will generate a more superior bioreactor that produce industrial scale of pure (purification-free) recombinant biopharmaceutical proteins or protein-based materials. Given that ZFNs are much more difficult to be customized to new targets, TALENs and CRISPR/Cas9 system are preferred to be used to fulfill the mission.
Methods
ZFN
ZFNs designed to cleave the N-terminal non-repetitive region of the BmFib-H gene were purchased from Sigma-Aldrich. These ZFNs had a five/six-zinc finger protein recognizing 15/18 bases (supplementary Fig. 2). The activities of two pairs of ZFNs for BmFib-H locus were measured by the yeast MEL-1 assay as described37.
mRNA synthesis
Plasmid DNA was prepared using a plasmid midi kit (QiaGen), digested with XbaI (Promega) and phenol/chloroform purified. mRNAs were in vitro transcribed and polyadenylated using the MessageMax™ T7 ARCA-Capped Message Transcription Kit and Poly(A) Polymerase Tailing Kit (Epicentre Biotechnologies). The resulting mRNA was purified using the MegaClear Kit (Ambion) before resuspension in RNase-free water (Sigma-Aldrich), quantitated using a NanoDrop-1000 (Thermo Scientific).
Designing and construction of art-Fib-H
The sequence of N terminal (contains the 1st repetitive region and 1st amorphous domain) and C terminal (contains the 11th amorphous domain and 12th repetitive region) regions of BmFib-H and EGFP (supplementary sequence) was synthesized and inserted into pUC57-T-simple using Genscript service, forming pUC57-T-art-Fib-H. The promoter sequence of BmFib-H was amplified from the genomic DNA of Dazao strains and inserted into pUC57-T-art-Fib-H using Hindlll and Spel digestion. The resulted plasmid was digested with Ascl and the fragment containing the BmFib-H promoter and art-Fib-H was inserted into a piggyBac transposon derived transformation vector pBac[3xp3-DsRed, af]4.
Microinjection of B. mori embryos
Two B. mori strains, which were reared on fresh mulberry leaves, were used in our study. Nistari (N4) is a non-diapausing strain and the preparation of embryo for microinjection is as Tamura et al. described38. Dazao is a diapausing strain and the embryos were prepared as Zhao et al. described39. mRNA samples or piggyBac transgenic vectors were microinjected into silkworm embryos within 2 h after oviposition. Microinjections were performed using TransferMan NK2 micromanipulator and Femto Jet 5247 microinjector (Eppendorf) under a microscopy SZX16 (Olympus). The injection opening was sealed with instant glue (Konishi co.) and injected embryos were incubated at 25°C and 90%RH. Larvae hatched from the injected embryos were collected and reared on fresh mulberry leaves. The resulted moths were sibling crossed to screen the mutant.
Sequencing of the mutations
The genomic DNA was extracted using nucleic acid isolation systems PI-1200 (Kurabo) and PCR amplified segments were inserted to a T-vector (TransgGen) and sequenced using commercial service (Beijing Genomics Institute). All the methods used for PCR and T-cloning were followed the standard molecular cloning protocols or instructions provided by the manufacturers.
Analysis of the cocoon proteins
25 mg cocoon shell from 10 randomly chosen cocoons was dissolved in 1 ml of 60% lithium thiocyanate (LiSCN). Protein concentration was measured by the Bradford protein assay and then subjected to SDS–PAGE (6% or 4–20%, Bio-Rad). The sample preparation and treatment protocol was as described by Zhao et al.16 The gel was visualized by Coomassie brilliant blue staining or silver staining. Western blotting was performed using polyclonal antibodies against the N terminal sequences of Fib-H (provided by Prof. Congzhao Zhou from University of Science and Technology of China), the fibroin light chain and fhx/P25 (provided by Prof. Chun Liu from our Lab).
Staining and imaging of silk gland
Silk glands of wandering larvae were dissected out in PBS. For whole silk gland imaging, the silk glands were placed on a glass slide and imaged under macro-scope (MVX10, Olympus). To avoid the autofluorescence of silk glands, all B. mori larvae used for fluorescent imaging were reared on artificial diet. For imaging of silk gland cells or subdivisions, silk glands were fixed in PBT (PBS with 1% Triton X-100) containing 4% paraformaldehyde, for 60 min. The nuclei were counterstained with DAPI and then visualized under fluorescence microscope (IX51, Olympus) or confocal microscope (FV1000, Olympus).
Microscopy and mechanical testing
The morphology of cocoons was observed using an SEM (JSM-5000, NeoScope). A piece of cocoon samples from each group were placed onto an SEM stub and coated with a thin layer of platinum (NeoCoater MP-19020NCTR). SEM micrographs were taken at room temperature. The tensile tests of cocoon shells (3 cm × 0.5 cm) were performed using the universal testing system (AGX-10, Shimadzu) at a speed of 2 mm/min in the air at room temperature and 60% relative humidity.
References
Wang, Y., Zhao, S., Bai, L., Fan, J. & Liu, E. Expression Systems and Species Used for Transgenic Animal Bioreactors. Biomed. Res. Int. 2013, 580463 (2013).
Askarieh, G. et al. Self-assembly of spider silk proteins is controlled by a pH-sensitive relay. Nature. 465, 236–238 (2010).
Xia, Q., Li, S. & Feng, Q. Advances in Silkworm Studies Accelerated by the Genome Sequencing of Bombyx mori. Annu. Rev. Entomol. 59, 513–536 (2014).
Tomita, M. et al. Transgenic silkworms produce recombinant human type III procollagen in cocoons. Nat. Biotechnol. 21, 52–56 (2003).
Adachi, T., Tomita, M., Shimizu, K., Ogawa, S. & Yoshizato, K. Generation of hybrid transgenic silkworms that express Bombyx mori prolyl-hydroxylase alpha-subunits and human collagens in posterior silk glands: Production of cocoons that contained collagens with hydroxylated proline residues. J. Biotechnol. 126, 205–219 (2006).
Adachi, T. et al. Production of a non-triple helical collagen alpha chain in transgenic silkworms and its evaluation as a gelatin substitute for cell culture. Biotechnol. Bioeng. 106, 860–70 (2010).
Royer, C. et al. Biosynthesis and cocoon-export of a recombinant globular protein in transgenic silkworms. Transgenic Res. 14, 463–472 (2005).
Hino, R., Tomita, M. & Yoshizato, K. The generation of germline transgenic silkworms for the production of biologically active recombinant fusion proteins of fibroin and human basic fibroblast growth factor. Biomaterials. 27, 5715–5724 (2006).
Ogawa, S., Tomita, M., Shimizu, K. & Yoshizato, K. Generation of a transgenic silkworm that secretes recombinant proteins in the sericin layer of cocoon: production of recombinant human serum albumin. J. Biotechnol. 128, 531–44 (2007).
Kurihara, H., Sezutsu, H., Tamura, T. & Yamada, K. Production of an active feline interferon in the cocoon of transgenic silkworms using the fibroin H-chain expression system. Biochem. Biophys. Res. Commun. 355, 976–980 (2007).
Tateno, M. et al. Production and characterization of the recombinant human mu-opioid receptor from transgenic silkworms. J. Biochem. 145, 37–42 (2009).
Iizuka, M. et al. Production of a recombinant mouse monoclonal antibody in transgenic silkworm cocoons. FEBS J. 276, 5806–5820 (2009).
Teulé, F. et al. Silkworms transformed with chimeric silkworm/spider silk genes spin composite silk fibers with improved mechanical properties. Proc. Natl. Acad. Sci. USA. 109, 923–928 (2012).
Zhu, Z. et al. Mechanical properties of regenerated Bombyx mori silk fibers and recombinant silk fibers produced by transgenic silkworms. J. Biomater. Sci. Polym. Ed. 21, 395–411 (2010).
Wen, H. et al. Transgenic silkworms (Bombyx mori) produce recombinant spider dragline silk in cocoons. Mol. Biol. Rep. 37, 1815–1821 (2010).
Zhao, A. et al. New and highly efficient expression systems for expressing selectively foreign protein in the silk glands of transgenic silkworm. Transgenic Res. 19, 29–44 (2010).
Tomita, M. et al. A germline transgenic silkworm that secretes recombinant proteins in the sericin layer of cocoon. Transgenic Res. 16, 449–465 (2007).
Wang, F. et al. An optimized sericin-1 expression system for mass-producing recombinant proteins in the middle silk glands of transgenic silkworms. Transgenic Res. 22, 925–938 (2013).
Iizuka, M., Tomita, M., Shimizu, K., Kikuchi, Y. & Yoshizato, K. Translational enhancement of recombinant protein synthesis in transgenic silkworms by a 50-untranslated region of polyhedrin gene of bombyx mori nucleopolyhedrovirus. J. Biosci. Bioeng. 105, 595–603 (2008).
Inoue, S. et al. A fibroin secretion-deficient silkworm mutant, Nd-sD, provides an efficient system for producing recombinant proteins. Insect Biochem. Mol. Biol. 35, 51–59 (2005).
Beumer, K. J. et al. Efficient gene targeting in Drosophila by direct embryo injection with zinc-finger nucleases. Proc. Natl. Acad. Sci. USA. 105, 19821–19826 (2008).
Carroll, D. Genome engineering with zinc-finger nucleases. Genetics. 188, 773–782 (2008).
Zhang, F. et al. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat. Biotechnol. 29, 149–153 (2011).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 337, 816–821 (2012).
Takasu, Y. et al. Targeted mutagenesis in the silkworm Bombyx mori using zinc finger nuclease mRNA injection. Insect Biochem. Mol. Biol. 40, 759–765 (2010).
Ma, S. et al. Highly efficient and specific genome editing in silkworm using custom TALENs. PLoS One. 7, e45035 (2012).
Zhou, C. Z. et al. Fine organization of Bombyx mori fibroin heavy chain gene. Nucleic Acids Res. 28, 2413–2419 (2000).
Xia, Q. et al. A draft sequence for the genome of the domesticated silkworm (Bombyx mori). Science. 306, 1937–1940 (2004).
Jones, D. et al. High-level expression of recombinant IgG in the human cell line per.c6. Biotechnol. Prog. 19, 163–168 (2003).
Shimura, K. Biochemical aspects on fibroin. Tanpakushitsu Kakusan Koso 24, 1324–1335 (1979).
Tomita, M. Transgenic silkworms that weave recombinant proteins into silk cocoons. Biotechnol. Lett. 33, 645–654 (2011).
Assenberg, R., Wan, P. T., Geisse, S. & Mayr, L. M. Advances in recombinant protein expression for use in pharmaceutical research. Curr. Opin. Struct. Biol. 23, 393–402 (2013).
Inoue, S. et al. Silk fibroin of Bombyx mori is secreted, assembling a high molecular mass elementary unit consisting of H-chain, L-chain and P25, with a 6:6:1 molar. J. Biol. Chem. 275, 40517–40528 (2000).
Ma, S. et al. CRISPR/Cas9 mediated multiplex genome editing and heritable mutagenesis of BmKu70 in Bombyx mori. Sci. Rep. 4, 4489 (2014).
Liu, Y. et al. Highly efficient multiplex targeted mutagenesis and genomic structure variation in Bombyx mori cells using CRISPR/Cas9. Insect. Biochem. Mol. Biol. 49C, 35–42 (2014).
Ma, S. et al. Multiplex genomic structure variation mediated by TALEN and ssODN. BMC Genomics. 15, 41 (2014).
Doyon, Y. et al. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat. Biotechnol. 26, 702–708 (2008).
Tamura, T. et al. Germline transformation of the silkworm Bombyx mori L. using a piggyBac transposon-derived vector. Nat Biotechnol. 18, 81–84 (2000).
Zhao, A. et al. Efficient strategies for changing the diapause character of silkworm eggs and for the germline transformation of diapause silkworm strains. Insect Sci. 19, 172–182 (2012).
Acknowledgements
This work was supported by grants from the National Basic Research Program of China (973 program, 2012CB114600), the National High-tech R&D Program (863 program, 2011AA100306) and the doctoral Innovation Fund of Southwest University (kb2010002).
Author information
Authors and Affiliations
Contributions
S.Y.M. and Q.Y.X. conceived and designed the study. S.Y.M., R.S., X.G.W., Y.Y.L., J.S.C., J.G., W.L., J.D.Z. and P.Z. performed and analyzed all the experiments. S.Y.M. R.S. and Q.Y.X. wrote the manuscript with support from all authors.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Ma, S., Shi, R., Wang, X. et al. Genome editing of BmFib-H gene provides an empty Bombyx mori silk gland for a highly efficient bioreactor. Sci Rep 4, 6867 (2014). https://doi.org/10.1038/srep06867
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep06867
This article is cited by
-
Transdermal peptide conjugated to human connective tissue growth factor with enhanced cell proliferation and hyaluronic acid synthesis activities produced by a silkworm silk gland bioreactor
Applied Microbiology and Biotechnology (2020)
-
New insight into the mechanism underlying the silk gland biological process by knocking out fibroin heavy chain in the silkworm
BMC Genomics (2018)
-
QTL analysis of cocoon shell weight identifies BmRPL18 associated with silk protein synthesis in silkworm by pooling sequencing
Scientific Reports (2017)
-
Increasing the yield of middle silk gland expression system through transgenic knock-down of endogenous sericin-1
Molecular Genetics and Genomics (2017)
-
Highly efficient and inducible DNA excision in transgenic silkworms using the FLP/FRT site-specific recombination system
Transgenic Research (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.