Abstract
Nasopharyngeal carcinoma (NPC) is a common form of malignant cancer, for which radiotherapy or chemotherapy are the main treatment methods. Cucurbitacin E (CuE) is a natural compound-based drug which from the climbing stem of Cucumic melo L (Guadi). Previously shown to be an antifeedant as well as a potent chemopreventive agent against several types of cancer. The present study, investigated anti-proliferation and cell cycle G2/M arrest induced by CuE in Detroit 562 cells (pharynx carcinoma) and HONE-1 (nasopharyngeal carcinoma) cells. Results indicate that the cytotoxicity is associated with accumulation in G2/M cell-cycle phases. CuE produced cell cycle arrest as well as the downregulation of cyclin B1 and CDC2 expression. In addition, treated cells with CuE and GADD45γ SiRNA that also coincided with GADD45γ gene activation in cell cycle arrest. Both effects increased proportionally with the dose of CuE; however, proliferation inhibition and mitosis delay was dependant on the amount of CuE treatment in the cancer cells.
Similar content being viewed by others
Introduction
Nasopharyngeal carcinoma (NPC) is a malignant tumor common in Southeast Asia and Taiwan. The age of NPC onset tends to be younger than that of other tumors, affecting most patients at approximately 30–50 years of age1. Infections with Epstein-Barr virus, genetic predisposition, as well as various dietary and environmental factors are believed to play important roles in the development of carcinogenesis2. Radiotherapy is the mainstay of treatment, for which the five-year survival rate is approximately 25%3.
Cucurbitacins are a group of tetracyclic triterpenes with medicinal properties derived from the climbing stem of Cucumic melo L4. They have been used extensively in traditional medicines throughout Asia and many cancer-preventive properties have previously been reported5. Interest in this herb has grown in recent years, due to its putative beneficial pharmacological effects as an anti-inflammatory6 and anticancer agent7,8. There have also been indications that cucurbitacins may help in the prevention and treatment of oxidative damage as well as the suppression of specific inflammatory factors9.
Cucurbitacin E (CuE, α-elaterin) is an active compound10, previously shown to be a strong antifeedant with the ability to disrupt cell actin11 and cell adhesion. Recent reports have demonstrated that CuE has an inhibitory effect on cancer cell proliferation, actin polymerization and permeability12. However, whether CuE inhibits NPC growth remains unknown. Furthermore, the mechanism underlying the anti-cancer effect of CuE has yet to be identified.
This present study was initiated to investigate whether CuE contributes to the anti-proliferation and apoptosis of NPC cells (HONE-1 and Detroit 562). It is expected that these experiments will provide scientific basis and technological support for further development of NPC therapy.
Results
CuE can mediate the survival of pharynx and nasopharyngeal cancer cells and thus inhibits their proliferation
To explore this anti-tumor activity, an in vitro study was conducted in which Detroit 562 and HONE-1 cell lines were subjected to increasing dosages of CuE (0, 6.25, 1.25 and 2.5 μM) for 24 to 72 hours. The proliferation of CuE-treated cancer cells was then measured using the MTT method (Figure 1). Our results indicate that the survival and proliferation of Detroit 562 (Figure 1A) and HONE-1 cells (Figure 1B) was decreased by CuE treatment in a dose- and time-dependent manner.
CuE mediates the survival of pharynx (Detroit 562) and nasopharyngeal (HONE-1) cancer cells, thereby inhibiting proliferation.
An in vitro study was initiated by treating each of the Detroit 562 and HONE-1 cell lines with increasing doses of CuE (0, 0.625, 1.25 and 2.5 μM) for 24 to 72 hours. The survival of CuE-treated cancer cells was then measured using the MTT method. Results were expressed as a percentage of control, which was considered 100%. All data were reported as the mean (±SEM) of at least three separate experiments. Statistical analysis was performed using a t-test, with significant differences determined at the level of *P < 0.05 versus the control group.
Non-CuE-induced apoptosis/necrosis of Detroit 562 and HONE-1 cells
To identify the role played by CuE in the apoptosis/necrosis of Detroit 562 and HONE-1 cells, we employed Annexin V-FITC and propidium iodide staining to reveal the formation of apoptotic cells following 4 hours of exposure to CuE. The percentage of apoptotic cells was assessed by flow cytometric analysis (Supplemental Figure S1A and Figure S1B). A dot-plot of Annexin V-FITC fluorescence versus PI fluorescence indicates a non-significant increase in the percentage of apoptotic cells treated with CuE, compared with untreated cells. No significant increase was observed in the percentage of cells undergoing necrosis, apoptosis (Supplemental Figure S1C) or caspase 3 activation at CuE concentrations of 0.625 to 2.5 μM (Supplemental Figure S2A, Figure S2B and Figure S2C). However, the results summarized in Supplemental Figure S1 and Figure S2 indicate that CuE may mediate the survival of Detroit 562 and HONE-1 cells. Thus, we hypothesize that the proliferation of these cells was inhibited by pathways other than apoptosis/necrosis.
CuE-induced accumulation of G2/M phase in CuE-treated cells
The cell-cycle distribution of CuE-treated cells was analyzed by flow cytometry. Cells were exposed to CuE for 24 hours prior to processing and analysis. As shown in Figure 2(A), exposure to CuE resulted in an increase in the number of G2/M phase, cells, which may imply that the Detroit 562 and HONE-1 cells underwent cell cycle arrest. Our results indicate that treatment with CuE increased the cell populations in G2/M phase, while simultaneously reducing the number of cells in the S and G1 phases (* p < 0.05 vs CuE 0 μM) (Figure 2B).
Influence of CuE on cell cycle progression/distribution in Detroit 562 and Hone-1 cells: (A) Cell cycle analysis of Detroit 562 and Hone-1 cells after being cultured with CuE for 24 h. (B) CuE induced an increase in G2/M phase cells (%). (C) MPM-2 (anti-phospho-Ser/Thr-Pro) expression in untreated and treated cancer cells. MPM-2 is an antibody that recognizes proteins which are only phosphorylated in mitosis. Cells were dually stained using propidium iodide to analyze DNA content and protein expression was quantified by flow cytometry. As a positive control, separate groups of cells were treated for 24 h with nocodazole (15 μg/mL), an anti-fungal agent known to induce metaphase arrest. Cell-cycle analysis and quantification of MPM-2 expression (gated cells) were performed by flow cytometry following treatment with CuE for 24 h. (D) CuE enhanced the level of MPM-2 in Detroit 562 and Hone-1 cells. Symbol (*) in each group of bars indicates that the difference resulting from treatment with CuE 0 μM is statistically significant at P < 0.05.
Effects of CuE on the mitotic index
To distinguish G2 arrest from mitotic arrest, we employed an additional marker, MPM-2 (anti-phospho-Ser/Thr-Pro). This antibody is capable of recognizing proteins whose epitopes are exclusively phosphorylated during mitosis, specifically from early prophase to metaphase13. MPM-2 is commonly used as an indicator of mitotic disturbance. To provide a positive control, we treated separate groups of Detroit 562 and HONE-1 cells with nocodazole (15 μg/mL), an inducer of metaphase arrest14. Treating the two types of cells with nocodazole for 24 hours resulted in synchronization of entire cell populations in the G2/M phase as well as an increase in MPM-2 labeling (Figure 2C and 2D). Among all cells treated with CuE, the MPM-2 level was elevated compared with control group (19% and 31% for Detroit 562 and Hone-1 cells treated with CuE, respectively) (Figure 2D). However, MPM-2 staining was not as strong as that achieved with nocodazole. This is likely because MPM-2 stained cells were in various stages of mitosis, some of which could not be identified using this early prophase marker. Specifically, the accumulated G2/M phase may not have been marked. Thus, although the elevated staining of MPM-2 suggests mitotic disturbance, it may underestimate it.
G2/M phase arrest in CuE-treated cells via CDC2 and cyclin B1 down regulation
Figure 3 illustrates mRNA gene expression and immunoblotting results of cellular proteins from Detroit 562 and Hone-1 cells treated with CuE. Gene expression and Western blot analysis revealed a decrease in cyclin B1 and CDC2 following incubation with CuE (Figure 3A and 3B). Western blotting (Fig. 3C) also showed an increase in GADD45γ protein expression (Supplemental Fig. S4A). Co-IP revealed that the GADD45γ/CDC2 complex was formed but the GADD45γ/Cyclin B1 complex was not (Supplemental Figures S4A and B). Cyclin B1 protein expression was quantified by measuring relative band intensities (Fig. 3D) This analysis further confirmed that cyclin B1, CDC2 and p-CDC2 (Fig. 3E) levels were significantly lower in cells incubated with CuE.
CuE represses cyclinB1 and CDC2 gene expression in Detroit 562 and HONE-1 cells.
The cells were treated with CuE (0, 0.625, 1.25 and 2.5 μM) for 24 h and cyclin B1 and CDC2 gene expression were subsequently detected by RT-PCR, Real time PCR and Western blot analysis. (A) The RT-PCR results of cyclin B1 and CDC2mRNAs in NPC cell lines following exposure to CuE. The panels (B) indicate quantitative RT-PCR (qPCR) analysis of mRNA expression in cyclin B1 and CDC2. (C and E) Representative blot from 3 independent experiments. (D) Quantification of band intensities. All data was reported as the mean (±SEM) of at least three separate experiments. Statistical analysis was performed using a t-test, with significant differences determined at the level of *P < 0.05 versus 0 μM control group.
Gene expression profile of cells following exposure to CuE
We then used SurePrint G3 Human Gene Expression Microarrays to study the genome-wide gene expression profiles of Detroit 562 cells that had been exposed to either the vehicle (DMSO) or CuE (1.25 μM) for four hours. The experiments were performed independently three times to enable comparative analysis between the Detroit 562 cells.
Principal component analysis (PCA) revealed that the microarray data derived from CuE-treated cells and DMSO-treated cells constituted two spatially-separated planes. This suggested that treatment with CuE had a far greater impact on the gene expression profile than could be reasonably attributed to technical errors. We therefore divided the expression levels in the CuE-treated cells by those in the vehicle-treated cells and considered changes greater than 2-fold to be substantial upregulation and changes smaller than 0.5-fold to be downregulation. To identify molecular networks of biological significance for these genes, we used distinct pathway analysis tools of bioinformatics, as well as the comprehensive knowledgebase of the Kyoto Encyclopedia of Genes and Genomes (KEGG) (www.kegg.jp). This enabled us to identify the KEGG pathway (Supplemental Table S1) as well as a battery of down-regulated (Supplemental Table S2) and up-regulated genes (Supplemental Table S3). Specifically, we determined that CuE treatment reduced the expression of cyclin B1 and CDC2 genes and increased the levels of GADD45 -α, -β and -γ genes (Supplemental Fig. S3).
Down-regulation of the CAGG45γ/CDC2 complex in NPC cell lines by silencing GADD45γ was reversed by CuE induced G2/M arrest
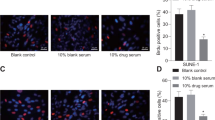
To confirm the relationship between GADD45γ expression and cell cycle arrest in NPC cell lines, we modulated cellular GADD45γ levels using Lipofectamine RNAiMAX gene delivery techniques. We then investigated the influence of cellular GADD45γ levels on anti-tumorigenic behavior in NPC cell lines. Co-IP analysis revealed a significant decrease in the levels of the GADD45γ/CDC2 complex after GADD45γ was silenced (Figure 4A). Findings from qPCR analysis also revealed higher GADD45γ mRNA expression levels in NPC cell lines treated with CuE (Fig. 4A). In addition, silencing GADD45γ significantly repressed CuE-induced cell cycle G2/M arrest in Hone-1 cells but not in Detroit 562 cells (Figures 4C and D). This data suggests that GADD45γ level regulated the tumorigenicity of NPC cells via CuE.
GADD45 family gene expression in NPC cell lines following exposure to CuE: (A) Quantitative RT-PCR (qPCR) analysis of GADD45-α, -β and -γ mRNA expression standardized against the levels of GAPDH in NPC cell lines exposed for 4 hours to DMSO (CuE 0 μM control) or CuE. (B) Co-IP of GADD45γ and CDC2 in NPC cell lines which treated with CuE 1.25 μM and/or GADD45γ SiRNA. (C) Reenter cell cycle which arrest by CuE in NPC cell lines via inhibition of GADD45γ gene expression. (D) Delay in mitosis in CuE-treated and/or GADD45γ SiRNA NPC cell lines.
These findings indicate that common molecular pathways are involved in inducing cell cycle G2/M arrest16. Findings from qPCR analysis (Fig. 4A) were further validated by a microarray analysis, which indicated substantial cyclin B1and CDC2downregulation (Figure 3) as well as notable upregulation of GADD45γ and the GADD45γ /CDC2 complexin NPC cell lines following exposure to CuE (Fig. 4). These results indicate that CuE may delay cancer cell growth in the G2/M phase via down-regulation of CDC2 gene expression, dissociation of the cyclin B1/CDC2 complex and up-regulation of GADD45 family proteins15,16.
Discussion
CuE has been used in traditional medicine throughout Asia for the treatment of tumors and many studies have attempted to elucidate the mechanism underlying the anticancer activity of this compound17,18.
In this study, CuE demonstrated anticancer activity as well as the ability to induce cell cycle G2/M arrest. We found that treating Detroit 562 and Hone-1 cell lines with CuE resulted in the down-regulation of CDC2 protein expression and phosphorylation. In addition, CDC2 proteins were shown to bind with GADD45γ, which mediated both the G1 and G2/M phases of the cell cycle16,17,19. Previous studies have also provided evidence to suggest that CDC2 binds with GADD45γ19 and Gadd45 gene family are important for anti-tumor immune responses20. Decreased mRNA and protein expression of GADD45α and GADD45γ but not GADD45β were found in 138 GCA tumor tissues21. The inhibition of JunD in prostate cancer cells results in GADD45α- and γ-dependent induction of cell death and inhibition of tumor growth22 and a nexus between DNA repair and epigenetic gene regulation23.
For example, Cdc2 tyrosine phosphorylation is maintained during HU-induced cell cycle arrest24 and phosphorylation of CDC2 on Thr14 and Thr 15 inhibits CDC2 activity25. CuE-induced G2/M arrest does not necessarily occur through tyrosine phosphorylation (Figure 3E), but rather through the down-regulation of CDC2 gene expression (Figure 3 A–D), the up-regulation of GADD45γ gene expression and the formation of the GADD45γ/CDC2 complex (Supplemental Figure S4) in NPC cell lines.
The results collected in this series of studies provide experimental evidence supporting the contention that CuE may irreversibly arrest the growth of Detroit 562 (human pharynx carcinoma) and HONE-1 (human nasopharyngeal carcinoma) cancer cells. The results of mechanistic analysis led to the conclusion that both the inhibition of proliferation and the induction of cell cycle arrest are highly dependent upon CuE accumulated in the cancer cells. However, a different dependency was observed between the Detroit 562 and HONE-1 cells.
In conclusion, this study demonstrates for the first time that CuE is an effective inhibitor of pharynx and nasopharyngeal carcinoma tumors. The role of CuE in the inhibition of tumor growth was highlighted by the delay of mitosis through the down-regulation of cyclin B1. These findings suggest the applicability of CuE as an anti tumor agent.
Methods
Materials
Cucurbitacin E, DMSO (dimethyl sulfoxide) and MTT [3-(4,5-dimethylthiazol-2-yl)- 2,5-diphenyltetrazolium bromide] were obtained from Sigma (St Louis, MO). Cell culture medium (minimum essential medium, RPMI 1640), fetal bovine serum, antibiotics, sodium pyruvate, trypsin and phosphate-buffered saline (PBS) were purchased from Gibco, BRL (Grand Island, NY). Polyvinylidene fluoride membrane (PVDF) (Millipore) and molecular weight marker were purchased from Bio Rad (USA). All other reagents and compounds were analytical grades.
Cells
The Detroit 562 and HONE-1 cells were from ATCC. Detroit 562 cells (Human pharynx carcinoma) were maintained on culture dishes, in 90% (v/v) minimum essential medium Eagle with 2 mM L-glutamine and Earle's BSS adjusted to contain 1.5 g/L sodium bicarbonate, 0.1 mM non-essential amino acid and 1 mM sodium pyruvate with 10% (v/v) fetal bovine serum (FBS). HONE-1 (human nasopharyngeal carcinoma) cells were cultured in RPMI 1640 supplemented with 10% (v/v) FBS. The cells were cultured in an atmosphere containing 5% CO2 at 37°C incubator.
Cell proliferation assay
The cells were seeded into 96-wells culture plate at 5000 cells/well. The cells were treated with 0, 0.625, 1.25 and 2.5 μM CuE, the CuE will complex with medium. Then the cell will incubate in 37 degree C for 24, 48 and 72 hours in the CO2 incubator. After incubate 24, 48 and 72 hours. The cell treatments MTT dye (1 mg/ml) at least 4 hours on each well. The reaction was stopped by the addition of DMSO and optical density was measured at 540 nm on a multi-well plate reader. Background absorbance of the medium in the absence of cells was subtracted. All samples were assayed in triplicate and the mean for each experiment was calculated. Results were expressed as a percentage of control, which was considered as 100%. Each assay was carried out in triplicate and the results were expressed as the mean (+/-SEM).
Measurement of Apoptosis
NPC cells were first seeded in 6-well plates (Orange Scientific, E.U.). Following treatment with CuE for four hours, the cells were harvested. The cells were re-centrifuged (the supernatant discarded) and resuspended/incubated in 1× annexin-binding buffer. Five μL of annexin V-FITC (BD Pharmingen, BD, USA) and 1 μL of 100 μg/mL PI working solution for 15 minutes. Following the incubation period, the stained cells were analyzed using flow cytometry (FACSCalibur, BD, USA). Data was analyzed using WinMDI 2.8 free software (BD, USA).
Caspase 3 activity assay
The caspase activity was assessed by the FITC rabbit anti-active caspase-3 (BD Pharmingen, USA). The cells were treated with CuE of 0, 0.625, 1.25 and 2.5 μM for 24 hours. The caspase activity was detected and inspected by the flow cytometry (FACSCalibur, BD, USA). Data was analyzed using WinMDI 2.8 free software (BD, USA).
Cell cycle analysis
For cell cycle analysis we used the fluorescent nucleic acid dye propidium iodide (PI) to identify the proportion of cells in each of the three interphase stages of the cell cycle. Cells were treated with CuE for 24 hours and then harvested and fixed in 1 ml cold 70% ethanol for at least eight hours at -20°C. DNA was stained in PI/RNaseA solution and the DNA content was detected using flow cytometry. Data was analyzed using WinMDI 2.8 free software.
Mitotic index analysis
The mitotic index was assessed by MPM-2 (anti-phospho-Ser/Thr-Pro) expression. After 24 h of treatment with CuE, cells were harvested and fixed in 70% ethanol overnight. Cells were then washed and suspended in 100 μl of IFA-Tx buffer (4% FCS, 150 nM NaCl, 10 nM HEPES, 0.1% sodium azide, 0.1% Triton X-100) with a primary MPM-2 antibody (1 μg/mL; Upstate Cell Signaling Solutions, Millipore, Watford, UK) at room temperature for 1 h. Cells were washed and resuspended in IFA-Tx buffer with a rabbit anti-mouse FITC-conjugated secondary antibody (1:50; Serotec, Oxford, UK) for 1 h at room temperature in darkness. Finally, cells were washed and resuspended in 500 μl of PBS with 20 μg/mL of PI (Sigma) for 30 min in the dark. MPM-2 expression was analyzed using flow cytometry (FACSCalibur, BD, USA). Data was analyzed using WinMDI 2.8 free software (BD, USA).
Western blot assay
A total of 50 μg of proteins were separated by 10% SDS-PAGE and transferred to PVDF membranes (Millipore, USA). The membranes were blocked with blocking buffer (Odyddey, USA) overnight and incubated with anti-β-actin (Sigma-Aldrich, St. Louis, MO, USA), anti-cyclin B1, anti-CDC2 (p34; sc-747), anti-p-CDC2 (p34 Tyr 15; sc-7989) (Santa Cruz BioTechnology, USA) and anti- GADD45γ (TA505437 OriGene Technologies, Rockville, MD, USA) antibodies for 1.5 ~ 2 hours. The blots were washed and incubated with a second antibody (IRDye Li-COR, USA) or conjugated with horseradish peroxidase (HRP) at a 1/20,000 dilution for 30 minutes. The antigen was then visualized using a near infrared imaging system (Odyssey LI-COR, USA) or chemiluminescence detection kit (ECL; Amersham Corp., Arlington Heights, IL, USA). The data was analyzed using Odyssey 2.1 software.
Co-immunoprecipitation (Co-IP)
Co-IP is an effective means of quantifying protein–protein interactions in cells. Briefly, 500 mg of cellular proteins were labeled using anti-CDC2 (Santa Cruz BioTechnology) and GADD45γ (TA505437 OriGene Technologies, Rockville, MD, USA) following overnight incubation at room temperature. The protein–antibody immunoprecipitates were collected by protein A/G plus-agarose (SC-2003 Santa Cruz BioTechnology). Following the final wash, the samples were boiled and centrifuged to pellet the agarose beads. Western blot analysis of the CDC2 protein in the supernatant was then conducted. Antigens were visualized using a near infrared imaging system (Odyssey LI-COR) and data were analyzed using Odyssey 2.1 software.
Gene expression profiling (GEP)
Briefly, the cells untreated or treated with CuE for 4 hours, were harvested and total RNA was isolated utilizing an RNasey kit (Qiagen) as described by the manufacturer. Total RNA was sent to Welgene Company for whole human genome SurePrint G3 arrays GEP analysis (Agilent Technologies).
RT-PCR
A reverse transcriptase system (Promega, Southampon, UK) was used to synthesize cDNA from 1 μg of total RNA. Between 2 and 4 μL of cDNA were used for PCR analysis. PCR (50 μL) reactions were performed using 100 ng of each primer16 and 1 unit of Dynazyme II (Flowgen, Lichfield, UK). Thermal cycling was conducted for 35 cycles at the following temperature/durations: 98°C for 10 s, 66°C for 30 s and 72°C for 1 min using a Progene thermal cycler (Cambridge, UK). A final extension of 72°C was performed for 10 min at the end of 35 cycles. The primers used for amplification of the target genes were checked against all other gene sequences for specificity. PCR reactions were analyzed on 1.5% agarose/TAE minigels and stained using 0.5 μg/mL ethidium bromide. Gels were visualized using an Apligene UV CCD camera system.
Real-time PCR
Real-time PCR was conducted using SYBR Green PCR MasterMix according to the manufacturer's instructions. Quantitative real-time PCR (qRT-PCR) was performed using approximately 200 ng of SYBR Green PCR MasterMix in an ABI 7300 system (Applied Biosystems, Foster City, CA, USA). PCR conditions were 95°C for 120 s, 60°C for 30 s and 72°C for 30 s for 40 cycles. Sample cells from three plates were run in duplicate, using the threshold suggested by the software for the instrument to calculate Ct. To normalize readings, we used Ct values from 18 s as internal controls for each run, obtaining a delta Ct value for each gene.
Small-interfering RNA
The specific small-interfering RNA of GADD45γ (Stealth RNAi siRNA Duplex Oligoribonucleotides) and Lipofectamine RNAiMAX gene transfection system were purchased from Invitrogen (Thermo Fisher Scientific, USA). Resuspend the Duplex siRNA in RNase-free buffer (10 mM Tris-HCl, pH 8.0, 20 mM NaCl, 1 mM EDTA). The transfection protocol was according Lipofectamine RNAiMAX's manual.
Statistical analysis
All data were reported as the mean (±SEM) of at least three separate experiments. A t-test or one-way ANOVA with post-hoc test was employed for statistical analysis, with significant differences determined as P < 0.05.
References
Lin, Y. T., Wang, L. F. & Hsu, Y. C. Curcuminoids suppress the growth of pharynx and nasopharyngeal carcinoma cells through induced apoptosis. J Agric Food Chem. 57, 3765–3770 (2009).
Chan, J. Y., Gao, W., Ho, W. K., Wei, W. I. & Wong, T. S. Overexpression of Epstein-Barr virus-encoded microRNA-BART7 in undifferentiated nasopharyngeal carcinoma. Anticancer Res. 32, 3201–3210 (2012).
Dawson, C. W., Port, R. J. & Young, L. S. The role of the EBV-encoded latent membrane proteins LMP1 and LMP2 in the pathogenesis of nasopharyngeal carcinoma (NPC). Semin Cancer Biol. 22, 144–153 (2012).
Ríos, J. L., Andújar, I., Escandell, J. M., Giner, R. M. & Recio, M. C. Cucurbitacins as inducers of cell death and a rich source of potential anticancer compounds. Curr Pharm Des. 18, 1663–1676 (2012).
Lee, D. H., Iwanski, G. B. & Thoennissen, N. H. Cucurbitacin: ancient compound shedding new light on cancer treatment. ScientificWorldJournal. 10, 413–418 (2010).
Recio, M. C., Andujar, I. & Rios, J. L. Anti-inflammatory agents from plants: progress and potential. Curr Med Chem. 19, 2088–2103 (2012).
Ayyad, S. E. et al. In vitro and in vivo study of cucurbitacins-type triterpene glucoside from Citrullus colocynthis growing in Saudi Arabia against hepatocellular carcinoma. Environ Toxicol Pharmacol. 33, 245–251 (2012).
Militão, G. C. et al. In vitro and in vivo anticancer properties of cucurbitacin isolated from Cayaponia racemosa. Pharm Biol. 50, 1479–1487 (2012).
Ríos, J. L. Effects of triterpenes on the immune system. J Ethnopharmacol. 128, 1–14 (2010).
Nakashima, S. et al. Cucurbitacin E as a new inhibitor of cofilin phosphorylation in human leukemia U937 cells. Bioorg Med Chem Lett. 20, 2994–2997 (2010).
Sörensen, P. M. et al. The natural product cucurbitacin E inhibits depolymerization of actin filaments. ACS Chem Biol. 7, 1502–1508 (2012).
Nakamura, S. Search for biofunctional constituents from medicinal foods-elucidation of constituents with anti-proliferation effects and the target molecule from Citrullus colocynthis. Yakugaku Zasshi. 132, 1063–1067 (2012).
Tapia, C. et al. Two mitosis-specific antibodies, MPM-2 and phospho-histone H3 (Ser28), allow rapid and precise determination of mitotic activity. Am J Surg Pathol. 30, 83–89 (2006).
Decordier, I., Cundari, E. & Kirsch-Volders, M. Survival of aneuploid, micronucleated and/or polyploid cells: crosstalk between ploidy control and apoptosis. Mutat Res. 651, 30–39 (2008).
Wang, X. W. et al. GADD45 induction of a G2/M cell cycle checkpoint. Proc Natl Acad Sci U S A. 96, 3706–3711 (1999).
Hsu, Y. C., Chen, M. J. & Huang, T. Y. Inducement of mitosis delay by cucurbitacin E, a novel tetracyclic triterpene from climbing stem of Cucumis melo L., through GADD45γ in human brain malignant glioma (GBM) 8401 cells. Cell Death Dis. 5, e1087; 10.1038/cddis.2014.22 (2014).
Hsu, Y. C., Huang, T. Y. & Chen, M. J. Therapeutic ROS targeting of GADD45γ in the induction of G2/M arrest in primary human colorectal cancer cell lines by cucurbitacin E. Cell Death Dis. 5, e1198; 10.1038/cddis.2014.151 (2014).
Hung, C. M., Chang, C. C., Lin, C. W., Ko, S. Y. & Hsu, Y. C. Cucurbitacin E as inducer of cell death and apoptosis in human oral squamous cell carcinoma cell line SAS. Int J Mol Sci. 14, 17147–17156 (2013).
Tamura, R. E. et al. GADD45 proteins: central players in tumorigenesis. Curr Mol Med. 12, 634–651 (2012).
Ju, S. et al. Gadd45b and Gadd45g are important for anti-tumor immune responses. Eur J Immunol. 39, 3010–3018 (2009).
Guo, W. et al. Methylation-mediated repression of GADD45A and GADD45G expression in gastric cardia adenocarcinoma. Int J Cancer. 133, 2043–2053 (2013).
Zerbini, L. F. et al. JunD-mediated repression of GADD45α and γ regulates escape from cell death in prostate cancer. Cell Cycle. 10, 2583–2591 (2011).
Schäfer, A. Gadd45 proteins: key players of repair-mediated DNA demethylation. Adv Exp Med Biol. 793, 35–50 (2013).
Boddy, M. N., Furnari, B., Mondesert, O. & Russell, P. Replication checkpoint enforced by kinases Cds1 and Chk1. Science. 280, 909–912 (1998).
Liu, C. et al. Ser 15 of WEE1B is a potential PKA phosphorylation target in G2/M transition in one-cell stage mouse embryos. Mol Med Rep. 7, 1929–1937 (2013).
Acknowledgements
The authors appreciate the funding support provided by the National Science Council (Taiwan) (grant NSC 101-2313-B-309 -001).
Author information
Authors and Affiliations
Contributions
C.M.H. conceived the idea. C.M.H., C.C.Chang. and Y.C.H. wrote the main manuscript text and C.W.L., C.C.Chen prepared figures and tables. All authors reviewed the manuscript
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/4.0/
About this article
Cite this article
Hung, CM., Chang, CC., Lin, CW. et al. GADD45γ induces G2/M arrest in human pharynx and nasopharyngeal carcinoma cells by cucurbitacin E. Sci Rep 4, 6454 (2014). https://doi.org/10.1038/srep06454
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep06454
This article is cited by
-
Identification and characterization of the Cucurbitacins, a novel class of small-molecule inhibitors of Tropomyosin receptor kinase a
BMC Complementary and Alternative Medicine (2019)
-
Inducement of apoptosis by cucurbitacin E, a tetracyclic triterpenes, through death receptor 5 in human cervical cancer cell lines
Cell Death Discovery (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.