Abstract
Six different types of cancer (i.e., breast, lung, colorectal, esophageal, gastric and thyroid cancer) have high rates of incidence or mortality worldwide. It has been shown that activation of de novo lipogenesis is an early and common event in the cancer microenvironment. In this study, we performed lipid imaging and profiling for 134 tissue samples from six different types of cancer using matrix-assisted laser desorption/ionization-Fourier transform ion cyclotron resonance mass spectrometry, with 2,5-dihydroxybenzoic acid and 1,8-bis(dimethyl-amino)naphthalene as matrices in the positive and negative ion modes, respectively. Multivariate statistical analysis coupled with lipid distribution images revealed that significantly increased levels of monounsaturated fatty acids and monounsaturated phosphatidylcholines relative to polyunsaturated fatty acids and polyunsaturated phosphatidylcholines were observed in the cancer microenvironment compared with the adjacent normal tissue. The immunohistochemical assay indicated that fatty acid synthase, stearoyl-CoA desaturase-1 and choline kinase α were up-regulated in the cancer microenvironment compared with the adjacent normal tissue. Our findings suggest that de novo lipogenesis was activated in six types of cancer to promote a biosynthesis of lipids with monounsaturated acyl chains and to suppress a biosynthesis of polyunsaturated lipids in the cancer microenvironment.
Similar content being viewed by others
Introduction
Cancer is a leading cause of mortality worldwide, accounting for around 13% of all deaths1. Therein, breast cancer in females and lung cancer in males are the frequently diagnosed cancers and the leading cause of cancer death; colorectal cancer is the third commonly diagnosed cancer in males and the second in females; gastric and esophageal cancer are followed by the high incidence and mortality rates2 and thyroid cancer incidence has also dramatically increased in the past 30 years3. All these cancers affect people's health seriously and great efforts have been made to identify the mechanism of their development and progression, as well as diagnostic biomarkers. Lipids as a kind of important metabolites participate in the self assembly of phospholipids to form biological membranes and are involved in cell differentiation and signal transduction as second messengers4. Emerging evidence indicates that cancer and diverse human diseases are strongly correlated with changes in the levels of lipids5,6.
Previous studies have noted that increased de novo lipogenesis is an early and common event in the development of cancer7,8 and cancer-related lipogenesis requires more lipid production to meet the requirements of rapid cell proliferation. During the above-mentioned process, a key lipogenic enzyme (i.e., fatty acid synthase, FASN) catalyzes the terminal steps in the de novo biosynthesis of saturated fatty acids (SFAs) from acetyl-CoA9. SFAs are partly converted into monounsaturated fatty acids (MUFAs) by stearoyl-CoA desaturase-1 (SCD1) and then generate monounsaturated phosphatidylcholines (MUPCs) mediated by a series of enzymes (e.g., choline kinase α (CKα) and lysophosphatidylcholine acyltransferase (LPCAT)4). On the basis of previous data about breast, lung and thyroid cancer, a significant up-regulation of MUPCs was observed in the cancer microenvironment. These findings indicate that cancer progression is associated with de novo lipogenesis10,11,12. However, no investigation of in situ fatty acids reveals the role of de novo lipogenesis in cancer microenvironment.
Matrix-assisted laser desorption/ionization-mass spectrometry imaging (MALDI-MSI) can simultaneously map multiple different lipids while preserving the morphological integrity of the analyzed tissue13,14. MALDI coupled with ultrahigh resolution and ultrahigh mass accuracy Fourier transform ion cyclotron resonance mass spectrometry (FTICR MS) can greatly improve reliability and validity of MSI15. In addition, matrix is an important factor for MALDI-MSI and the most commonly used matrix for detecting lipids in the positive ion mode is 2,5-dihydroxybenzoic acid (DHB). Previous studies have noted that 1,8-bis(dimethyl-amino)naphthalene (DMAN) is an ideal matrix for studying low-molecular-weight metabolites in the negative ion mode without any interference from the matrix, which could be used for the detection of fatty acids in tissue16,17. Both matrices could be coated on tissue sections by a sublimation apparatus to generate high-purity and homogeneous matrix film, resulting in a significant improvements in lipid signals18. Furthermore, a strategy of merging both negative and positive ion MSI data from serial tissue sections was reported to provide an augmentation in the amount of useful information and multivariate statistical analysis would screen biomarkers to distinguish cancer and normal regions of tissue19. All these MSI improvements will be of benefit for the study of changes in lipid distribution in tissue.
In the present study, we investigated direct effects of de novo lipogenesis, especially the distribution of lipid species, on tissue samples from 134 patients with six types of cancer (i.e., breast, lung, colorectal, esophageal, gastric and thyroid cancer, Table 1). Both DHB and DMAN matrices were used to acquire mass spectra in the positive and negative ion modes, respectively, which contribute to an increase in the number of lipids detected. A combination of MSI and immunohistochemical assay was performed to reveal the link between changes in the levels of lipids and the expression of lipogenic-related enzymes in cancer microenvironment. Our findings suggest that de novo lipogenesis has been activated in six different types of cancer and that increased activities of FASN, SCD1, or CKα may be associated with increased levels of lipids with monounsaturated acyl chains and decreased levels of polyunsaturated species.
Results
MSI of six different types of tissue samples
To investigate the effect of cancer-related lipogenesis in tissue, MSI was performed in this study for direct analysis of lipids in six different types of cancer (i.e., breast, colorectal, esophageal, lung, gastric and thyroid cancer). DHB matrix was used to ionize phosphatidylcholines (PCs) and sphingomyelins (SMs) in the positive ion mode and DMAN was used to ionize fatty acids (FAs), phosphatidylethanolamines (PEs) and phosphatidylinositols (PIs) in the negative ion mode. In addition, a sublimation method for matrix deposition was adopted to improve repeatability and sensitivity of MSI, with the best signal intensity. For DMAN matrix, time-dependent changes in the intensities of metabolites in tissue under high vacuum condition were observed in Supplementary Fig. S1A and S1B. It was found that the absolute intensities of the metabolites almost remained unchanged in less than 25 min under high vacuum condition. The sublimation time and temperature for the different amount of the matrices were also systematically optimized to obtain a homogeneous coating on the surface of ITO slides (see Supplementary Fig. S1C online).
Representative mass spectra of two adjacent tissue sections from breast cancer tissue were acquired in the positive (left column, Fig. 1A) and negative (right column, Fig. 1A) ion modes, respectively. It should be noted that significant differences (e.g., PC(34:1) at m/z 798.5426, PC(38:4) at m/z 832.5818 and C18:1 at m/z 281.2484) were observed between the cancerous area and the adjacent normal area in the positive and negative ion modes, respectively.
MSI of six different types of cancer.
(A) Representative mass spectra from the cancerous area and the adjacent normal area of breast cancer tissue in the positive (left panel) and negative (right panel) ion modes. (B) Ion images of lipids from breast, colorectal, esophageal, lung, gastric and thyroid cancer in the negative (right panel) and positive (middle panel) ion modes. Optical images of H&E stained sections adjacent to the section used for MSI are shown in left panel and the cancerous area was circled with red dotted line and the adjacent normal area was circled with blue dotted line. MSI data were acquired with the spatial resolution of 200 μm and 1000 μm in the positive and negative ion modes, respectively.
Representative ion images of MSI from six different types of cancer in the positive and negative ion modes, with their individual corresponding H&E staining, are shown in Fig. 1B. For the H&E staining, the cancerous areas and adjacent normal areas of six cancer tissues are circled by red dotted line and blue dotted line, respectively. The intensities of the cancer-related MUPCs (i.e., [PC (32:1) + K]+ at m/z 770.5103, [PC (34:1) + K]+ at m/z 798.5426 and [PC (36:1) + K]+ at m/z 826.5732) in the cancerous area were significantly increased compared with those in the adjacent normal area, whereas the intensities of polyunsaturated PC (PUPC, i.e., [PC(38:4) + K]+ at m/z 832.5818) and phosphatidic acid (PA, i.e., [PA(40:5) + K]+ at m/z 789.4818) in the cancerous area were significantly decreased compared with those in the adjacent normal area. As shown in Fig. 1B (right column), differentially expressed lipids obtained by MSI in the negative ion mode are FAs, PEs and PIs. It should be noted that different kinds of FAs have shown different biological behaviors in the cancerous area. MUFAs (i.e., C16:1 at m/z 253.2174 and C18:1 at m/z 281.2484) in the cancerous area presented high intensities compared with those in the adjacent normal area, whereas PUFAs, PE and PI with polyunsaturated acyl chains (i.e., C22:4 at m/z 331.2645, [PE(38:4)-H]− at m/z 750.5441 and [PI(38:4)-H]− at m/z 885.5499) in the cancerous area had a lower level than those in the adjacent normal area. However, some exceptions were also observed for the alteration in the levels of these lipids, for example, the levels of C20:4 and PI(38:4) were increased in thyroid cancer, whereas no difference in their levels was detected between the cancerous area and the adjacent normal area for esophageal and gastric cancer. Detailed information on the identification of these lipids of interest is shown in Supplementary Table S1.
Lipid profiling of tissue samples
To further assess changes in the levels of the above-mentioned lipids, lipid profiling was performed on the basis of 134 tissue samples from six different types of cancer. Five mass spectra in the cancerous area and the adjacent normal area at five different positions, respectively, for each of tissue samples were acquired to screen cancer-related lipids and to assess tissue heterogeneity. Finally, a total of 164 metabolites (or variables), corresponding to the m/z values after isotopic deconvolution and a combination of [M+H]+, [M+Na]+ and [M+K]+ as one m/z value, were enrolled in the final data set for multivariate statistical analysis.
PLS-DA score plot revealed the obvious differences between the cancerous area and the adjacent normal area from the patients with breast, colorectal, esophageal, lung, gastric and thyroid cancer, with the predicted residual sum of square (PRESS) of 0.4603, 0.5379, 0.3757, 0.5069, 0.4125 and 0.4018, respectively (Fig. 2A). Y-scatterplot of VIP values versus FC values (cancer vs normal) of the variables was also used to select important variables. For breast cancer, as shown in Fig. 2B, the important variables with extremely large VIP and FC values can be easily found in the upper left or upper right hand quadrant. As exemplified by PC(32:1) and SM(22:0), both of FC values were 3.06 and −3.56, with VIP values of 1.55 and 1.75, respectively, indicating that both were significantly increased and decreased in the cancerous area compared with those in the adjacent normal area. All important variables with VIP >1 for six types of cancer were extracted on the basis of six Y-scatterplots (Fig. 2B). Finally, 18 common variables labeled in blue were extracted, which were strongly associated with six types of cancer and they are C16:1, C18:1, C20:1, PC(32:1), PC(34:1), PC(36:1), C20:4, C22:4, C22:5, PC(38:4), PC(38:6), PA(36:2), PA(38:3), PA(40:5), PE(38:4), PI(38:4), SM(22:0) and SM(24:1). The change trends of these 18 lipids between the cancerous area (red) and the adjacent normal area (blue) are shown in Fig. 2C. It is worth noting that the levels of MUPCs (i.e., PC(32:1), PC(34:1) and PC(36:1)) and MUFAs (i.e., C16:1, C18:1 and C20:1) in the cancerous area were significantly increased compared with those in the adjacent normal area (Wilcoxon-Mann-Whitney test, p <0.05), whereas polyunsaturated lipids (i.e., PC(38:4), PC(38:6), PA(38:3), PA(40:5), PE(38:4) and PI(38:4)) and PUFAs (i.e., C20:4, C22:4 and C22:5) in the cancerous area were remarkably decreased compared with those in the adjacent normal area (Wilcoxon-Mann-Whitney test, p <0.05) except thyroid cancer. Detailed values of VIP, FC and p are listed in Supplementary Table S2. It should be noted that elevated levels of some polyunsaturated lipids in thyroid cancerous area may be attributed to different pathological phenotype compared with other cancer. Interestingly, remarkably decreased levels of some monounsaturated and saturated SMs (i.e., SM(24:1) and SM(22:0)) were observed in the cancerous area compared with the normal area.
Metabolite profiling and feature selection of six types of cancer.
(A) PLS-DA score plots of metabolites from the cancerous area (red dots) and the adjacent normal area (blue dots) of six types of cancer. (B) Y-scatterplots of VIP values versus FC values of metabolites from the cancerous area and the adjacent normal area of six types of cancer. 18 common variables labeled in blue have significant differences in their levels between the cancerous area and the adjacent normal area for six types of cancer. (C) Change trends of these 18 lipids in the cancerous area (red bar) and the adjacent normal area (blue bar) of six types of cancer (*, p <0.05; **, p <0.01).
Correlation of characteristic lipids in six different types of cancer
To further confirm the association of the levels of lipids with specific fatty acyl chain length and saturation with cancer, Spearman correlation analysis was performed to identify their relationships. As shown in Supplementary Fig. S2, for breast, colorectal and esophageal cancer, MUFAs (i.e., C16:1, C18:1 and C20:1) were strongly and positively correlated with MUPCs (i.e., PC(32:1), PC(34:1) and PC(36:1)), whereas both were negatively correlated with polyunsaturated lipids. PUFAs (i.e., C20:4, C22:4 and C22:5) were strongly and positively correlated with polyunsaturated lipids (i.e., PC(38:4), PC(38:6), PA(38:3), PA(40:5), PE(38:4) and PI(38:4)); for lung and gastric cancer, the above-mentioned correlations became slightly lower; and for thyroid cancer, there were no obvious correlations among the above-mentioned lipids in the cancerous area.
Association of the ratios of MUFAs/SFAs, MUPCs/SPCs and the sum of FAs (or PCs) with cancer
It has been found that during de novo lipogenesis in vitro, SFAs are partly and endogenously converted into MUFAs by the rate-limiting enzyme of SCD1 and then the resulting products are converted by a series of enzymes to generate MUPCs. To confirm these phenomena in vivo, the ratios of MUFAs/SFAs (i.e., C16:1/C16:0 and C18:1/C18:0) and MUPCs/SPCs (i.e., PC(32:1)/PC(32:0), PC(34:1)/PC(34:0) and PC(36:1)/PC(36:0)), as well as the sum of FAs (i.e., C16:1, C16:0, C18:1 and C18:0) and PCs (i.e., PC(32:1), PC(32:0), PC(34:1), PC(34:0), PC(36:1) and PC(36:0)) in the cancerous area and the adjacent normal area for six types of cancer were investigated, respectively. As shown in Supplementary Fig. S3, most ratios of MUFAs/SFAs in the cancerous area were significantly higher than those in the adjacent normal area except the ratio of C16:1/C16:0 in thyroid cancer. The ratio of PC(32:1)/PC(32:0) in breast, colorectal, esophageal, lung and gastric cancerous area was significantly increased compared with that in their corresponding adjacent normal area, whereas this ratio in thyroid cancerous area was decreased. The ratio of PC(34:1)/PC(34:0) in lung or thyroid cancerous area was significantly increased or decreased compared with that in their individual corresponding adjacent normal area, whereas no difference in this ratio was detected between the cancerous area and the adjacent normal area for breast, colorectal, esophageal and gastric cancer. The ratio of PC(36:1)/PC(36:0) in breast, esophageal, lung, gastric and thyroid cancerous area was significantly increased, whereas no difference in this ratio was observed between colorectal cancerous area and adjacent normal area. In addition, it is worth noting that a common phenomenon for six types of cancer is that sums of FAs and PCs in the cancerous area were significantly increased compared with those in the adjacent normal area. These findings may imply that de novo fatty acid synthesis was increased in the cancerous area for six types of cancer and cancer-related pathway converting MUFAs to MUPCs may was also activated during cancer development.
Expression of FASN, SCD1 and CKα
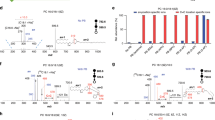
We have performed immunohistochemical assays to examine the expression of FASN, SCD1 and CKα for a total of 134 tissue samples, including 105 cancerous regions and 62 normal regions. Immunohistochemical images from six representative tissue sections adjacent to the tissue sections analyzed by MSI are shown in Fig. 3A. As shown in Fig. 3B, it is found that three enzymes were significantly overexpressed in the cancerous areas except for CKα in esophageal and lung cancer and SCD1 in lung cancer. It is worth noting that immunohistochemical images of overexpression of FASN, SCD1 and CKα in the cancerous areas were strongly correlated with elevated levels of MUPCs and MUFAs in the cancerous areas detected by MS images (Fig. 1B) or profiles (Fig. 2C).
Immunohistochemical localization of the expression of FASN, SCD1 and CKα in six types of cancer tissues.
(A) Representative immunohistochemical staining of FASN, SCD1 and CKα in six types of cancer tissues. Immunohistochemical assays of three enzyme were performed based on three serial tissue sections adjacent to their individual tissue section used for MSI and H&E staining in Figure 1, with two zoomed optical images (250×) from the cancerous area and the adjacent normal area, respectively. (B) Scatterplots of the mean optical density of FASN, SCD1 and CKα expression. Quantitative analysis of the expression of three enzymes in the cancerous area and the adjacent normal area is shown as mean ± Standard deviation (*, p < 0.05; **, p < 0.01).
Distinguish of six different types of cancer
PLS-DA score plot further revealed the difference in the levels of these lipids in six types of cancer. As shown in Supplementary Fig. S4A, breast and thyroid cancer could be successfully differentiated from colorectal, esophageal, lung and gastric cancer based on the first two principal components (t1 and t2), suggesting that the expression of some lipids is significantly different between the former two cancers and the latter four cancers. To test the variable importance in discriminating six types of cancer, the top 18 significantly changed lipids are shown in Supplementary Fig. S4B based on their VIP projection (VIP>1). As shown in the heatmap of VIP projection (right panel, see Supplementary Fig. S4B online), the lipids (i.e., PC(36:3), PC(32:1) and SM(34:1)) presented high level in colorectal, esophageal and gastric cancer. For breast cancer, the levels of PC(38:3), PC(38:4), C22:4, C20:2, C20:3, PC(36:0), PC(38:5) and PC(36:1) were increased. The lipids of PC(38:2) and PC(34:1) in lung cancer and lipids of C22:3, PA(40:5), SM(36:2), PA(36:3) and PA(38:5) in thyroid cancer were also up-regulated. Taken together, our findings suggest that changes in the levels of the above-mentioned lipids may have cancer-specific metabolic features.
Discussion
Recently, emerging evidence indicates that the progression of cancer is usually associated with increased de novo lipogenesis20,21,22. In this study, we combined MSI and immunohistochemical assays to investigate the associations of cancer-related lipogenesis with the expression of lipids based on 134 clinical tissue samples from six different types of cancer. A MALDI-FTICR MS was employed to obtain the reliable distribution and accurate mass of lipids in clinical tissues. The use of both DHB and DMAN matrices provided an opportunity to obtain global analysis of lipid species on the basis of two adjacent tissue sections in both negative and positive ion modes, respectively. Based on the sublimation of DMAN, which had no matrix- related interference in negative ion mode, we have successfully performed fatty acid imaging or profiling for clinical tissues with well-preserved morphological features of tissues, suggesting that the different ionization modes can provide detailed information on metabolites in the cancer microenvironment. This is the first time that lipidomic changes (e.g., FAs, PCs, PAs, PEs, PIs and SMs) have been systematically characterised in conjunction with multiple lipogenic enzymes in six types of cancer tissue. It is worth noting that a consistent increase in the levels of the lipids with monounsaturated acyl chains where observed to be associated with the increased expression of FASN, SCD1 and CKα and a relative decrease in the levels of polyunsaturated lipids in the cancerous area of clinical samples were observed.
As shown in Fig. 1, MUPCs (i.e., PC(32:1), PC(34:1) and PC(36:1)) and MUFAs (i.e., C16:1 and C18:1) were significantly up-regulated and species with polyunsaturated fatty acyl chains (i.e., PC(38:4), PA(40:5), PE(38:4), PI(38:4)) and PUFAs (i.e., C20:4 and C22:4) were down-regulated in the cancerous area of six different types of cancer compared with those in the adjacent normal area. These trends were further confirmed on the basis of multivariate statistical analysis (Fig. 2A) and Y-scatterplots (Fig. 2B) of lipid profile data from a total of 134 tissue samples. Finally, 18 common lipids (i.e., C16:1, C18:1, C20:1, PC(32:1), PC(34:1), PC(36:1), C20:4, C22:4, C22:5, PC(38:4), PC(38:6), PA(36:2), PA(38:3), PA(40:5), PE(38:4), PI(38:4), SM(22:0) and SM(24:1)) with larger VIP and FC values were selected for six different types of cancer (Fig. 2C). These are consistent with previous studies about the overexpression of MUPCs in lung, breast and colorectal cancer10,12,23. To further examine the relationship of these lipid abundances, Spearman correlation analysis of 22 lipids indicated a highly positive correlation between MUFAs and MUPCs and negative correlation between monosaturated and polyunsaturated lipids existed. These findings suggest that there may be similar lipogenic mechanisms to generate the above-mentioned lipids for six different types of cancer, but it should be noted that less correlation in thyroid cancer was observed, which may be associated with its lower aggressive behaviors compared with other cancers.
Activation of de novo lipogenesis in cancer tissues that we focused on in this study was mediated by multiple lipogenic enzymes. FASN catalyzes the synthesis of palmitate (C16:0) and stearic (C18:0) acids from acetyl-CoA, with the overexpression in lung, breast and prostate cancer8,24,25, which is consist with our results. As shown in Fig. 3 and Supplementary Fig. S2, increase in the sum of FAs (C16:1, C16:0, C18:1 and C18:0) was positively correlated with the up-regulation of FASN in six types of cancer, indicating that FAs synthesis may play an important role in cancer pathogenesis. In addition, increase in the ratios of MUFAs/SFAs (C16:1/C16:0, C18:1/C18:0) and MUPCs/SPCs (PC(32:1)/PC(32:0), PC(34:1)/PC(34:0), PC(36:1)/PC(36:0)) was also positively correlated with the overexpression of SCD1, which catalyzes the biosynthesis of MUFAs from SFAs, which are consistent with previous studies about lung, hypopharyngeal, prostate, ovarian, gastric, breast and colorectal cancer26,27,28.
Although the overexpression of CKα for the synthesis of PCs in the cancerous areas of breast, colorectal, gastric and thyroid cancer was only observed, increase in the sum of PCs (PC(32:1), PC(32:0), PC(34:1), PC(34:0), PC(36:1) and PC(36:0)) was detected in all six types of cancer. These results may suggest that other enzymes may be involved in the synthesis of PC, such as LPCAT423, the expression of which was not determined due to sample limitations. In addition, PUFAs and polyunsaturated lipids were down-regulated in the cancerous areas compared with the adjacent normal area. Taken together, the common lipogenesis pathway for six types of cancer was summarized in Fig. 4. Our results suggest that the use of in situ MSI detection coupled with immunohistochemical assay might provide a new perspective to explore the common mechanisms of different types of cancer.
The lipid metabolic network of significantly changed lipids in six types of cancer tissues.
The key enzyme of FASN catalyzes the synthesis of SFAs from acetyl-CoA. SFAs are endogenously converted into MUFAs through the part influence of SCD1 and then MUPCs are generated from MUFAs mediated by a series of enzymes (e.g., CKα). PUFAs could be produced by desaturation and chain elongation, followed by involving in the synthesis of polyunsaturated lipids (e.g., PUPCs). Red and blue arrows show up- and down-regulated expressions, respectively, in the cancerous areas of six types of cancer. Red and blue background indicates the up- and down-regulated levels of lipids, respectively. Ion images from breast cancer tissue are shown in the up panel.
The development and progression of cancer can be viewed as a multistep process, with the complex interactions between deregulated lipogenesis and the cancer microenvironment29. It was found that increase in the degree of saturation could make cell membrane less fluid in most aggressive cancers22,30. Based on our observation that de novo lipogenesis would provide a consistent increase in the levels of lipids including monounsaturated acyl chains in the cancerous tissue, MUFAs and MUPCs replenish the membrane components for rapid cell proliferation and also pack the membrane more densely. This alteration of membrane fluidity may promote the carcinogenesis in six types of cancer. On the other hand, polyunsaturated acyl chains are more susceptible to peroxidation and oxidized lipid species may be ultimately degraded into smaller reactive products such as 4-hydroxyalkenal and malondialdehyde, which can cause cell damage31,32, which are consistent with our observation that modulation of de novo lipogenesis has increased the ratio of monounsaturated lipids to polyunsaturated lipids in the cancerous tissue compared with that in the adjacent normal tissue, cancer cells are protected from oxidative stress–induced cell death. More importantly, different lipid metabolism may exist in thyroid cancer, resulting in its less aggressive behavior.
The development and progression for the six different types of cancer may involve different processes. As shown in Supplementary Fig. S4, changes in the levels of 18 common lipids (i.e., PC(38:3), C22:3, PC(38:4), C22:4, C20:3, C20:2, PC(32:1), PA(40:5), SM(36:2), PC(36:3), SM(34:1), PC(38:2), PC(36:0), PC(38:5), PA(36:3), PA(38:5), PC(34:1) and PC(36:1)) for six different types of cancer were strongly correlated with the types of cancer. Polyunsaturated lipids in breast, colorectal and thyroid cancer were significantly increased compared with those in esophageal, lung and gastric cancer, while the levels of monosaturated lipids in esophageal and gastric cancers were increased compared with those in breast, colorectal and thyroid cancer. For lung cancer, however, elevated levels of PC(34:1) and PC(38:2) were observed. These findings indicate that some different lipogenesis mechanisms among six different types of cancer may be involved.
In summary, in the present study, based on the integrated strategy of MSI (or mass spectrometry profiling) in positive and negative ion modes using both DHB and DMAN matrices, respectively, we successfully detected several kinds of lipids (i.e., FAs, PCs, PAs, PEs, PIs and SMs) and investigated direct effects of de novo lipogenesis in the six types of cancer. After merging ion images with immunohistochemical staining results, a significant change in the abundance of monosaturated lipids relative to polyunsaturated lipids and the overexpression of FASN, SCD1 and CKα were observed in the cancer microenvironment. The alteration in the degree of lipid unsaturation generated by de novo lipogenic enzymes in the cancer microenvironment may related to carcinogenesis. The improved MSI platform may be powerful tool to study cancer mechanisms and to screen novel diagnostic biomarkers.
Methods
Tissue samples
All specimens were remaining tissues after clinical pathological experiments and informed consent was obtained from each patient. The approval was given by the Ethics Committee of Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences and all experiments were performed in accordance with relevant guidelines and regulations. In this study, a total of 134 primary tumor tissues were recruited, which were from 50 patients with breast cancer (invasive ductal and adenocarcinoma), 6 with colorectal cancer (adenocarcinoma), 15 with esophageal cancer (squamous carcinoma), 26 with lung cancer (squamous carcinoma and adenocarcinoma), 19 with gastric cancer (adenocarcinoma) and 18 with thyroid cancer (papillary and follicular). Cancerous area and adjacent normal area for each of tissue samples were carefully distinguished by the microscopic examination. The detailed information is listed in Table 1.
Sample preparation and matrix deposition
All tissue samples were snap-frozen in liquid nitrogen and stored at −80°C until use. A 12 μm thin tissue section was cut and then thaw-mounted onto a glass slide coated by indium-tin-oxide (ITO) in a freezing microtome. After the resulting slides were rewarmed at room temperature, DHB or DMAN (Sigma-Aldrich) was coated using the sublimation apparatus (a homebuilt device). The sublimation was operated under the fixed vacuum (5 mTorr) at 120°C for 5 min. Finally, approximately 11 mg of DHB or 25 mg of DMAN was coated on each glass slide and then recrystallization was performed to enhance signal intensity as described previously33.
MSI or mass spectrometry profiling
MSI or mass spectrometry profiling of tissue sections was performed using an 9.4 T Apex-ultraTM hybrid Qh-FTICR MS (Bruker Daltonics, Billerica, MA, USA) equipped with a 355 nm Nd:YAG Smartbeam II laser (200 Hz, spot diameter 50 μm). The experimental condition was optimized to obtain the best signal-to-noise ratio and good reproducibility in both positive and negative ion modes. For the MSI data acquisition, two adjacent tissue sections were analyzed at the spatial resolution of 200 μm and 1000 μm in the positive and negative ion modes, respectively. A mass spectrum (or one pixel) was accumulated with three full scans with 50 and 70 laser shots each over a mass range of 400 ~ 1000 Da and 100 ~ 1000 Da at the resolutions of 530,000 and 130,000 at m/z 400 in the positive and negative ion modes, respectively. As DMAN is easily sublimed under high vacuum conditions, low spatial resolution was used to ensure the reproducibility of MSI for an entire tissue section. Shown in Supplementary Fig. S1 indicates that DMAN matrix can generate reproducible signals during the experimental time period of less than 25 min. External calibration was performed using a lipid mixture (m/z 622.44423, m/z 734.56943, m/z 790.63203 and m/z 898.72593, which were from Avanti Polar Lipids, Inc. and identified by mass spectrometry) in positive ion mode and a tuning mixture (Part No. G2432A, Agilent Technology, Inc.) in negative ion mode over the above-mentioned mass ranges. For the profiling data acquisition, the experimental parameters were the same as those of the MSI data acquisition. Five mass spectra at different positions of the cancerous area and the adjacent normal area were randomly acquired to confirm data reliability and to assess the heterogeneity of tissues.
Structure identification of differentially expressed lipids of interest
Identification of lipid species of interest was first performed based on the Lipidmaps (http://www.lipidmaps.org/tools/index.html), with a mass tolerance of <2.5 ppm. Differentially expressed lipids were further identified on the basis of their measured accurate molecular masses, reliable isotope distributions and tandem mass spectral data (see Supplementary Table S1 online). The collision-induced dissociation experiments were selected to get tandem mass spectra of the lipids of interest.
Data handing and statistical analysis
Molecular images were visualized using the FlexImaging software (version 2.1, Bruker Daltonics) with the absolute intensity. Peaks with signal-to-noise ratio of >3, relative intensity of >0.1% and absolute intensity thresholds of 10,000 were selected as reliable variables, along with reliable isotope distributions using DataAnalysis 4.0 software (Bruker Daltonics). After isotopic deconvolution, the metabolites among different samples and different pixels were aligned within a narrow mass tolerance window (±0.001 Da) as one metabolite and [M+H]+, [M+Na]+ and [M+K]+ ions were also combined as one metabolite (or one variable). The intensities of all variables from one mass spectrum were normalized to a constant number of 1000 and the half of the baseline strength in each spectrum was adopted as their intensity of missing metabolites. Resulting datasets in the positive and negative ion modes from two adjacent tissue sections were combined into one dataset and then transfer into Microsoft Excel for the following statistical analysis. SAS software (version 9.2, SAS Institute Inc., USA) was used for partial least squares discriminant analysis (PLS-DA), Wilcoxon-Mann-Whitney test and Spearman correlation analysis. Metabolites with p value of less than 0.05 (Wilcoxon-Mann-Whitney test) were considered to be statistically significant and variable importance in the projection (VIP) of greater than 1.0 had differential reliability. GraphPad software was used to generate box-chart. Important variables were further selected using a Y-scatterplot of VIP values versus fold change (FC) values.
Immunohistochemical assay
Three lipogenic-related enzymes (i.e., FASN, SCD1 and CKα) were chosen for analysis by immunohistochemical staining on the basis of the fact that they were related to synthesis of SFAs, MUFAs and MUPCs, respectively. Briefly, three serial frozen tissue sections, which were adjacent to the tissue sections analyzed by mass spectrometry, were warmed at room temperature for 30 min. After quenching of endogenous peroxidase activity with 3% H2O2 for 10 min, theses sections were blocked with bovine serum albumin for 60 min at room temperature. These resulting sections were first incubated at room temperature with antibodies against FASN (1:250, Abcam, Cambridge, MA), SCD1 (1:200, Abcam) and CKα (1:200, Abcam), respectively and then stored at 4°C overnight followed by rewarming at 37°C for 45 min. After washing with PBS three times, these sections were allowed to react with a secondary antibody against rabbit IgG (1:800), mouse IgG (1:200), mouse IgG (1:200), respectively. Detection was performed with the EnVision horseradish peroxidase system. To reduce nonspecific staining, phosphate buffer was used as a negative control. Adjacent normal area from tissue samples was used as an internal negative control and the cancerous area was used as an internal positive control for immunohistochemical staining.
All immunohistochemical photographs were analyzed by using Image ProPlus (IPP, version 6.0, Media Cybernetics, Silver Spring, MD, USA) as described previously34. In each area, five views were randomly selected under a microscope (250×). The mean optical density, as a quantitative measurement of stain intensity, was computed to determine average enzyme expression.
References
Ferlay, J. et al. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer 127, 2893–917 (2010).
Jemal, A. et al. Global cancer statistics. CA Cancer J. Clin. 61, 69–90 (2011).
Ito, Y., Nikiforov, Y. E., Schlumberger, M. & Vigneri, R. Increasing incidence of thyroid cancer: controversies explored. Nat. Rev. Endocrinol 9, 178–84 (2013).
Santos, C. R. & Schulze, A. Lipid metabolism in cancer. FEBS J. 279, 2610–23 (2012).
Glaysher, J. Lipid metabolism and cancer. Curr. Opin. Lipidol 24, 530–1 (2013).
Zhang, Y. et al. Ammonia-treated N-(1-naphthyl) ethylenediamine dihydrochloride as a novel matrix for rapid quantitative and qualitative determination of serum free fatty acids by matrix-assisted laser desorption/ionization-Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chim. Acta 794, 82–9 (2013).
Swinnen, J. V. et al. Overexpression of fatty acid synthase is an early and common event in the development of prostate cancer. Int. J. Cancer 98, 19–22 (2002).
Piyathilake, C. J. et al. The expression of fatty acid synthase (FASE) is an early event in the development and progression of squamous cell carcinoma of the lung. Hum. Pathol. 31, 1068–73 (2000).
Menendez, J. A. & Lupu, R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer 7, 763–77 (2007).
Lee, G. K. et al. Lipid MALDI profile classifies non-small cell lung cancers according to the histologic type. Lung Cancer 76, 197–203 (2012).
Ishikawa, S. et al. Increased expression of phosphatidylcholine (16:0/18:1) and (16:0/18:2) in thyroid papillary cancer. PLoS One 7, e48873 (2012).
Ide, Y. et al. Human Breast Cancer Tissues Contain Abundant Phosphatidylcholine(36ratio1) with High Stearoyl-CoA Desaturase-1 Expression. PLoS One 8, e61204 (2013).
Seeley, E. H. & Caprioli, R. M. MALDI imaging mass spectrometry of human tissue: method challenges and clinical perspectives. Trends Biotechnol. 29, 136–43 (2011).
Chughtai, K. & Heeren, R. M. Mass spectrometric imaging for biomedical tissue analysis. Chem. Rev. 110, 3237–77 (2010).
Smith, D. F. et al. High mass accuracy and high mass resolving power FT-ICR secondary ion mass spectrometry for biological tissue imaging. Anal. Bioanal. Chem. 405, 6069–76 (2013).
Shroff, R. & Svatos, A. Proton sponge: a novel and versatile MALDI matrix for the analysis of metabolites using mass spectrometry. Anal. Chem. 81, 7954–9 (2009).
Shroff, R., Rulisek, L., Doubsky, J. & Svatos, A. Acid-base-driven matrix-assisted mass spectrometry for targeted metabolomics. Proc. Natl. Acad. Sci. U S A 106, 10092–6 (2009).
Hankin, J. A., Barkley, R. M. & Murphy, R. C. Sublimation as a method of matrix application for mass spectrometric imaging. J. Am. Soc. Mass Spectrom. 18, 1646–52 (2007).
Thomas, A. et al. Histology-driven data mining of lipid signatures from multiple imaging mass spectrometry analyses: application to human colorectal cancer liver metastasis biopsies. Anal. Chem. 85, 2860–6 (2013).
Hilvo, M. et al. Novel theranostic opportunities offered by characterization of altered membrane lipid metabolism in breast cancer progression. Cancer Res. 71, 3236–45 (2011).
von Roemeling, C. A. et al. Stearoyl-CoA desaturase 1 is a novel molecular therapeutic target for clear cell renal cell carcinoma. Clin. Cancer Res. 19, 2368–80 (2013).
Rysman, E. et al. De novo lipogenesis protects cancer cells from free radicals and chemotherapeutics by promoting membrane lipid saturation. Cancer Res. 70, 8117–26 (2010).
Kurabe, N. et al. Accumulated phosphatidylcholine (16:0/16:1) in human colorectal cancer; possible involvement of LPCAT4. Cancer science 104, 1295–302 (2013).
Yoon, S. et al. Up-regulation of acetyl-CoA carboxylase alpha and fatty acid synthase by human epidermal growth factor receptor 2 at the translational level in breast cancer cells. J. Biol. Chem. 282, 26122–31 (2007).
Swinnen, J. V. et al. Selective activation of the fatty acid synthesis pathway in human prostate cancer. Int. J. Cancer 88, 176–9 (2000).
Roongta, U. V. et al. Cancer cell dependence on unsaturated fatty acids implicates stearoyl-CoA desaturase as a target for cancer therapy. Mol. Cancer Res. 9, 1551–61 (2011).
Hess, D., Chisholm, J. W. & Igal, R. A. Inhibition of stearoylCoA desaturase activity blocks cell cycle progression and induces programmed cell death in lung cancer cells. PLoS One 5, e11394 (2010).
Mason, P. et al. SCD1 inhibition causes cancer cell death by depleting mono-unsaturated fatty acids. PLoS One 7, e33823 (2012).
Baenke, F., Peck, B., Miess, H. & Schulze, A. Hooked on fat: the role of lipid synthesis in cancer metabolism and tumour development. Dis. Model Mech. 6, 1353–63 (2013).
Ollila, S., Hyvonen, M. T. & Vattulainen, I. Polyunsaturation in lipid membranes: dynamic properties and lateral pressure profiles. J. Phys. Chem. B 111, 3139–50 (2007).
Deigner, H. P. & Hermetter, A. Oxidized phospholipids: emerging lipid mediators in pathophysiology. Curr. Opin. Lipidol 19, 289–94 (2008).
Schneider, C., Porter, N. A. & Brash, A. R. Routes to 4-hydroxynonenal: fundamental issues in the mechanisms of lipid peroxidation. J. Biol. Chem. 283, 15539–43 (2008).
Yang, J. H. & Caprioli, R. M. Matrix Sublimation/Recrystallization for Imaging Proteins by Mass Spectrometry at High Spatial Resolution. Anal. Chem. 83, 5728–34 (2011).
Yin, X. et al. DACT1, an antagonist to Wnt/beta-catenin signaling, suppresses tumor cell growth and is frequently silenced in breast cancer. Breast Cancer Res. 15, R23 (2013).
Acknowledgements
This study was funded by the grant (No. 91029701) from the National Natural Science Foundation of China (to Z. Li).
Author information
Authors and Affiliations
Contributions
S.G. and Z.L. provided the original concept for the research and designed the study. S.G. performed the experiments with the assistance of D.Z., Y.W. collected tissue samples. S.G. and Z.L. discussed the data and wrote the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Guo, S., Wang, Y., Zhou, D. et al. Significantly increased monounsaturated lipids relative to polyunsaturated lipids in six types of cancer microenvironment are observed by mass spectrometry imaging. Sci Rep 4, 5959 (2014). https://doi.org/10.1038/srep05959
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep05959
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.