Abstract
Bacterial biofilms are particularly problematic since they become resistant to most available antibiotics. Hence, novel potential antagonists to inhibit biofilm formation are urgent. Here the influences of two natural products, ursolic acid and resveratrol, on biofilm of the clinical methicillin-resistant Staphylococcus aureus (MRSA) isolate were investigated using RNA-seq and the differentially expressed genes were analyzed using Cuffdiff. The results showed that ursolic acid inhibition of biofilm formation may reduce amino acids metabolism and adhesins expression and resveratrol may disturb quorum sensing (QS) and the synthesis of surface proteins and capsular polysaccharides. In addition, the transcriptome analysis of resveratrol and the combination of resveratrol with vancomycin inhibition of established biofilm revealed that resveratrol would disturb the expression of genes related to QS, surface and secreted proteins and capsular polysaccharides. These findings suggest that ursolic acid and resveratrol could be useful to be adjunct therapies for the treatment of MRSA biofilm-involved infections.
Similar content being viewed by others
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) is an important human pathogen that causes abscesses in many organ tissues, septicemia and even life-threatening infections1. Many reports have shown that staphylococcal infections were associated with biofilm formation2,3. Bacterial biofilms are particularly problematic because sessile bacteria can often withstand a host's immune response and the bacteria within biofilms become resistant to most available antibiotics4. Hence, the research and development of novel potential antagonists to inhibit biofilm formation or remove established biofilm is both urgent and necessary.
Increasing evidence suggests that quorum sensing (QS) is important for the construction and/or dissolution of biofilm communities5. As such, QS inhibitors (QSIs) have the potential to be used to be adjuvants in antimicrobial therapy. In addition, some QSIs could be used to suppress the emergence and spread of bacteria-forming biofilm6.
To develop less or non-toxic natural products (such as QSIs) that inhibit MRSA biofilm formation or remove established biofilm, hundreds of natural compounds isolated from traditional Chinese medicines were tested by our group. Ursolic acid inhibited MRSA biofilm formation but has no impact on established biofilm (Supplementary Fig. S1a online, Supplementary Table S1 online), whereas resveratrol inhibited MRSA biofilm formation and removed partially established biofilm (Supplementary Fig. S1b online, Supplementary Table S2 online).
Studies on global S. aureus biofilm transcriptional profiles suggested that planktonic and biofilm cultures showed distinct patterns of gene expression using microarray4,7,8. However, few papers have reported on MRSA biofilm treated with drugs at the genetic level using microarray9, let alone using RNA-seq strategies. In the current study, we quantitatively compared the abundance of each gene under different conditions and pooled and normalized fragments of each gene and sample data size. The resulting number, fragments per kilobase of transcript per million mapped fragments (FPKM), allows for comparisons between differently expressed genes and growth conditions10. Here, to further investigate the mechanisms of inhibiting biofilm formation or removing established biofilm at the genetic level, we used high-throughput Illumina sequencing of cDNA (Illumina RNA-seq) to study the differential gene expression of MRSA by the addition of these two compounds.
Our data show that the use of ursolic acid to inhibit biofilm formation may reduce the metabolism of some amino acids as well as the expression of adhesins. However, the mechanisms of using resveratrol to inhibit biofilm formation and remove established biofilm are likely to interfere with quorum sensing (QS) system and synthesis of surface proteins and capsular polysaccharides. This study used RNA-seq strategies to examine the effects of natural products on bacterial biofilm.
Results
MICs and MBCs determination
The susceptibilities of the MRSA planktonic cells to ursolic acid and resveratrol were determined in vitro by methods recommended by the CLSI. The MICs amd MBCs are given in Supplementary Table S3. The MIC of ursolic acid against the MRSA was lower (37 μg/mL) than resveratrol (350 μg/mL). In addition, the MBC of resveratrol against the MRSA was higher (>800 μg/mL) than ursolic acid (175 μg/mL).
Resveratrol and ursolic acid inhibit MRSA biofilm formation
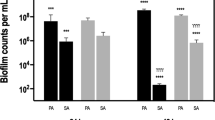
The in vitro effects of resveratrol and ursolic acid on MRSA biofilm formation were investigated using crystal violet staining semi-quantitative assays and scanning electron microscopy (SEM) images. MRSA was able to form biofilm on 96-well plates after 18 h incubation. The SEM images also showed that MRSA formed thick, heterogenous clumps on the coverslip (Fig. 1a). Crystal violet staining assays revealed that the inhibiting effect of ursolic acid on MRSA biofilm formation was stronger (66.3%) than that of resveratrol (39.85%) (Table 1, Supplementary Fig. S2 online). Moreover, the SEM images further qualitatively showed that resveratrol and ursolic acid could inhibit MRSA biofilm formation (Fig. 1b,c).
Scanning electron microscopy images showing the structure of methicillin-resistant Staphylococcus aureus in different conditions at 18 h (a–c) or 36 h (d–g).
Magnifications, ×3000. (a) Control without ethanol (at 18 h) (M18), (b) 100 μg/mL resveratrol (MR100), (c) 30 μg/mL ursolic acid (MU30), (d) control without ethanol (36 h) (M36), (e) 8 μg/mL vancomycin (MV), (f) mixture with 8 μg/mL vancomycin and 150 μg/mL resveratrol (MVR) and (g) 150 μg/mL resveratrol (MR150).
Resveratrol, vancomycin and their combination remove established MRSA biofilm
The in vitro effects of resveratrol, vancomycin and their combination on established MRSA biofilm were also investigated using the methods described above. The semi-quantitative assay results demonstrated that their combination had the strongest effect on established MRSA biofilm (inhibitory rate, 55.43%). However, the inhibitory rates of resveratrol and vancomycin alone removing established MRSA biofilm were decreased to 23.42% and 27.71%, respectively (Table 1, Supplementary Fig. S2 online). The SEM images also showed that MRSA biofilm became thinner after treatment with resveratrol, vancomycin, or their combination than no treatment (Fig. 1d–g).
Sequencing of the MRSA transcriptome
The number of mapped cDNA reads, 12.1–30.0 million (100 bp each) per sample, totaled 17,693,687,500 bases of sequenced cDNA (Table 2). Using the Staphylococcus aureus subsp. aureus COL genome as a reference, all of the MRSA transcripts were assignable to the genome. Each sample yielded a high number of mRNA reads (up to 90.8%). A total of 15,773,232,900 bp sequences of cDNA from mRNA transcripts (10.8–26.9 million reads per sample) were generated. The distribution of the reads in each sample could be randomly mapped along the relative position through the genome without bias (Supplementary Fig. S3 online).
Quantitative analysis of the global genes expression after treatment MRSA biofilm
In this study, the expression level distribution of all genes revealed that samples in the inhibitory biofilm formation conditions (MR100 & MU30) showed more differences than those samples in the removed established biofilm conditions (MR150 & MVR) (Supplementary Figs. S4–S5 online). The global expression level distribution profiles for mRNA isolated from all the test conditions were shown in Fig. 2 (Supplementary Tables S4–S6 online). The analyses of differentially expressed genes revealed that most genes were uniformly up-regulated or down-regulated in the removed conditions (Fig. 2c, fifth and sixth circles) but there were a few differences in the inhibitory conditions (Fig. 2a,b, third circles). The chemical structure of resveratrol had no correlation with that of ursolic acid, so we performed the differential expression analyses independently between MR100 and MU30 (Fig. 2a,b).
Distribution of differentially expressed genes in inhibitory and removed conditions Red, yellow and blue correspond to genes with high, medium and low expression, respectively.
(a) Distribution of differentially expressed genes in the resveratrol inhibiting methicillin-resistant Staphylococcus aureus (MRSA) biofilm formation condition. From outside to inside, the three circles in each plot correspond to the following: (1 and 2) log2 of the fragments per kilobase of transcript per million mapped fragments (FPKM) of each gene in MR100 and M18C06; (3) genes differentially expressed in MR100 compared with M18C06. (b) Distribution of the differentially expressed genes in ursolic acid inhibitory MRSA biofilm formation condition. From outside to inside, the three circles in each plot correspond to the following: (1 and 2) log2 of FPKM each gene in MU30 and M18C06, respectively; (3) genes differentially expressed in MU30 compared with M18C06. (c) Distribution of the differentially expressed genes in the resveratrol and its combination with vancomycin removed established MRSA biofilm conditions. From outside to inside, the six circles in each plot correspond to the following: (1, 2 and 3) log2 of FPKM of each gene in MVR, MR150 and M36C05, respectively; (4) genes differentially expressed in MVR compared with MR150; (5 and 6) genes differentially expressed in MVR and MR150 compared with M36C05, respectively.
The variance analysis package Cuffdiff was used to systematically search the transcriptome data for the subset of genes with statistical significance (P < 0.05, false discovery rate (FDR) < 0.001; >2-fold differential expression change in treatment conditions versus controls)11. Although the expression levels of most genes did not differ markedly when the cells were obtained after treatment (Supplementary Fig. S6 online), the significant differences for the expression levels of some genes were established by the use of Cuffdiff on FPKM values in both conditions. In the inhibiting MRSA biofilm formation conditions, 193 genes were up-regulated and 48 genes were down-regulated in the resveratrol inhibited MRSA biofilm formation sample (MR100) (Fig. 3a, Supplementary Table S7 online), while 89 genes were up-regulated and 94 genes were down-regulated in the ursolic acid inhibited MRSA biofilm formation sample (MU30) compared with the levels of gene expression in cells within control biofilm (M18C06) (Fig. 3b, Supplementary Table S8 online). Moreover, 32 of 241 genes differentially expressed in MR100 were found in MU30 (Supplementary Fig. S7a online).
Volcano plots of the distribution of gene expression for methicillin-resistant Staphylococcus aureus (MRSA) in the inhibitory and removed conditions.
Cuffdiff analysis was performed to show the differentially expressed genes. Green, red and gray correspond to genes with >2 log2-fold differential expression, >1 log2-fold differential expression and <1 log2-fold differential expression, respectively. (a) Distribution of differentially expressed genes in MU30 compared with M18C06, (b) distribution of differentially expressed genes in MR100 compared with M18C06, (c) distribution of differentially expressed genes in MVR compared with M36C05 and (d) distribution of differentially expressed genes in MR150 compared with M36C05.
A total of 201 genes were up-regulated and 243 genes were down-regulated in the resveratrol removed established biofilm sample (MR150), while 165 were up-regulated and 176 were down-regulated in the combination resveratrol with vancomycin removed established biofilm sample (MVR) compared with the gene expression levels in controls (M36C05) (Fig. 3c,d, Supplementary Tables S9–S10 online). Of the 444 genes that were differentially expressed in MR150, 296 were found in MVR (Supplementary Fig. S7b online). The lack of complete overlap between the sets of genes differentially expressed in the different samples may reflect the effects of the different compounds on MRSA biofilm in different conditions. We also obtained differential gene expression in MVR compared with MR150. The results showed that 37 genes were up-regulated and three were down-regulated (Supplementary Fig. S8 online, Supplementary Table S10 online).
The differentially expressed genes were classified into functional categories based on clusters of orthologous gene (COG) designations. The functional profiles of these gene sets differed between both conditions (Fig. 4). In the inhibiting MRSA biofilm formation conditions, 167 genes were up-regulated and 25 were down-regulated in MR100 compared with the expression levels of genes in the control (M18C06) and the 75 up-regulated and 70 down-regulated genes in MU30. The functional classes generally contained more up-regulated and less down-regulated genes in MR100, the finding was inconsistent with the functional classes in MU30 (Fig. 4).
Functional classification of differentially expressed genes in both conditions.
Profiles of the functional classes are shown as the number of differentially expressed genes in each condition. a) Inhibitory condition and (b) removed condition. Clusters of orthologous gene designations are described in Supplementary Table S12 online.
In the removing established biofilm conditions, 375 of 444 genes differentially expressed in MR150 were classified into COG functional categories, which included 175 up-regulated and 200 down-regulated genes. In addition, 299 of 341 genes differentially expressed in MVR were classified into COG functional categories, which contained 151 up-regulated and 148 down-regulated genes. Interestingly, in COG functional categories, the tendency toward up-regulated or down-regulated genes differentially expressed in MR150 is consistent with those of MVR (Fig. 4b).
Key genes associated with MRSA biofilm formation
Notably, the sets of genes that were highly up-regulated and down-regulated in both conditions included some key known genes encoding virulence factors, surface proteins, capsular polysaccharides and others related to biofilm formation of S. aureus (Fig. 5, Supplementary Tables S13–S14 online).
Heatmap of differentially expressed genes associated with methicillin-resistant Staphylococcus aureus (MRSA) biofilm formation and virulence.
(a) Resveratrol inhibiting MRSA biofilm formation condition, (b) ursolic acid inhibiting MRSA biofilm condition and (c) resveratrol and its combination with vancomycin removing established MRSA biofilm condition. Chemically, resveratrol had no correlation with ursolic acid, so we performed separate differential expression analyses.
In the inhibiting biofilm formation conditions, hld encoding δ-hemolysin was up-regulated in two samples, while rsbU encoding sigma factor B regulator protein was only up-regulated in MR100. Both genes could secrete virulence factors in S. aureus. Staphylococcal protein A (spa), which encoded the protein A, was up-regulated in MR100. Genes (cap5ABCFG) mediating the synthesis of capsular polysaccharides were down-regulated in MR100. However, genes encoding adhesins12 (isdB, srtB, ebh and sdrC) were down-regulated in MU30. In addition, genes related to metabolism (arcA, arcB2, arcD and aur), which are important to biofilm survival, were also down-regulated in the MU30 except for genes associated with purine metabolism (purN, purH and purD) (Fig. 5, Supplementary Table S13 online).
In the removing established biofilm conditions, genes associated with the QS systems of S. aureus (agrA, agrB, agrC, hld and sarA) were down-regulated in MR150 and MVR (Fig. 5c, Fig. 6, Supplementary Table S14 online). Moreover, genes encoding surface and secreted proteins (spa, sek, sdrD and efb) were up-regulated in both samples. All cap genes were highly down-regulated in both samples, but cap5M and cap5N were not differentially expressed in MVR. Meanwhile, genes associated with biofilm survival (arcA, carA, carB, pyrB, pyrC, pyrE, pyrF, pyrG and pyrR) were up-regulated in both samples. We also found that most differentially expressed genes related to virulence and biofilm in MR150 were consistent with those in MVR except for the genes associated with purine metabolism (purA, purD, purH and purN). In addition to above key genes, other key genes, such as clfA and spoVG genes related to virulence and biofilm formation, were differentially expressed in MR150 and MVR (Fig. 5c, Fig. 6, Supplementary Table S14 online).
Coverage changes of differentially expressed genes associated with methicillin-resistant Staphylococcus aureus (MRSA) biofilm and virulence in removing established MRSA biofilm conditions.
Red, blue and green lines represent M36C05, MVR and MR150, respectively. For the x-axis, the position of the genome increases from left to right and the length of each gene is proportional to the width of the position in the x-axis. The use of “..” indicates discontinuous gene segments.
In addition, in the removing established MRSA biofilm conditions, the expression of six genes associated with S. aureus biofilm that were up- or down-regulated by more than twofold in the RNA-seq experiment were determined by real-time RT-PCR. In MR150 sample compared to M36C05 sample, the levels of expression of sdrD and spa increased by 2.55- and 1.45-fold, respectively. The levels of expression of hld, agrA and cap5C changed slightly. The level of expression of cap5B decreased by 10-fold. In MVR sample compared to M36C05 sample, the level of expression of sdrD increased by 7.13-fold. The levels of expression of hld, cap5B and cap5C decreased by 17.87-, 16.24- and 5.67-fold, respectively (Supplementary Fig. S9 online). In general, the trends in regulating gene expression by RNA-seq technique are consistent with the results of real-time RT-PCR.
Discussion
Staphylococcus aureus subsp. aureus COL (S. aureus COL), a MRSA strain, is also resistant to several antibiotics including penicillin and tetracycline13 and shares the same phenotype as the MRSA strain. In addition, all of the MRSA transcripts were assignable to the S. aureus COL genome, which illustrated the suitability of using the S. aureus COL genome as a reference here. In addition, the next generation sequencing approach resulted in a massive amount of mRNA informative reads to a level at which all genes could be covered. The distribution of reads could be randomly mapped along the relative position through the genome in each sample (Supplementary Fig. S3 online), which illustrated the suitability of using RNA-seq for bacterial transcriptomic studies without bias and for the following comparative transcriptome analysis.
It has become clear that there are at least two mechanisms of biofilm formation in S. aureus14: one requires the production of the polysaccharide intercellular adhesion proteins, which is encoded by the ica locus15 and the other is an ica-independent mechanism16. Alternatively, we did not find any icaADBC genes that showed significant changes in inhibitory or removed conditions. As such, the biofilm formation of the clinical MRSA isolate studied here may belong to an ica-independent mechanism. Meanwhile, some of the best studied factors involved in the ica-independent biofilm processes are accessory gene regulator (agr) and staphylococcal accessory regulator A (sarA) global regulatory systems17. The agr locus of S. aureus is one QS cluster of five genes (hld, agrB, agrD, agrC and agrA) that up-regulates the production of secreted virulence factors, including the α-, β- and δ-hemolysins18. Of these virulence factors, δ-hemolysin, which is encoded by hld and derived from translation of RNA III, is uniquely regulated by agr operon19. Moreover, Sakoulas et al. indicated that δ-hemolysin production could be used as a maker of agr function20.
In the inhibiting biofilm formation conditions, we only found that hld of the QS systems was up-regulated, which indicates that resveratrol and ursolic acid may enhance MRSA agr function at the RNA level to inhibit biofilm formation. Pratten et al. discovered that hld expression was the highest at the base of the biofilm, where the highest numbers of bacteria were also observed21. However, Resch et al. reported that toxins were up-regulated in planktonic growth conditions4. In addition, Vuong et al. proposed that δ-hemolysin might also serve as a surfactant in vivo that prevents the adherence of staphylococcal cells to surfaces22. Surfactants may also decrease the adherence of staphylococci to biomaterials. Based on the results in this study, the conclusion that hld expression was up-regulated in MR100 and MU30 is appropriate. Moreover, rsbU encoding virulence factor was up-regulated in MR100, which also corresponds to the discovery of Resch et al. The expression level of spa was up-regulated in MR100, which was consistent with the microarray and real-time polymerase chain reaction comparison results23. In addition, cap genes, which mediate the synthesis of capsular polysaccharides inducing human infections24, are important to S. aureus biofilm formation9. In MR100, cap genes were partially down-regulated. As such, the above analysis suggested that resveratrol inhibited MRSA biofilm formation by disturbing QS systems and the synthesis of surface proteins and capsular polysaccharides, while ursolic acid inhibited MRSA biofilm formation by reducing the metabolism of some amino acids as well as the expression of adhesins. We also speculated that the mechanism by which ursolic acid inhibited MRSA biofilm formation could differ from that by which it inhibited Escherichia coli biofilm formation, which may affect sulfur metabolism (through cysB)25,26.
To date, two staphylococcal QS (SQS) systems have been described. Here we provide a relationship graph of SQS in S. aureus according to some studies27,28 (Supplementary Fig. S10 online). In the removing established MRSA biofilm conditions, the genes associated with QS were differentially expressed. Therefore, the use of resveratrol or its combination with vancomycin could reduce MRSA virulence and biofilm by impacting on the QS systems. Korem et al. demonstrated that in the absence of target of RNAIII-activating protein expression or phosphorylation, the gene expression levels for biofilm survival were reduced in arcABC, pyrR, pyrP, pyrB, pyrC, carA and carB using microarray27. In this study, however, arcA, carA, carB, pyrB, pyrC, pyrE, pyrF, pyrG and pyrR were up-regulated in both MR150 and MVR (Supplementary Table S14 online), a finding that was contradictory to the microarray results, which may result from different phenotype between S. aureus. The expression levels of genes encoding surface and secreted proteins were up-regulated, a finding that is consistent with the microarray results23. Reduced capsule gene expression may render an organism more sensitive to the host's immune response, namely phagocytosis, which may result in more rapid clearance of infection29. In MR150 and MVR, the reduction expression levels of genes encoding the synthesis of capsular polysaccharides is consistent with the microarray results9,23. Therefore, the mechanisms of resveratrol and its combination with vancomycin inhibiting the development of established MRSA biofilm may impact on the expression levels of genes related to QS, surface proteins, secreted proteins as well as the synthesis of capsular polysaccharides. Moreover, there are some genes associated with virulence and biofilm, such as clfA and spoVG12. In this study, clfA expression was dramatically down-regulated, which illustrated that resveratrol and its combination with vancomycin may reduce bacterial virulence. Inactivation of spoVG in a methicillin-resistant Staphylococcus epidermidis drastically decreased methicillin resistance and biofilm formation30.
We found that the expression level of spoVG was down-regulated in S. aureus after treatment with resveratrol or its combination with vancomycin. Therefore, the mechanisms of using resveratrol and its combination with vancomycin to inhibit the development of established biofilm may be similar. However, the effect of the combination on established biofilm was more obvious than the effects of resveratrol and vancomycin alone. The transcriptome analyses indicated that (i) the expression levels of the genes associated with purine metabolism (purADHN) were induced in MVR and vancomycin (MV) but not in MR150; and (ii) vancomycin inhibited the development of established MRSA biofilm by disturbing the expression of genes related to metabolism instead of those associated with QS, surface proteins, secreted proteins as well as the synthesis of capsular polysaccharide (Supplementary Table S15 online). As such, we speculated that these induced genes related to purine metabolism (purADHN) may be one reason why the use of combination resveratrol and vancomycin more effectively eliminates established biofilm.
In conclusion, the transcriptome analysis in this study suggested that ursolic acid and resveratrol may have different mechanisms of inhibiting MRSA biofilm formation. Resveratrol inhibiting biofilm formation may disturb QS, the synthesis of surface protein and capsular polysaccharides, whereas ursolic acid-induced inhibition of biofilm formation may reduce the metabolism of some amino acids and the expression of adhesins. These differences may explain why resveratrol inhibited the further development of established biofilm whereas ursolic acid only inhibited biofilm formation. In addition, the MRSA isolate is sensitive to vancomycin (See Methods section), but once biofilm was established, the effect of vancomycin against bacteria was sharply reduced. In contrast, the combination of resveratrol and vancomycin markedly increased efficacy against established biofilm (Table 1). Meanwhile, the transcriptome analysis showed that the tendency toward differential genes expression in MR150 was consistent with those of MVR (Fig. 4b).
In addition to affecting proteins synthesis in biofilm-associated cells, the presence of resveratrol and ursolic acid altered the expression of a number of genes encoding key virulence factors involved in the pathogenesis of S. aureus. Studies on the effects of two natural compounds on MRSA biofilm using RNA-seq suggested that ursolic acid could be used to prevent S. aureus biofilm formation but that amounts of the corresponding toxin (δ-hemolysin) may be increased, which is obviously counterproductive. Therefore, the combination of ursolic acid and QS blockers may be used to inhibit S. aureus biofilm formation. The combination of resveratrol with vancomycin may be used to inhibit the development of established biofilm. Further investigation with a large panel of MRSA isolates would be required to confirm these findings and determine whether ursolic acid and resveratrol may prove to be therapeutically useful in reducing the morbidity and mortality linked with S. aureus biofilm-mediated infections.
Methods
Strain and growth conditions
The clinical MRSA isolate used in this study was provided by the Clinical Laboratory Department of the First Affiliated Hospital, Nanjing Medical University, Nanjing, China. The strain was isolated from blood and was resistant to many antibiotics except for linezolid, teicoplanin and vancomycin (Supplementary Tables S16 online). Moreover, the isolate was identified using a MicroFlex LT instrument (Bruker Daltonics) according to the manufacturer's recommendations. Spectra were analyzed by Flexcontrol 3.0 software and Biotyper 2.0 database (Bruker Daltonics)31. Blood agar medium was used to culture the colonies. Nutrient broth (NB) medium was used for routine culturing of the strain, while trypticase soy broth (TSB) medium was used to study the effects of natural compounds on MRSA biofilm in 96-well flat-bottom polystyrene plates (Costar 3599; Corning; USA). The resveratrol and ursolic acid used in this study were isolated from natural products in our group and their purities were confirmed to be >98% using high performance liquid chromatography methods. They were dissolved in ethanol at 30 mg/mL and 5 mg/mL, respectively. The vancomycin used in this study was purchased from Sigma and dissolved in water at 5 mg/mL. All compounds were filtered with a 0.22-μm filter in sterile conditions and then stored at 4°C.
Minimal inhibitory concentration (MIC) and minimal bactericidal concentration (MBC) assays
MICs and MBCs were determined by a microtitre broth dilution methods as recommended by the Clinical and Laboratory Standards Institute (CLSI) with a few modifications. Briefly, the test medium was TSB and the density of bacteria was 5 × 105 colony forming units (CFU)/mL. Cell suspensions (200 μL) were inoculated into the wells with ursolic acid or resveratrol at different final concentrations (25, 30, 33, 35, 37 and 40 μg/mL for ursolic acid, 37.5, 75, 150, 300, 350 and 400 μg/mL for resveratrol). These concentrations were selected owing to the solubility reducing of ursolic acid and resveratrol in TSB medium as their concentrations increasing. The inoculated microplates were incubated at 37°C for 18 h before being read. The MIC was defined as the lowest concentration of the drug that inhibited the growth of the test microorganism by >90%32.
The MBC was obtained by subculturing 100 μL from each well from the MIC assay onto TSA plates. The plates were incubated at 37°C for 24 h and the MBC was defined as the lowest concentration of substance that produced subcultures growing no more than five colonies on each plate33. Experiments were carried out on three separate occasions.
Resveratrol and ursolic acid inhibiting MRSA biofilm formation assays
This assay was performed as previously described with a few modifications34. Briefly, one MRSA colony was used to inoculate 5 mL of NB medium (pH 7.2) and was grown in an orbital shaker for 24 h at 37°C. The culture was then diluted (1:100) in fresh TSB supplement with resveratrol or ursolic acid of which final concentrations were 100 μg/mL and 30 μg/mL, respectively, according to supplementary table S1 and table S2. Next, 200 μL samples were added to the wells of 96-well plates. The plates were incubated at 37°C for 18 h with shaking. We also prepared controls with or without ethanol to observe the effects of solvent (each well contained 0.6% [v/v] ethanol) on biofilm formation. For quantification of the total biofilm mass, the suspension cultures were decanted and the plates were washed three times with phosphate buffered saline (PBS) (pH 7.2). The remaining biofilm was fixed in 200 μL of methanol for 15 min and then dried at 60°C. The biofilm was stained with 50 μL of 0.1% crystal violet (w/v) for 15 min. The wells were also washed three times with PBS to remove unbound crystal violet dye and dried for 2 h at 60°C. After the addition of 200 μL of 95% ethanol (v/v) to each well, the plates were shaken for 1 h to release the stain from the biofilm and the absorbance at OD570 nm was measured with a microplate reader (BioTek, USA). All assays were performed in triplicate at least and repeated three times starting from new cultures. The inhibitory rates were then calculated using the following formula:

Treatment of MRSA biofilm with resveratrol, vancomycin, or their combination
After 18 h of biofilm growth, the suspension cultures were removed from each well. The plates were washed twice with sterile PBS. A total of 200 μL of fresh TSB with resveratrol (final concentration, 150 μg/mL,according to supplementary table S2), vancomycin (final concentration, 8 μg/mL), or combination (resveratrol 150 μg/mL + vancomycin 8 μg/mL) was added to the wells. Controls were prepared to observe the effects of the solvent (each well contained 0.5% [v/v] ethanol) on the established biofilm. The plates were incubated at 37°C for 18 h with shaking. The quantitative methods of their effects on established biofilm were the same as the methods of inhibiting biofilm formation. All assays were performed in triplicate at least and repeated three times starting from new cultures.
SEM measurements assay
SEM was performed on biofilm formed on glass coverslips (0.2 mm thick and 6 mm in diameter) by dispensing 700 μL of the cell suspensions into the wells of 24-well flat bottomed polystyrene plates (Costar 3524; Corning; USA). The plates were incubated at 37°C with shaking. The coverslips were then washed three times with water and fixed using 2.5% glutaraldehyde in 0.1 M sodium phosphate buffer (pH 7.2) for 4 h at 4°C. Next the coverslips were washed three times with 0.1 M sodium phosphate buffer for 15 min each, followed by three 50% ethanol (v/v) washed for 10 min each. The samples were freeze-dried for 5 h and ultimately coated with gold and palladium in an evaporator. The observations were usually performed with a scanning electron microscope (FEI Quanta 200; USA).
Total RNA isolation
All samples used for the RNA sequencing were prepared in the 24-well flat bottomed polystyrene plates. After the biofilms were rinsed, the compound-free and treated biofilm cells were scraped with pipettor and placed in the RNAprotect Bacteria Reagent (QIAGEN GmbH, Germany). The sessile cell suspension was then transferred to a microcentrifuge tube and incubated for 5 min at room temperature to stabilize the mRNA. Next, the cell suspensions were centrifuged at 8,000 × g for 5 min to pellet the cells and the supernatant was decanted. Total RNA was purified using RNeasy Mini Kit (Qiagen) according to the manufacturer's recommended protocol with some modifications (Supplementary materials online). Each total RNA sample was suspended in 30 μL of RNA storage solution and the quality of total RNA obtained was determined using Agilent 2100 bioanalyzer.
Enrichment and sequencing of mRNA
A total of 10 μg of each RNA sample was subjected to further purification to enrich the mRNA using a MICROBExpress Kit (Ambion) according to the manufacturer's instructions. Each mRNA sample was suspended in 25 μL of RNA storage solution and the quality of mRNA obtained was determined using Agilent 2100 Bioanalyzer. Bacterial mRNA was fragmented using a RNA fragmentation kit (Ambion) and the yield fragments were in the size range of 200–250 bp. Double-stranded cDNA was generated using the SuperScript Double-Stranded cDNA Synthesis Kit (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. An Illumina Paired End Sample Prep kit was used to prepare RNA-seq library according to the manufacturer's instructions. All of the samples were sequenced using the Hiseq2000 (Illumina, CA) sequencer at Beijing Genomics Institute at Shenzhen.
Analysis pipeline
Reads were aligned to Staphylococcus aureus subsp. aureus COL (S. aureus COL) (RefSeq accession number NC_002951.2) using the Burrows-Wheeler Alignment tool (BWA)35. The RNA-seq data analysis included the following stages. (1) If the pair-end reads satisfied N > 2% and low quality (quality value < 20) >50%, the reads were removed. In addition, if the terminal 20 bp consisted of N or was of low quality, the reads were removed. Using the above quality control (QC) standards, clean data were produced. (2) The clean data were aligned to S. aureus COL using BWA. (3) When some reads were unmappable or had incorrect alignment, they were removed. Based on the above standard, the QC of alignment was produced. (4) The commonly used fragments per kilobase of transcript per million mapped fragments (FPKM) incorporate normalization steps to ensure that expression levels for different genes and transcripts can be compared across runs11. Based on FPKM normalization, we performed analyses of coverage, distribution and differentially expressed genes (workflow shown in Supplementary Fig. S11 online).
Identification of differentially expressed genes
Differentially expressed genes were identified using Cuffdiff, a separate program in Cufflinks10, which calculated expression in two or more samples and tested the statistical significance of each observed expression changes between them11. Genes with an adjusted P value < 0.05, FDR < 0.001 and fold change > 2 were identified as being differentially expressed. Finally, CummeRbund11 was used to visualize and integrate all of the data produced by Cuffdiff analysis.
Real-time RT-PCR
Six genes (sdrD, spa, hld, agrA, cap5B and cap5C) that were up- or down-regulated by more than twofold in the presence of resveratrol and the combination with vancomycin under the removing established biofilm condition were selected to validate the data generated from RNA-Seq study by real-time RT-PCR. These genes were selected because they encoded a range of virulence factors, including those associated with biofilm formation and capsule synthesis.
The primer pairs used in real-time RT-PCR are listed in Supplementary Table S17. Total RNA was reverse transcribed into cDNA using the Takara RNA PCR kit (AMV) ver.3.0 kit (TaKaRa, Japan), according to the manufacturer's protocol. The resulting cDNAs were stored at −20°C until they were required. The real-time RT-PCR was carried out in a 20 μL volume using SYBR® Premix Ex Taq™ (Tli RNaseH Plus) kit (TaKaRa, Japan) as recommended by the manufacturer. These reactions were performed using the Applied Biosystems 7300 Real-time PCR System by using the following cycle parameters: 95°C for 30 sec, followed by 40 cycles of 95°C for 5 sec, 60°C for cap5C gene and 57°C for other genes for 30 sec and 72°C for 30 sec; and one dissociation step of 95°C for 15 sec, 60°C for 30 sec and 95°C for 15 sec. All measurements were independently conducted 3 times on 2 separate biological isolates. The specificity of primer sets used for real-time RT-PCR amplification was evaluated by melting curve analysis. The standard curve method was used for quantification against a known concentration of plasmid DNA, pMD18T. All primers used and sequences are listed in Supplementary Table S17.
Statistical analysis
At least three independent replicates of each 96-well plate experiment were performed. The biofilm inhibition results and real-time RT-PCR results were statistically analyzed using SPSS software version 18.0 (SPSS, Chicago, IL, USA). P values ≤ 0.05 were considered significant.
References
Mazmanian, S. K., Ton-That, H. & Schneewind, O. Sortase-catalysed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol Microbiol 40, 1049–1057 (2001).
Reid, G. Biofilms in infectious disease and on medical devices. Int J Antimicrob Agents 11, 223–226, 237–239 (1999).
Brackman, G., Cos, P., Maes, L., Nelis, H. J. & Coenye, T. Quorum sensing inhibitors increase the susceptibility of bacterial biofilms to antibiotics in vitro and in vivo. Antimicrob Agents Chemother 55, 2655–2661 (2011).
Resch, A., Rosenstein, R., Nerz, C. & Gotz, F. Differential gene expression profiling of Staphylococcus aureus cultivated under biofilm and planktonic conditions. Appl Environ Microbiol 71, 2663–2676 (2005).
Parsek, M. R. & Greenberg, E. P. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol 13, 27–33 (2005).
Kiran, M. D. et al. Discovery of a quorum-sensing inhibitor of drug-resistant staphylococcal infections by structure-based virtual screening. Mol Pharmacol 73, 1578–1586 (2008).
Resch, A. et al. Comparative proteome analysis of Staphylococcus aureus biofilm and planktonic cells and correlation with transcriptome profiling. Proteomics 6, 1867–1877 (2006).
Baker, J. et al. Copper stress induces a global stress response in Staphylococcus aureus and represses sae and agr expression and biofilm formation. Appl Environ Microbiol 76, 150–160 (2010).
Smith, K. et al. Influence of tigecycline on expression of virulence factors in biofilm-associated cells of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 54, 380–387 (2010).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28, 511–515 (2010).
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7, 562–578 (2012).
Clarke, S. R. & Foster, S. J. Surface adhesins of Staphylococcus aureus. Adv Microb Physiol 51, 187–224 (2006).
Gill, S. R. et al. Insights on evolution of virulence and resistance from the complete genome analysis of an early methicillin-resistant Staphylococcus aureus strain and a biofilm-producing methicillin-resistant Staphylococcus epidermidis strain. J Bacteriol 187, 2426–2438 (2005).
O'Gara, J. P. ica and beyond: biofilm mechanisms and regulation in Staphylococcus epidermidis and Staphylococcus aureus. Fems Microbiol Lett 270, 179–188 (2007).
Tu, Q. P. et al. Isolation and characterization of biofilm formation-defective mutants of Staphylococcus aureus. Infect Immun 75, 1079–1088 (2007).
Fitzpatrick, F., Humphreys, H. & O'Gara, J. P. Evidence for icaADBC-independent biofilm development mechanism in methicillin-resistant Staphylococcus aureus clinical isolates. J Clin Microbiol 43, 1973–1976 (2005).
Lauderdale, K. J., Boles, B. R., Cheung, A. L. & Horswill, A. R. Interconnections between Sigma B, agr and proteolytic activity in Staphylococcus aureus biofilm maturation. Infect Immun 77, 1623–1635 (2009).
Ji, G., Beavis, R. & Novick, R. P. Bacterial interference caused by autoinducing peptide variants. Science 276, 2027–2030 (1997).
Morfeldt, E., Tegmark, K. & Arvidson, S. Transcriptional control of the agr-dependent virulence gene regulator, RNAIII, in Staphylococcus aureus. Mol Microbiol 21, 1227–1237 (1996).
Sakoulas, G. et al. Accessory gene regulator (agr) locus in geographically diverse Staphylococcus aureus isolates with reduced susceptibility to vancomycin. Antimicrob Agents Chemother 46, 1492–1502 (2002).
Pratten, J., Foster, S. J., Chan, P. F., Wilson, M. & Nair, S. P. Staphylococcus aureus accessory regulators: expression within biofilms and effect on adhesion. Microbes Infect 3, 633–637 (2001).
Vuong, C., Saenz, H. L., Gotz, F. & Otto, M. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J Infect Dis 182, 1688–1693 (2000).
Beenken, K. E. et al. Global gene expression in Staphylococcus aureus biofilms. J Bacteriol 186, 4665–4684 (2004).
Lowy, F. D. Staphylococcus aureus infections. N Engl J Med 339, 520–532 (1998).
Ren, D. et al. Differential gene expression for investigation of Escherichia coli biofilm inhibition by plant extract ursolic acid. Appl Environ Microbiol 71, 4022–4034 (2005).
Lynch, A. S. & Abbanat, D. New antibiotic agents and approaches to treat biofilm-associated infections. Expert Opin Ther Pat 20, 1373–1387 (2010).
Korem, M., Gov, Y., Kiran, M. D. & Balaban, N. Transcriptional profiling of target of RNAIII-activating protein, a master regulator of staphylococcal virulence. Infect Immun 73, 6220–6228 (2005).
Laverty, G., Gorman, S. P. & Gilmore, B. F. Biomolecular mechanisms of staphylococcal biofilm formation. Future Microbiol 8, 509–524 (2013).
O'Riordan, K. & Lee, J. C. Staphylococcus aureus capsular polysaccharides. Clin Microbiol Rev 17, 218–234 (2004).
Bischoff, M. et al. Microarray-based analysis of the Staphylococcus aureus sigmaB regulon. J Bacteriol 186, 4085–4099 (2004).
Szabados, F., Woloszyn, J., Richter, C., Kaase, M. & Gatermann, S. Identification of molecularly defined Staphylococcus aureus strains using matrix-assisted laser desorption/ionization time of flight mass spectrometry and the Biotyper 2.0 database. J Med Microbiol 59, 787–790 (2010).
Oo, T. Z., Cole, N., Garthwaite, L., Willcox, M. D. & Zhu, H. Evaluation of synergistic activity of bovine lactoferricin with antibiotics in corneal infection. J Antimicrob Chemother 65, 1243–1251 (2010).
Wang, X. et al. Effect of berberine on Staphylococcus epidermidis biofilm formation. Int J Antimicrob Agents 34, 60–66 (2009).
O'Toole, G. A. & Kolter, R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol 28, 449–461 (1998).
Li, H. & Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Acknowledgements
The study was supported by the National Natural Science Foundation of China (No. 31170131, 31070312) and Jiangsu Qinglan Project.
Author information
Authors and Affiliations
Contributions
N.Q., X.T. and Y.J. contributed equally to this work. A.J. and N.Q. conceived and designed the study. X.T. and W.Z. performed experiments. N.Q., L.L. and S.Y. performed sequencing. Y.J. designed the analysis and performed comparative genomics. X.T. and Y.J. analyzed the differentially expressed genes. X.T., N.Q. and A.J. wrote and revised the paper.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Qin, N., Tan, X., Jiao, Y. et al. RNA-Seq-based transcriptome analysis of methicillin-resistant Staphylococcus aureus biofilm inhibition by ursolic acid and resveratrol. Sci Rep 4, 5467 (2014). https://doi.org/10.1038/srep05467
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep05467
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.