Abstract
Beta-lactams are used as major therapeutic agents against a number of infectious agents. Due to widespread use of β-lactams, β-lactamases have evolved at a rapid pace leading to treatment failures. Yersinia enterocolitica causes many gastrointestinal problems. It is an extremely heterogeneous species comprising more than fifty serotypes and six biotypes which differ in their ecological niches, geographical distribution and pathogenic potential. Though biotype 1A strains have been associated with outbreaks of Yersiniosis, there has been a controversy regarding their pathogenicity. The strains of Y.enterocolitica isolated from India belonged to biotype 1A and possessed genes for two β-lactamases namely, blaA and blaB. An earlier study by us reported differential expression of blaA by strains of Y. enterocolitica biotype 1A. The present study has been carried out to understand the molecular bases which regulate the expression of blaA in Y. enterocolitica biotype 1A. We concluded that six types of blaA variants were present in strains of biotype 1A. Neither amino acid substitutions in blaA nor mutations in promoter regions of blaA contributed to differential expression of blaA in Y. enterocolitica biotype 1A. Rather, the secondary structures attained by mRNA of blaA might underlie the differential expression of blaA in Y. enterocolitica.
Similar content being viewed by others
Introduction
Beta-lactamases and their mutated forms such as extended spectrum β-lactamases (ESBLs), AmpC β-lactamases, carbapenemases and metallo β-lactamases represent a major cause of multidrug resistance among enteric bacteria. Following selective pressure due to widespread use of β-lactam antibiotics in clinical medicine, genes for β-lactamases have evolved at a very rapid pace with over 1300 unique β-lactamases reported in clinical isolates1. Yersinia enterocolitica, a member of the family Enterobacteriaceae is an important zoonotic pathogen. It is an extremely heterogeneous species comprising fifty serotypes and six biotypes viz. 1A, 1B, 2, 3, 4 and 5 which differ not only in their geographical distribution and ecological niches but also in pathogenic potential2. On the basis of pathogenicity, Y. enterocolitica can be divided into highly pathogenic biotype 1B strains, moderately pathogenic biotype 2–5 strains and supposedly non-pathogenic biotype 1A strains. Batzilla et al.3 proposed that biotype 1A strains represent a potential group of emerging pathogens which share known and putative virulence-associated features with the pathogenic bioserotypes. Studies have shown that strains classified as biotype 1A, in fact, represent more than one subspecies3. Despite the lack of classical determinants of virulence in biotype 1A strains4 these have been frequently isolated from clinical samples across the globe5,6,7,8.
In India, the first food-borne outbreak due to Y. enterocolitica was reported from Tamil Nadu in 19979. Since then, this pathogen has been isolated from a variety of sources – wastewater, pork, pigs, diarrheic human patients, ice cream, dairy products and even traditional fast food6,9,10,11,12,13. The strains of Y.enterocolitica isolated from India6,11 principally belonged to biotype 1A. The strains of Y. enterocolitica are known to possess two β-lactamases, namely β-lactamase A (blaA) and β-lactamase B (blaB)14,15. BlaA is a broad-spectrum constitutively expressed Ambler class A enzyme where as blaB is an inducible Ambler Class C enzyme16.
The most common mode of microbial resistance to β-lactams is their enzymatic hydrolysis by β-lactamases. Over the time, β-lactamases have expanded their substrate spectrum with simple mutations in the gene or in the genetic environment of β-lactamases. Modifications in the regulatory regions, mutations in the promoter sequences and integration of insertion sequences containing efficient promoters have frequently been associated with high-level expression of chromosomal β-lactamases17. Studies on bla genes of Y. enterocolitica have reported differences in β-lactamase expression and β-lactam susceptibility among different biotypes, focusing primarily on ‘pathogenic biotypes’18,19,20,21. The previous study on molecular characterization of bla genes and differences in β-lactamase expression in biotype 1A strains was reported from our laboratory14,15. Previous studies from our laboratory also reported that the expression of blaB in biotype 1A strains was quite similar among the strains, while the expression of blaA was quite variable among different strains14,15. To the best of our knowledge, till date no study has been carried out to understand the differential regulation of expression of blaA in Y. enterocolitica biotype 1A strains. The present study is an extension of our previous observations and was undertaken to understand the molecular basis of differential expression of β-lactamase genes of biotype 1A strains of Y. enterocolitica. This study also provides insights into the blaA variants in biotype 1A strains of Y. enterocolitica. Thus, the promoter regions and the structural regions of blaA genes in biotype 1A strains of Yersinia enterocolitica isolated from India and from other parts of the world were PCR-amplified, sequenced and compared. Three dimensional structures of blaA variants were modeled in-silico to co-relate the role of specific substitutions in differential expression of β-lactamases.
Results
Minimal Inhibitory Concentrations of β-lactam antibiotics
Y.enterocolitica strains showed variable susceptibilities to β-lactam antibiotics of different generations (Table 1). All biotype 1A strains of Indian and non-Indian origin except Y.enterocolitica strain E3 were resistant to amoxicillin and amoxicilln-clavulanate, antibiotic-inhibitor combination. The MIC of all biotype 1A strains to amoxicillin exceeded the upper range of E-test i.e > 256 mg/L. For six Indian strains, the MIC of amoxicilln-clavulanate combination also exceeded the upper range of E-test. The susceptibility of Y.enterocolitica to the third generation cephalosporins, cefpodoxime and cefotaxime was also tested by E-test. Antibiotic susceptibility of Y.enterocolitica strains to the third generation cephalosporin cefpodoxime varied and ranged from 0.125–8 mg/L. While most of the biotype 1A strains were either sensitive or intermediate to cefpodoxime, four strains were resistant to this antibiotic. Further, all Y. strains were sensitive to third generation cephalosporin, cefotaxime. For biotype 1A strains the MIC was lowest for cefotaxime and ranged between 0.032–0.64 mg/L. Strain E3 showed lowest MIC to all antibiotics including amoxicillin and amoxicillin-clavulanate.
Specific activities of β-lactamases
The mean specific activities of the blaA variants produced by Y. enterocolitica biotype 1A strains expressed as amount of nitrocefin hydrolyzed (μmol/min/mg of protein) are summarized in Table 2. The specific activity of the variant blaA2 was quite high than the mean specific activities of other variants, while that of blaA4 was the least. The mean specific activities of the variants blaA1 and blaA3 were similar to each other.
Promoter sequences and CCDS of blaA gene
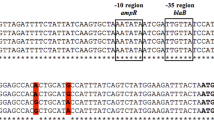
The amplification of blaA gene using primers blaA11 and blaA12 resulted in the expected amplicon of 1.49 kb in 68 out of 81 strains of Y. enterocolitica. The band was excised from the agarose gel and sequenced after purification using QIAGEN purification kit. The BLAST analysis of the sequences confirmed these to be part of the blaA gene. Multiple sequence alignment of the promoter regions revealed that promoter sequences of all biotype 1A strains were identical (Fig. 1). The amplification of blaA gene using published primers15 resulted in the amplification of desired product of 896 bp in all the strains. The band was excised from gel, purified using QIAGEN purification kit and sequenced. CCDSs of blaA were translated into their corresponding amino acid sequences and analyzed. Comparison of amino acid sequences of blaA using multiple sequence alignment revealed that though the sequence was conserved, some substitutions were identified. Since the initial 30 amino acids constitute the signal sequence and are cleaved before the mature enzyme is released in the periplasmic space, these were excluded from comparative studies. P32S and G60C was observed in 12 of 35 strains viz.Y. enterocolitica strains 12, 52, 8, 9, 42, 47, 48, 109, 111, E8, 10, 11; G58A in Y. enterocolitica strains E2, E4, E3, 29, 30, E7; R69Q in Y. enterocolitica strains E6, E2, E4, E3, 29, 30, E7; L98T and N101D in Y. enterocolitica strain 19. I113V and T225S in Y. enterocolitica strain E4; L144V in Y. enterocolitica strains E2 and E4. The mutation sites in individual strains are shown in Table 3 and summarized in Fig. 2.
Phylogenetic analysis of blaA of Y. enterocolitica strains
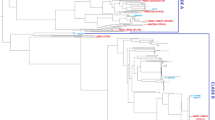
The phylogenetic analysis of blaA of 35 strains of Y. enterocolitica biotype 1A was carried using the Maximum Likelihood method of MEGA5.5. The tree with the highest log likelihood (−939.3021) is shown (Fig. 3). The tree is drawn to scale, with branch lengths represented by the number of substitutions per site. The MEGA analysis clustered isolates into three major groups.
Modeling analysis
Based on the identity percentage and E-values (expectation values), β-lactamase of Burkholderia multivorans (PDB code 3W4Q_A; Uniprot ID A9ANW2) was selected as the template protein due its high sequence similarity, identity and low E-value. Of the 20 models built using MODELLER, the 3D model with the lowest modeler objective function was selected as the final model.
Protein structure validation by PROCHECK and VERIFY3D
The results of PROCHECK as represented in Ramachandran contour plot showed that ca. 90% of the residues lay in the most favoured regions and only 1% of residues in disallowed regions. Verify3D profiles which were used to identify unreliable regions that had been modeled improperly showed that 90% region of the protein model of each blaA variant scored > 0.2 which was highly significant.
Molecular docking
Evaluation of binding affinity of β-lactam antibiotics with class A β-lactamases was performed using AutoDock Vina. The binding poses for each ligand molecule (amoxicillin, clavulanic acid) into the structures were determined and different poses were generated based on the total score (Dock score). The negative low free energy of binding of docked complexes and low inhibition constants indicated high affinity of β-lactamase for these antibiotics (Table 4 & 5, Fig. 4).
Molecular docking analysis of the six blaA variants with amoxicillin and clavulanic acid.
Molecular interactions of docked blaA1 (a & b), blaA2 (c & d), blaA3 (e & f), blaA4 (g & h), blaA5 (i & j), blaA5 (k & l) and blaA6 (m & n) with amoxicillin and clavulanic acid respectively. The two antibiotics are represented by stick and the interacting amino acids are shown in dark color.
mRNA secondary structure analysis
The secondary structures of mRNA of the six blaA variants were analyzed. These formed branched secondary structures. The structure of blaA2 showed very high level of branching, with a number of bulge loops, interior loops and a smaller region of double stranded RNA helix. On the other hand the mRNA secondary structure of blaA4 was mostly composed of a double-stranded RNA helix of stacked base pairs with little branching (Fig. 5).
Discussion
The MIC of different β-lactam antibiotics for Y. enterocolitica biotype 1A strains using E-test revealed that all strains were resistant to amoxicillin and amoxicillin-clavulanate. Y. enterocolitica strains have been reported to be ‘naturally resistant’ to ampicillin30,31. Although the MIC of amoxicillin-clavulanate was perceptibly less than that of amoxicillin, it did not change the resistance phenotype, implying the existence of inhibitor resistant blaAs in Y. enterocolitica biotype 1A. Earlier studies have shown that Y. enterocolitica strains of serotypes O: 3, O: 5 and O: 931 were all resistant to ampicillin and a few were resistant to amoxicillin-clavulanate. Szych et al.32 reported that though all strains of bioserotypes 4/O: 3 and 1B/O: 8 were resistant to ampicillin, yet 90% of these were sensitive to amoxicillin-clavulanate. They also reported that strains of bioserotype 2/O: 9 were resistant to both ampicillin and amoxicillin-clavulanate. Recent studies8,33 also showed that strains of Y.enterocolitica bioserotype 4/O: 3 were sensitive to amoxicillin-clavulanate where as that of bioserotypes 2/O: 5, 27 and 2/O: 9 were resistant. From the available information it is apparent that both the biotype and serotype play role in affecting the susceptibility of Y. enterocolitica to amoxicillin-clavulanate. Since all strains in the present study belonged to biotype 1A, it is not possible to comment on the influence of biotype on antibiotic susceptibility; but based on our findings it may be concluded that serotype had no effect on the susceptibility of Y. enterocoloitica biotype 1A strains to amoxicillin-clavulanate. The biotype 1A strains were either sensitive or showed intermediate susceptibility to the third generation cephalosporin, cefpodoxime and only a few strains were resistant. This is the same as observed in an earlier study8. Y. enterocolitica strain E3 was sensitive to all the β-lactam antibiotics tested and showed lowest MIC to all the β-lactam antibiotics tested including amoxicillin and amoxicillin-clavulanate combination.
The mean specific activities of variants blaA1 and blaA3 were almost similar and the mutations/substitutions which lead to these variants had no effect on the enzyme activity. Similarly mutations that generated variants blaA5 and blaA6 did not affect the enzyme activity significantly. However, the mutations leading to variants blaA2 and blaA4 significantly increased or decreased the specific activities of the enzyme respectively.
Bacteria have been reported to hyper produce β-lactamases by different mechanisms viz. mutation in the promoters, integration of insertion sequences containing efficient promoters or point mutations in the structural gene17. In the family Enterobacteriaceae, mutations and insertions, mostly in the promoter regions of the β-lactamase genes, have been associated with hyper production of β-lactamases34,35,36. In the present study, we attempted to understand the molecular basis of differential expression of chromosomal blaA of biotype1A strains of Y. enterocolitica. For this, the nucleotide sequences of promoter regions of blaA of biotype 1A strains were PCR-amplified and analyzed. Genetic events that affect the transcription efficacy of bacterial promoters usually occur in the −35 or −10 regions or sometimes in the space between these regions. It is well known that promoter strength is linked to the sequence of the −35 and −10 boxes, creating the 70 bp contact region for RNA polymerase. The −10 sequence TATAAT and the −35 sequence GTGGTG of blaA of Y. enteocolitica were similar to the canonical E.coli sequence indicating these were strong promoters. Alignment and comparison of blaA of biotype 1A strains by Clustal Ώ revealed that the ribosomal binding site (RBS), the −10 region and −35 regions were identical in all strains indicating that the promoter region had no role in differential expression of β-lactamases. Thus it was argued that the differential expression of class A beta-lactamase enzyme in Y. enterocolitica biotype 1A might be due to the changes in gene sequence of blaA.
To investigate the role of changes in gene sequence of blaA in differential expression of enzyme, blaA genes of biotype 1A strains were PCR-amplified and their CCDSs translated into corresponding amino acid sequences. The four significant motifs conserved across the Ambler class A β-lactamase viz. SXXK, SDN, EXXLN and KTG, (where X represents any amino acid) were present in blaA of all Y. enterocolitica strains, their respective sequence and position being 75STFK78, 136SDN138, 171EPDLN175 and 239KTG241. One of the four conserved domains 171EPDLN175 was located in the omega loop, a structural domain constituting a part of active-site pocket. Clustal Ώ alignment and comparison of amino acid sequences of structural β-lactamase blaA of biotype 1A strains revealed that amino acid substitutions were present at some places. Since the initial 30 amino acids constitute the signal sequence and are cleaved before the mature enzyme is released in the periplasmic space, these were excluded from the comparative studies. The most frequent point substitutions observed in 12 of the 35 Y. enterocolitica strains were a substitution from P32S and G60C. As proline and serine are both polar amino acids, substitution of one with the other will affect only the bulkiness of the side chain of the protein. Glycine is a simple nonpolar amino acid its replacement with cysteine, a polar sulfur containing amino acid may affect the properties of the side chain of the protein. The substitution of glycine with alanine G58A in Y. enterocolitica strains would also affect only the bulkiness of the side chain because both are nonpolar aliphatic amino acids. As arginine is a positively charged amino acid, while glutamine is a polar uncharged amino acid, substitution of R69N in Y. enterocolitica strains may also affect enzyme activity. Lysine is a positively charged amino acid while threonine is a polar uncharged amino acid; substitution K98T is expected to affect the side chain of the protein. Asparagine is a polar uncharged amino acid, while aspartate is a negatively charged amino acid; substitution of N101D in Y. enterocolitica strain 19 may influence enzyme activity due to change in the properties of the side chain. Substitution of I111V and T225S in Y. enterocolitica strain E4 and L144V in Y. enterocolitica strains E2 and E4 would also affect only the bulkiness of the side chain of the protein without any profound effect on enzyme activity because of the similarity in the chemical nature of the substituent and the substitute. For example isoleucine and valine are nonaliphatic amino acids, threonine and serine are polar amino acids and leucine and valine both nonpolar amino acids.
Phylogenetic analysis based on amino acid sequences of blaA clustered the biotype 1A strains into 3 major groups. In our earlier study also the Y. enterocolitica biotype 1A strains clustered into three groups based on the profiles generated by the restriction analysis of blaA15.
The analysis of amino acid sequences of blaA of Y. enterocolitica biotype 1A strains showed that although none of the substitution/mutation occurred in the four major motifs conserved across class A β-lactamases, or in the omega-loop region of blaA, yet there were differences in the specific activities of blaA. Amino acid substitutions which do not occur in the active site of the enzyme may create diverse local changes in the 3D structure of the enzyme and increase its conformational flexibility which may in turn affect the binding affinity with the substrate. To evaluate the role of such amino acid substitutions on binding affinity we classified the blaA of biotype 1A strains of Y. enterocolitica into six variants based on the mutation/substitution in the amino acid sequence. Three dimensional modeling of the blaA variants was carried out to co-relate structural changes with the higher enzyme specific activities and MIC of different strains. In-silico docking of the blaA variants was carried out with amoxicillin and clavulanic acid to analyze if amino acid substitutions affected the residues involved in hydrogen bonding and hydrophobic interactions in the enzyme-ligand complex. It is well known that hydrogen bonds and hydrophobic interactions not only play a crucial role in molecular recognition and the overall stability of the protein structure; these also stabilize the protein-ligand complexes. Amoxicillin and clavulanic acid were chosen for docking experiments as both are penicillins and blaA is a penicillinase. Intermolecular hydrogen bonds formed between the modeled structures of blaA variants and amoxicillin/clavulanic acid were determined. Pymol analysis revealed that each blaA variant had its own drug-interacting active-site amino acids. In all blaA variants S-135, A-137 and S-242 participated in hydrogen bonding with amoxicillin and S-135 in hydrogen bonding with clavulanic acid. Kumar et al.37 reported that the number of the residues involved in hydrogen bond interactions was an indicator of binding affinity of the enzyme with the ligand which might be decreased in mutant forms of class A β–lactamase. However, in our study no co-relation was observed between the number of residues involved in hydrogen bond interactions in blaA variants and their MIC for amoxicillin or clavulanic acid. Yi et al.38 also observed that the cetadazidime MIC and the relative frequency of substitutions in PenA, a class A β–lactamase of Burkholderia thailandensis did not co-relate well. It may be worthwhile to mention that the Y. enterocolitica isolates used in the present work were isolated from environmental as well as clinical settings. Variations/mutations associated with their respective blaAs might have evolved over time either in nature or in clinical settings in patients treated with antibiotics. Salverda et al.39 have also reported that most of the mutations that evolved in β-lactamases in clinical settings do not clearly co-relate with the stability or enhanced hydrolytic capability of the enzyme.
Variation in amino acid sequence is not the only factor that might contribute to differences in specific activities of β-lactamases. Enzyme activity may also be affected at the level of translation. In this regard, the secondary structure of mRNA plays an important role. When the secondary structures of mRNA of blaA variants were predicted using the web server mfold it was found that except for blaA4, all variants formed branched secondary structures. The mRNA secondary structure of blaA2 showed extensive branching and a lesser region of double stranded RNA helix compared to other blaA variants. On the other hand the mRNA secondary structure of blaA4 comprised mainly of double-stranded RNA helix of stacked base pairs with little branching. Several studies have shown that longer and stronger-paired zones in mRNA secondary structure tend to be more stable, preventing the ribosome from breaking the pairing and helping translation to proceed40,41. A lower translation rate means lesser number of copies of protein and hence a lower enzyme specific activity. Thus the secondary structures attained by mRNA of blaA2 and blaA4 might be the probable reasons for the higher and lower specific activities of their respective β-lactamases.
Overall the present work revealed that variations in promoter regions were not responsible for differential expression of blaA in biotype 1A strains of Y. enterocolitica. Analysis of amino acid sequences revealed that six types of blaA variants were present among biotype 1A strains of Y. enterocolitica.In-silico analysis of the blaA variants showed that amino acid substitutions did not contribute to the differential expression of the enzyme activities either. Rather the secondary structures attained by mRNA of blaA might underlie the differential expression of blaA in Y. enterocolitica. Further studies on secondary structures of mRNA of blaA gene will be helpful in better understanding the mechanisms that might underlie the differential expression of β-lactamases in Y. enterocolitica.
Methods
Bacterial strains
A total of 81 strains of Y. enterocolitica belonging to biotype 1A maintained in our laboratory (University of Delhi South Campus) were examined. Of these, sixty-five strains were isolated from India, which comprised 35 clinical, 18 wastewater, 7 swine and 5 pork isolates. All isolates were authenticated by and have been deposited with Yersinia National Reference Laboratory and WHO Collaborating Center, Pasteur Institute, Paris (France). All these strains have also been deposited with the National repository i.e. Microbial Type Culture Collection (MTCC) and Gene Bank located at Institute of Microbial Technology, Chandigarh. Ten strains of Y. enterocolitica biotype 1A belonging to European and American origin were obtained from E. Carniel (Yersinia National Reference Laboratory and WHO Collaborating Center, Pasteur Institute, Paris, France). Another six biotype 1A strains of European origin were provided by J. Heesemann (Max von Pattenkofer Institute, Munich, Germany). The serovars, source of isolation, country of origin and reference laboratory accession numbers of these strains have been reported previously22. All strains were maintained on trypticase soy agar at 4°C.
Determination of the Minimal Inhibitory Concentration (MIC)
MIC of amoxicillin, amoxicillin-clavulanate, cefotaxime and cefpodoxime was determined for 25 strains of Y. enterocolitica biotype 1A using E-test (bioMerieux Inc., MO, USA) following the manufacturer's instructions. Briefly, each strain was grown on Muller-Hinton agar plates at 25°C for 18–24 h. Single colonies from the agar plates were suspended in 1 ml of normal saline until a turbidity of 0.5 McFarland standard was reached (equivalent to ca.1–2 × 108 CFU/ml). Sterile glass spreader was used to spread 100 μl of the cell suspension on the surface of Muller-Hinton agar plates. The E-test strips were placed on the surface of inoculated Muller-Hinton agar plates and incubated at 28°C for 16–18 h. The MIC values were recorded as the minimum concentration of the antibiotic (μg/ml) where no visible growth of the test organism was observed and were interpreted according to the guidelines of Clinical and Laboratory Standards Institute23.
Spectrophotometeric assay of β-lactamases
Thirty-five strains of Y. enterocolitica biotype 1A were used for assaying β-lactamase activity as reported by Stock and Weidmann18 with slight modification. A loopful of bacterial growth was inoculated in 10 ml of LB broth and incubated overnight at 28°C. The bacteria were sedimented by centrifugation at 4°C, washed thrice with chilled phosphate buffer (0.05 M, pH 7.0) and finally resuspended (1 g wet weight) in 5 ml of sonication buffer. The cells were disrupted by sonication on ice with three pulses of 30 sec each. The clear cell lysate, prepared by centrifugation at 12,000 g for 30 min at 4°C was used to assay the β-lactamase activity. The protein content of the lysates was estimated by Bradford's method24. β-lactamase activity was assessed spectrophotometrically by hydrolysis of nitrocefin. The assay mixture contained 110 μl nitrocefin (0.5 μg/μl), 30 μg of crude cell lysates containing β-lactamase in a final volume of 1.0 ml of 50 mM phosphate buffer (pH 7.0). β-lactamase activity was monitored by measuring decrease in the optical density at 486 nm for 10 min at 28°C. The specific activity of the enzyme was expressed as μmol of nitrocefin hydrolyzed/min/mg of protein and was calculated based on the molar extinction coefficient of 20,500 M−1cm−1 for nitrocefin25.
Preparation of genomic DNA
Each strain was grown overnight in trypticase soy broth at 28°C. One ml of the culture was pelleted at 8,000 × g for 10 min. The genomic DNA was prepared from each strain by using DNeasy Tissue kit (Qiagen, Hilden, Germany) with modifications for Gram-negative bacteria according to the manufacturer's recommendations. Purified DNA was eluted in sterile water and quantitated spectrophotometrically at 260 nm.
Primer designing for PCR amplification of blaA gene including the promoter and downstream region
For amplification of partial coding sequence of blaA including its promoter, primers A11f 5′CAAATCGCCAGCAGAACAGA 3′ and A12r 5′GGGCAGCGAGCAGGGGTAAA 3′ were designed by retrieving the whole genome sequence of Y. enterocolitica biotype 1B and the corresponding sequence of blaA along with the 350 bp each upstream and downstream of the gene. The primers were synthesized from Microsynth GmbH (Balgach, Germany).
PCR amplification of the promoter region and partial coding sequence of blaA was carried out for 81 strains of Y. enterocolitica in a Thermal Cycler (My Cycler™, Bio-Rad, CA, USA). The PCR reaction mixture comprised of 1× PCR buffer (10 mM Tris-HCL, 1.5 mM MgCl2, 1.5 mM KCl and 0.1% Triton X-100), 200 μM of each of the four dNTPs (MBI Fermentas GmbH, Germany), 10 pmol each of forward and reverse primers, 2 U of Taq DNA polymerse (NewEngland Biolabs, MA, USA) and 50–100 ng of genomic DNA in a total volume of 25 μl. For PCR, the initial denaturation was performed at 95°C for 5 min followed by 25 consecutive cycles of 30 sec at 95°C, 30 sec at 56°C and 90 sec at 72°C and a final extension for 10 min at 72°C. The PCR products were analyzed by electrophoresis on 1% (w/v) agarose gel. The gels were stained with ethidium bromide (0.5 μg/ml) and visualized under UV transilluminator.
PCR amplification of blaA gene
PCR amplification of complete coding sequence (CCDS) of blaA was performed for 35 strains using published primers15 in a Thermal Cycler (My Cycler™, Bio-Rad, CA, USA). The PCR conditions were same as that for partial coding region of blaA and its flanking regions.
Sequencing of CCDS of blaA and its upstream and downstream regions
PCR amplicons were excised from gel, purified using QIA Quick Gel Extraction Kit (Qiagen, Hilden, Germany). The purified products were sequenced using the Big Dye Terminator Cycle Sequencing Ready Reaction kit and analyzed by capillary electrophoresis on an ABI PRISM 310 Genetic Analyzer (Applied Biosystems, CA, U.S.A). The nucleotide sequences amplified using published primers15 were aligned with those sequences which were amplified using primer pair A11f and A12r. The sequences which were common to the amplicons obtained using the two sets of primers and the extra sequences which were not common were used to construct the full sequence of blaA including the upstream and downstream sequences. CCDSs of blaA of 35 strains of Y. enterocolitica biotype 1A were translated into their corresponding amino acid sequences using the software expasy (www.expasy.org/translate). The amino acid sequences of blaA were aligned by Clustal Ώ. (www.ebi.uc.ak/clustal Ώ).
Phylogenetic analysis of blaA of Y. enterocolitica strains
The amino acid sequences of blaA of 35 strains of Y. enterocolitica strains biotype 1A were also analyzed from evolutionary perspectives using the software MEGA5.2 (Molecular Evolutionary Genetic Analysis). The evolutionary history was inferred by using the Maximum Likelihood method based on the JTT matrix-based model (www.megasoftware.net).
Selection of Y. enterocolitica strains for modeling of blaA
To investigate the role of point substitutions in amino acid sequence in differential expression of blaA, analysis of protein models of blaA variants was carried out. After analysis by ClustalΏ, amino acid sequences of blaA of Y. enterocolitica strains representing type/(s) of substitution/(s) were selected for modeling of their β-lactamases. The blaA of Y. strains 8, 19, 20, 29, E2 and E4 were selected for modeling and accordingly named as blaA1, blaA2, blaA3, blaA4, blaA5 and blaA6.
Homology modeling of β-lactamases
Since the three dimensional (3D) structure of blaA of Y. enterocolitica is not known, the 3D structures of β-lactamase variants blaA1, blaA2, blaA3, blaA4, blaA5 and blaA6 were predicted by homology modeling. The pairwise alignment between the target and template sequences was performed with PDB-BLAST. Three-dimensional structures of blaA1, blaA2, blaA3, blaA4, blaA5 and blaA6 were built using the software MODELLER 9.12 (http://salilab.org/modeller/). Twenty models satisfying the spatial restraints based on target-template alignment were built for each protein, of which the 3D model with the lowest modeler objective function was selected. Following the modeling process, the structure was validated by PROCHECK and VERIFY3D. The PROCHECK program performs a detailed analysis of the stereochemistry of the modeled protein structure in the form of Ramachandran plot by plotting the phi-psi torsion angles. Population of phi-psi torsion angles in the plot gives an assessment of the overall quality of the modeled structure and also highlights regions that may need further investigation26. The quality of the three-dimensional models was also assessed by VERIFY3D, which tests the compatibility of a protein structure with its amino acid sequence. For high-resolution, experimentally determined structures, the VERIFY3D scores are positive and consistently high i.e > 0.227.
Molecular docking
Molecular docking analysis of the modeled protein structures was carried out to identify if substitutions in the amino acid sequences of blaA resulted in variations in their binding affinity to β-lactam antibiotic amoxicillin and the β-lactamase inhibitor clavulanic acid. The structures of amoxicillin and clavulanic acid were retrieved from the PubChem compound database (http://pubchem.ncbi.nlm.nih.gov/)28. Subsequently the downloaded SDF file was converted to PDB format using iBabel. Protein structures were pre-processed for docking using latest AutoDockTools (ADT) version by adding polar hydrogen atoms, removing all non-protein molecules from the modeled structure, adding Kollman charges to the protein and converting it to PDBQT format. Ligand structures were also prepared using ADT, using the default method for preparing ligands for docking that added hydrogen and charges. The default rotatable bonds were accepted as well and the structure was converted to PDBQT format. The search space for docking was determined visually by centering the Grid Box to the known binding site of the ligand and expanding the dimensions of the cubic search space to completely encompass the site. Docking was performed using AutoDock Vina (http://vina.scripps.edu/index.html) with default parameter settings other than the search space specification described above and the mean predicted binding affinity from the set of predicted binding poses was accepted as the true binding affinity for each docking run. The maximum number of poses per ligand was set to 10. The predicted binding of the ligand was compared to the known binding sites of β-lactamase in order to make predictions about differential responses with respect to each of the drugs. Interaction between ligand and target protein was analyzed by PyMOL29.
Analysis of mRNA secondary structure
The secondary structure of mRNA of blaA variants was predicted using the mfold webserver at default parameters (http://mfold.rna.albany.edu/). The mfold predicts energetically optimal secondary structure of an RNA molecule taking into account many realistic physical parameters that affect the RNA folding, such as pH, temperature and local biases of RNA.
References
Bush, K. 2013. Proliferation and significance of clinically relevant β-lactamases. Ann. N. Y. Acad. Sci. 1277, 84–90 (2013).
Bottone, E. J. Yersinia enterocolitica: overview and epidemiologic correlates. Microbes Infect. 1, 323–333 (1999).
Batzilla, J., Heesemann, J. & Rakin, A. The pathogenic potential of Yersinia enterocolitica 1A. Int. J. Med. Microbiol. 301, 556–561 (2011).
Sihvonen, L. M. et al. Clinical isolates of Yersinia enterocolitica biotype 1A represent two phylogenetic lineages with differing pathogenicity-related properties. BMC Microbiol. 12, 208.
Bhagat, N. & Virdi, J. S. The enigma of Yersinia enterocolitica biovar 1A. Crit. Rev. Microbiol. 37, 25–39 (2011).
Singh, I., Bhatnagar, S. & Virdi, J. S. Isolation and characterization of Yersinia enterocolitica from diarrheic human subjects and other sources. Curr. Sci. 84, 1353–1355 (2003).
MacDonald, E. et al. Yersinia enterocolitica outbreak associated with ready-to-eat salad mix, Norway, 2011. Emerg. Infect. Dis. 18, 1496–1499 (2012).
Fredriksson-Ahomaa, M., Cernela, N., Hächler, H. & Stephan, R. Yersinia enterocolitica strains associated with human infections in Switzerland 2001–2010. Eur. J. Clin. Microbiol. Infect. Dis. 31, 1543–1550 (2012).
Abraham, M. et al. An outbreak of food poisoning in Tamil Nadu associated with Yersinia enterocolitica. Indian J. Med. Res. 106, 46–468 (1997).
Singh, I. & Virdi, J. S. Isolation, biochemical characterization and in vitro tests of pathogenicity of Yersinia enterocolitica isolated from pork. Curr. Sci. 77, 1019–1021 (1999).
Sinha, I., Choudhary, I. & Virdi, J. S. Isolation of Yersinia enterocolitica and Yersinia intermedia from wastewaters and their biochemical and serological characteristics. Curr. Sci. 79, 510–513 (2000).
Ramesh, A., Padmapriya, B. P., Chrashekar, A. & Varadaraj, M. C. Application of a convenient DNA extraction method and multiplex PCR for the direct detection of Staphylococcus aureus and Yersinia enterocolitica in milk samples. Mol. Cell. Probes. 16, 307–314 (2002).
Divya, K. H. & Varadaraj, M. C. Prevalence of very low numbers of potential pathogenic isolates of Yersinia enterocolitica and Yersinia intermedia in traditional fast foods of India. Indian J. Microbiol. 51, 461–468 (2011).
Sharma, S., Ramnani, P. & Virdi, J. S. Detection and assay of β-lactamases in clinical and non-clinical strains of Yersinia enterocolitica biotype 1A. J. Antimicrob. Chemother. 54, 401–405 (2004).
Sharma, S., Mittal, S., Mallik, S. & Virdi, J. S. Molecular characterization of β-lactamase genes (blaA and blaB) of Yersinia enterocolitica biotype 1A. FEMS Microbiol. Lett. 257, 319–327 (2006).
Pham, J. N., Bell, S. M., Martin, L. & Carniel, E. The β-lactamases and β-lactam antibiotic susceptibility of Yersinia enterocolitica. J. Antimicrob. Chemother. 46, 951–957 (2000).
Pfeifer, Y., Cullik, A. & Witte, W. Resistance to cephalosporins and carbapenems in Gram-negative bacterial pathogens. Int. J. Med. Microbiol. 300, 371–379 (2010).
Stock, I. & Wiedemann, B. J. An in-vitro study of the antimicrobial susceptibilities of Yersinia enterocolitica and the definition of a database. Antimicrob. Chemother. 43, 37–45 (1999).
Stock, I., Heisig, P. & Wiedemann, B. Beta-lactamase expression in Yersinia enterocolitica biovars 1A, 1B and 3. J. Med. Microbiol. 49, 403–408 (2000).
Bent, Z. W. & Young, G. M. Contribution of BlaA and BlaB beta-lactamases to antibiotic susceptibility of Yersiniaenterocolitica biovar 1B. Antimicrob. Agents Chemother. 54, 4000–4002 (2010).
Bonke, R. et al. Antimicrobial susceptibility and distribution of β-lactamase A (blaA) and β-lactamase B (blaB) genes in enteropathogenic Yersinia species. Microb. Drug Resist. 17, 575–581 (2011).
Sachdeva, P. & Virdi, J. S. Repetitive elements sequence (REP/ERIC)-PCR based genotyping of clinical and environmental strains of Yersinia enterocolitica biotype 1A reveal existence of limited number of clonal groups. FEMS Microbiol. Lett. 240, 193–201 (2004).
Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. 20th informational supplement M100–S20 (Clinical and Laboratory Standards Institute, Wayne, PA 2010).
Bradford, M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem. 72, 248–254 (1976).
Pérez-Llarena, F. et al. The bla gene of the cephamycin cluster of Streptomyces clavuligerus encodes a class A beta-lactamase of low enzymatic activity. J. Bacteriol. 179, 6035–6040 (1997).
Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. PROCHECK - a program to check the stereochemical quality of protein structures. J. App. Cryst. 26, 283–291 (1993).
Lüthy, R., Bowie, J. U. & Eisenberg, D. Assessment of protein models with three-dimensional profiles. Nature. 356, 83–85 (1992).
Li, Q., Cheng, T., Wang, Y. & Bryant, S. H. PubChem as a public resource for drug discovery. Drug Discov. Today. 15, 1052–1057 (2010).
Lill, M. A. & Danielson, M. L. 2011. Computer-aided drug design platform using PyMOL. J. Comput. Aided Mol. Des. 25, 13–19 (2011).
Aarestrup, F. M. et al. Resistance to antimicrobial agents used for animal therapy in pathogenic-, zoonotic- and indicator bacteria isolated from different food animals in Denmark: a baseline study for the Danish Integrated Antimicrobial Resistance Monitoring Programme (DANMAP). APMIS. 106, 745–770 (1998).
Baumgartner, A., Küffer, M., Suter, D., Jemmi, T. & Rohner, P. 2007. Antimicrobial resistance of Yersinia enterocolitica strains from human patients, pigs and retail pork in Switzerland. Int. J. Food Microbiol. 115, 110–114 (2007).
Szych, J., Jakubczak, A., Wardak, S. & Madajczak, G. Antimicrobial susceptibility of Yersinia enterocolitica and Yersinia pseudotuberculosis strains isolated from humans in Poland during 2004–2009. Med. Dosw. Mikrobiol. 61, 311–319 (2009).
Terentjeva, M. & Bērziņs, A. J. Prevalence and antimicrobial resistance of Yersiniaenterocolitica and Yersinia pseudotuberculosis in slaughter pigs in Latvia. Food Prot. 73, 1335–1338 (2010).
Siu, L. K., Ho, P. L., Yuen, K. Y., Wong, S. S. & Chau, P. Y. Transferable hyperproduction of TEM-1 beta-lactamase in Shigella flexneri due to a point mutation in the pribnow box. Antimicrob. Agents Chemother. 41, 468–470 (1997).
Nelson, E. C., Segal, H. & Elisha, B. G. Outer membrane protein alterations and blaTEM-1 variants: their role in beta-lactam resistance in Klebsiella pneumoniae. J. Antimicrob. Chemother. 52, 899–903 (2003).
Siu, L. K., Lu, P. L., Chen, J. Y., Lin, F. M. & Chang, S. C. High-level expression of ampC beta-lactamase due to insertion of nucleotides between −10 and −35 promoter sequences in Escherichia coli clinical isolates: cases not responsive to extended-spectrum-cephalosporin treatment. Antimicrob. Agents Chemother. 47, 2138–2144 (2003).
Kumar, K. M., Lavanya, P., Anbarasu, A. & Ramaiah, S. Molecular dynamics and molecular docking studies on E166A point mutant, R274N/R276N double mutant and E166A/R274N/R276N triple mutant forms of class A β-lactamases. J. Biomol. Struct. Dyn. (10.1080/07391102.2013.847804) (2013).
Yi, H. et al. Twelve positions in a β-lactamase that can expand its substrate spectrum with a single amino acid substitution. PLoS One. 7, e37585 (2012).
Salverda, M. L., De Visser, J. A. & Barlow, M. Natural evolution of TEM-1 beta-lactamase: experimental reconstruction and clinical relevance. FEMS Microbiol.Rev. 34, 1015–1036 (2010).
Kozak, M. Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc. Natl. Acad. Sci. (USA). 83, 2850–2854 (1986).
Studer, S. M. & Joseph, S. Unfolding of mRNA secondary structure by the bacterial translation initiation complex. Mol. Cell. 22, 105–115 (2006).
Acknowledgements
We thank Mr. Rajesh Kumar Mahto for technical help. NS is supported by UGC-Dr. D.S. Kothari Postdoctoral Fellowship. The project was supported by Indian Council of Medical Research grant (Grant no. AMR/17/2011-ECD-1), DU-DST-PURSE grant and the UGC-Special Assistance Programme (DRS-I) to the Department of Microbiology, University of Delhi South Campus.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: N.S. and J.S.V. Bioinformatic analysis: M.K. Wrote the paper: N.S., J.S.V. and M.K.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article's Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Singhal, N., Kumar, M. & Virdi, J. Molecular Analysis of β-Lactamase Genes to Understand their Differential Expression in Strains of Yersinia enterocolitica Biotype 1A. Sci Rep 4, 5270 (2014). https://doi.org/10.1038/srep05270
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep05270
This article is cited by
-
Exploring the genetic determinants underlying the differential production of an inducible chromosomal cephalosporinase - BlaB in Yersinia enterocolitica biotypes 1A, 1B, 2 and 4
Scientific Reports (2020)
-
Virulence-associated traits and in vitro biofilm-forming ability of Escherichia coli isolated from a major river traversing Northern India
Environmental Science and Pollution Research (2019)
-
Antifungal Signature: Physicochemical and Structural In Silico Analysis of Some Antifungal Peptides
International Journal of Peptide Research and Therapeutics (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.