Abstract
X-linked severe combined immunodeficiency (SCID-X1) caused by mutations in interleukin 2 receptor gamma (IL2RG) gene threatens the survival of affected boys during the first year of life unless hematopoietic stem cell transplantation is provided. Although viral vector-mediated gene therapy has been successfully performed in patients with no HLA-matched donors, leukemia caused by vector-mediated insertional mutagenesis has been reported in some individuals. Transcription activator-like effector nuclease (TALEN) is an artificial sequence-specific endonuclease that is expected to revolutionize the precise correction of disease-causing mutations and eliminate the risk of insertional mutagenesis. Here, we report TALEN-mediated genome editing of the IL2RG locus. We transfected TALENs along with a targeting vector into Jurkat cells and we confirmed the precise introduction of the exogenous gene into the IL2RG locus. In addition, we found that the length of homology arm in the targeting vector influenced the efficiency of TALEN-mediated homologous recombination.
Similar content being viewed by others
Introduction
X-linked severe combined immunodeficiency (SCID-X1) is the most common form of combined immunodeficiency; it is caused by mutations in the X chromosome-linked interleukin 2 receptor gamma gene (IL2RG)1. IL2RG, also known as CD132, is a common cytokine receptor subunit for IL-2, IL-4, IL-7, IL-9, IL-15 and IL-21. Lack of IL2RG function results in loss of multiple cytokine activities, leading to absence of T and natural killer (NK) lymphocytes and nonfunctional B lymphocytes. Most SCID-X1 patients die during infancy unless reconstitution of the immune system is achieved through hematopoietic stem cell (HSC) transplantation or gene therapy2.
Bone marrow transplantation (BMT) is a curative treatment for SCID-X1. In spite of improvements in transplantation protocols and the introduction of other sources of stem cells such as umbilical cord blood, BMT is associated with serious risks as a result of chemotherapy conditioning regimens, graft-versus-host disease (GVHD) and related infections. Furthermore, only a small percentage of SCID-X1 patients receive BMT owing to a shortage of human leukocyte antigen (HLA)-matched donors.
Gene therapy using viral vectors to transduce therapeutic IL2RG into autologous CD34+ bone marrow cells has been successful for immune reconstitution for some individuals, resulting in long-term correction of the SCID-X1 phenotype3,4,5. However, some patients developed T-cell acute lymphoblastic leukemia within 2.5–5 years after gene therapy, one of whom died6,7,8,9. Patients' blast cells showed retrovirus vector integration in close proximity to the proto-oncogenes LMO2, CCND2 and BMI1, thus leading to aberrant transcription and expression of these genes10,11,12. Therefore, gene targeting is needed to eliminate the risks posed by random integration of viral vectors.
Gene targeting based on homologous recombination has been widely used in genetic research; however, its efficiency remains very low. Similarly, therapeutic genes do not undergo homologous recombination at the mutated locus13. Homologous recombination is primarily involved in the repair chromosomal double-strand breaks (DSBs), a process known as homology directed repair (HDR)14. In the absence of homologous donor DNAs, DSBs are repaired by non-homologous end-joining (NHEJ), which frequently results in small insertions and/or deletions (indels). Induction of artificial DSB would significantly improve the efficiency of homologous recombination.
Recently, genome editing technologies such as zinc-finger nuclease (ZFN) and transcription activator-like effector nuclease (TALEN) have enabled the introduction of site-specific DSBs into DNA. ZFNs consist of a DNA-binding domain and a FokI endonuclease domain. Each DNA-binding domain recognizes 3–4 DNA bp (nucleotide triplets). Pairs of ZFNs are required to induce DSB in a target locus because the FokI cleavage domain is only enzymatically active as a dimer15. ZFN-induced DSBs were initially shown to enhance gene targeting frequencies in Drosophila16. This was soon followed by endogenous gene correction of IL2RG by ZFN in SCID-X1 cells17. However, ZFN have not become widely used, as some nucleotide triplets lack specific DNA recognition domains and engineering of ZFN is time consuming, laborious and intricate18.
TALEN technologies have also been developed to generate site-specific DSBs19. TALEN is a sequence-specific endonuclease that consists of a transcription activator-like effector (TALE) and a FokI endonuclease. TALE is a DNA-binding protein that has a highly conserved central region with tandem repeat units of 34 amino acids. The base preference for each repeat unit is determined by two amino acid residues called the repeat-variable di-residue (RVD), which recognizes one specific nucleotide in the target DNA. Arrays of DNA-binding repeat units can be customized for targeting specific DNA sequences. As with ZFNs, dimerization of two TALENs on targeted specific sequences in a genome results in FokI-dependent introduction of DSBs, stimulating HDR and NHEJ repair mechanisms20,21,22. In contrast to ZFNs, the design and construction of TALENs is rapid, affordable and efficient.
Gene therapy without the risk of vector-related insertional mutagenic events is desired for patients who lack HLA-matched donors. Genome editing is an ideal technique to correct mutations in disease-causing genes. Here, we report TALEN-mediated genome editing and introduction of a reporter gene into the IL2RG locus in Jurkat cells.
Results
TALEN design for inducing DSBs at the endogenous human IL2RG gene
To induce mutations in IL2RG, we targeted the translational start codon of IL2RG (Figure 1a). Nucleotide sequences targeted by TALENs were designed using the ZiFit program and DNA recognition modules were assembled with the Golden Gate method23,24. Assembled modules were cloned into three different expression vectors (pcDNA-TAL, pcDNA-TAL-NC and pcDNA-TAL3), which differed on the basis of the N- and C-terminal backbone regions of the TALE protein. We examined the nuclease activity of TALENs with different backbones by using the SSA assay24. All three TALENs showed increased luciferase activity induced by DSB-directed nucleotide repair (Figure 1b). Among these, TALENs with the TAL-NC backbone showed highest activity compared to those with TAL and TAL3 backbones. This result suggested that N- and/or C-terminal domains of TALEN are crucial for activity and that the TAL-NC backbone, which is the shortest one tested, was suitable to induce mutation. Therefore, we used the TALEN with the TAL-NC backbone targeting the IL2RG start codon for our experiments.
TALEN design for inducing DSBs in the endogenous IL2RG gene in Jurkat cells.
(a) Schematic of the TALEN target site at the IL2RG start codon in Jurkat cells. IL2RG contains eight exons. The binding sites for the TALEN pair used in this study are shown in capital letters, the spacer sequence is indicated in small letters, the start codon (ATG) is in bold and the TALEN target designed by ZiFiT in the spacer sequence is underlined. (b) Functional evaluation of engineered TALENs using the SSA assay. HEK293T cells were transfected with the SSA reporter vector, pRL-TK reference vector and TALEN expression vectors. After 48 h, a luciferase assay was performed. Data are shown as mean ± SD (n = 3).
Targeted genomic insertion-deletion at the IL2RG locus in Jurkat cells
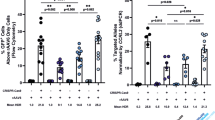
To examine whether DSB and subsequent NHEJ-directed mutations could be induced in the genome, Jurkat cells, which are established as a cell line derived from a 14-year-old boy with acute lymphoblastic leukemia and have only a single copy of X chromosome-linked IL2RG25, were transfected with plasmids expressing TALENs by electroporation. Five days after electroporation, genomic DNA was extracted for the T7 endonuclease assay. As shown in Figure 2a, cleaved DNAs appeared in Jurkat cells transfected with TAL-NC TALENs, but not in cells transfected with TAL3 TALENs. This finding indicated that TALENs with the TAL-NC backbone were able to induce mutations around the translational initiation site of the IL2RG locus. Next, we examined mutations in individual clones that had been isolated by limiting dilution. Sequencing analysis of cloned Jurkat cells revealed that nucleotide deletions were present near the translational initiation site of the IL2RG locus and that individual clones had different mutations (Figure 2b). Mutation ratios ranged from 4.8% to 13.3% (Table 1). Furthermore, flow cytometric analysis demonstrated that IL2RG protein expression on the cell surface was absent in clones del-2, del-3 and del-4, all of which had deletions in the region containing the translational start codon (Figure 2c). Although clone del-1 contained a deletion of 2-nucleotides, the translational initiation codon remained intact, resulting in similar expression of IL2RG compared to wild-type cells. To evaluate the response to IL-2 in the mutated Jurkat cells, we stimulated the cells with PMA and ionomycin in the presence of exogenous IL-2 and analyzed the expression of BCL2, which is known to be a downstream target of IL-2 signaling26. The expression of BCL2 increased by more than two fold after stimulation in wild-type Jurkat cells (Figure 2d). On the other hand, BCL2 expression did not increase in the mutated cells (Figure 2d). No significant difference in endogenous IL2 expression of each clone was observed after stimulation (data not shown). In summary, we successfully induced mutation in the IL2RG locus by TALEN-mediated genome editing and showed that the mutated cells lost IL2RG protein expression and response to IL-2.
TALEN-induced genomic mutation in IL2RG.
(a) T7 endonuclease I assay using TALENs for Jurkat cells. Jurkat cells were transfected with TALEN expression vectors by electroporation. After 5 days culture, genomic DNA was isolated and the TALEN target locus was amplified by PCR. A T7 endonuclease I assay was performed using purified PCR products. The arrowhead indicates the expected position of the digested products in the agarose gel. (b) Sequencing results of the PCR fragments, revealing different mutations in the TALEN target site. Jurkat cells were cultured for 5 days after electroporation and cloning was performed by limiting dilution. Genomic DNA was isolated from cloned Jurkat cells and DNA sequencing was performed. Sequences for wild-type (WT) and deletion mutants (del1–4) are shown. (c) Functional analysis of genome-modified Jurkat cells. The level of IL2RG expression in genome-modified Jurkat cells was analyzed using flow cytometry. Cells were incubated with APC-conjugated-anti-hCD132 antibody for IL2RG and APC-IgG2b antibody as an isotype control. MFI, Mean Fluorescence Intensity of CD132. (d) qPCR analysis of BCL2. BCL2 expression was examined 48 hr after the PMA and ionomycin stimulation in the presence of exogenous IL-2. Data are shown as mean ± SD (n = 3).
Targeted knock-in at the IL2RG locus
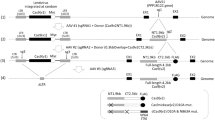
Because TALENs targeting the start codon were shown to induce DSBs in the IL2RG locus, we attempted to induce the HDR. Long and short homology arms were cloned into the targeting vector, which harbored the gene for the Venus fluorescent protein (a brighter variant of the yellow fluorescent protein)27 and a loxP-flanked Neomycin resistance cassette. The 5′ end of the DT-A negative selectable marker was also inserted downstream of exon 6 (Figure 3a and Figure S1). Thus, it was expected that fluorescence from Venus expression, driven by the IL2RG promoter, would be detectable by TALEN-mediated DSB and subsequent HDR. TALENs and linearized targeting vector were co-transfected into Jurkat cells by electroporation. Transfected cells were selected in G418-containing medium and single-cell cloning was performed by limiting dilution. Flow cytometric analysis of individual clones revealed that the majority of Jurkat cells co-transfected with TALENs and targeting vector expressed Venus (clones KI 1–4), while only one clone (KI 5) showed high levels of Venus (Venushi). Southern blot analysis clearly indicated that the gene encoding for Venus was knocked-in at the IL2RG locus (Figure 3c). However, clone KI 5 (Venushi) showed a band corresponding to wild-type cells, as well as additional bands. These data suggested that targeting vectors were randomly integrated into the genome and that Venus expression in the Venushi clone was driven by adjacent transcriptional activity. We then tested whether the targeting vector with shorter homology arms could also induce HDR. We constructed two novel targeting vectors (pVenus-M and pVenus-S) with shorter homology arms. pVenus-M has a 3000 bp 5′ homology arm and a 2000 bp 3′ homology arm, while pVenus-S has homology arms of 1000 bp each. Jurkat cells were co-transfected with linearized pVenus-M or –S vectors and TALEN plasmids by electroporation and single-cell clones were isolated by G418 selection and limiting dilution. Expression of Venus was analyzed by flow cytometry and knock-in efficiency was estimated. Venus expression was detected in Jurkat cells transfected with both of the shorter targeting vectors as well as with pVenus-L. However, the frequency of Venus-positive cells among the isolated clones was lower with vectors containing shorter homology arms. Furthermore, the ratio of Venushi Jurkat cells, which were considered to have random integrations, was increased in cells transduced with shorter targeting vectors. Therefore, the results suggested that the recombination frequency was determined by the length of the homology regions.
TALEN-mediated genome editing.
(a) Top: Schematic of the endogenous IL2RG locus. HindIII, HindIII restriction sites used for Southern blot analysis; TM, transmembrane domain. Middle: Schematic of the targeting vector. The targeting vector contained Venus cDNA, lox-P-flanked Neomycin resistance cassette and DT-A negative selectable marker. The 5′ homology arm upstream of the IL2RG start codon was cloned upstream of Venus cDNA and the 3′ homology arm downstream of the IL2RG transmembrane sequence (exon 6) was cloned downstream of the Neomycin resistance cassette (Neor). 5′ probe, Probe used for Southern blot analysis. Bottom: Schematic of the targeted IL2RG locus. A novel HindIII restriction site would introduced when the targeted knock-in was successful. (b) Flow cytometric analysis of Venus in Jurkat cells with targeted knock-in of IL2RG. Each Jurkat cell clone was analyzed for YFP fluorescence expressed from knocked-in Venus cDNA. WT, wild-type Jurkat cells; KI 1-4, individual clones with targeted knock-in; KI 5, Venushi; MFI, Mean Fluorescence Intensity of Venus. (c) Southern blot analysis of Jurkat cells with targeted knock-in of IL2RG. HindIII digestion resulted in a 3788 bp band from the WT endogenous IL2RG locus and a 3515 bp band (containing Venus cDNA and the Neomycin resistance cassette) from the targeted knock-in. Targeting vector was used as positive control for targeted knock-in and genomic DNA isolated from WT Jurkat cells was used as negative control.
Discussion
In this study, we showed that TALEN-mediated genome editing technology could replace an endogenous target gene by using exogenous artificial nucleotides. TALEN-mediated DSB at the target locus has been reported to facilitate HDR by exogenous nucleotides in somatic cells and embryonic stem cells21,22,28. We showed that exogenous nucleotides containing the Venus-Neo cassette were specifically integrated at the IL2RG locus (Figure 3). Furthermore, we demonstrated that the efficiency of HDR was dependent on the length of the homology regions used in the gene targeting vector (Table 2).
By using a gene targeting vector along with TALENs, we have succeeded in isolating the knock-in cells with high efficiency. Generation of knock-in cell lines would accelerate the in vitro analysis of gene function in the cell and would be expected to be applied in various fields. Thus, generation of the cells harboring mutations found in patients would promote the understanding of the disease pathology of each patient.
Gene therapies using viral vectors to introduce therapeutic genes into the genome have not been able to uniquely target the site of gene insertion. Because of this, activation of proto-oncogenes caused by vector-mediated insertional mutagenesis has occurred in some patients, sometimes resulting in the development of leukemia. To reduce the risk of leukemogenesis, site-directed recombination induced by TALEN and vectors containing homology regions of the therapeutic gene is a superior method for replacing a mutant endogenous gene with a functional exogenous cDNA in somatic stem cells. Moreover, expression of the therapeutic gene would be regulated by an endogenous promoter. We found that longer homology regions were required for higher recombination efficiencies (Table 2). The efficiency of recombination at the IL2RG locus was quite high (up to 76%) and fewer numbers of cells with random integrations appeared when the longest targeting vector was used (Table 2). The incidence of random integration into the genome indicates that this technology has some limitations that need to be overcome. Some diseases are caused by loss-of-function mutations in regulatory genes, resulting in uncontrolled gene expression that can lead to another disorder. These kinds of diseases are not suitable candidates for vector-mediated gene therapy. However, physiological expression of therapeutic genes achieved by genome editing-mediated gene therapy could treat these diseases. SCID-X1 is thought to be suitable for gene therapy, as lymphocyte progenitor cells with functional IL2RG show a selective advantage over defective progenitors. This phenomenon was first observed in a SCID-X1 patient who had a reverse mutation in IL2RG that restored gene function29. Gene therapy for diseases in which there is no selective advantage conferred by the transgene, such as chronic granulomatous disorder, is thought to require pre-conditioning of bone marrow to engraft the gene-corrected progenitors30. Therefore, it is important to carefully select candidate diseases for gene therapy.
Disease-causing mutations of IL2RG in SCID-X1 patients are not evenly distributed31. Precise corrections of the mutation at the single nucleotide level have been reported32. This therapeutic strategy requires the design and construction of custom-made TALENs and targeting vectors for each patient. Although personalized medicine is an ideal strategy, this approach is costly and time consuming, also increases the risk of medical errors. By comparison, our strategy could replace mutations by functional cDNA with a single set of TALENs and gene targeting vector (Figure 3).
We showed that the N- and/or C- terminal regions of TALEN are crucial for activity. As shown in Figure 2b and 2c, mutations near the start codon of the IL2RG locus resulted in decreased or diminished IL2RG expression in TAL-NC-transfected cells. Moreover, mutant cells showed decreased expression of BCL2 by PMA and ionomycin treatment in the presence of IL-2, indicating that TALEN-mediated loss of IL2RG expression leads to functional alteration triggered by IL-2 (Figure 2d). Cloned Jurkat cells transfected with TAL3 TALENs induced mutations to the same extent as that observed in TAL-NC TALEN-transduced cells, while bands from T7 endonuclease digests were barely detectable (data not shown). However, it should be noted that endonuclease FokI fused with the TAL3 TALEN differed from that of TAL and TAL-NC. Although normal FokI endonuclease functions as a homodimer, mutant FokI fused with the TAL3 backbone functions as an obligate heterodimer33. Thus, it is possible that the lower rate of mutation in TAL3 TALEN-transfected cells was due to reduced nuclease activity. On the other hand, it is assumed that the number of spacer nucleotides (which are inserted between both TALENs) also influenced the induction of mutations, since luciferase activity was higher in TAL-NC TALENs than in TAL TALENs (Figure 1b).
In conclusion, we showed that genome editing by artificial nuclease along with gene targeting vector could be a powerful tool to modify endogenous gene. To carry out clinical applications of genome editing-mediated gene therapy, some problems need to be solved, such as random integration events. For gene replacement in hematopoietic stem cells, this issue could be addressed with a screening system that removes random knock-in clones and off-target mutated clones, or a system based on positive-selection of targeted knock-in clones.
Methods
TALEN and targeting-vector construction
All of TALEN vectors used in this study were obtained from Addgene. The pair of TALENs recognizing human IL2RG was designed using ZiFiT34. TALE repeats were constructed using the Golden Gate TALEN assembly method as described previously, with some modifications24. The module plasmids were BsaI digested and fragments encoding RVD-specific sequences were purified in advance by gel electrophoresis. RVDs were cloned into array plasmids by using a DNA Ligation Kit (Takara, Shiga, Japan). Screening of assembled clones by colony PCR and cloning of the constructed array plasmid and the appropriate last repeat into mammalian expression vectors (pcDNA-TAL, pcDNA-TAL-NC, or pcDNA-TAL3) by using the Golden Gate method were performed as described previously24. Each plasmid or vector was transformed into Escherichia coli DH5α competent cells.
The pVenus vector was used as the backbone to construct the IL2RG-targeting vector pVenus-L. A 5662 bp 5′ homology arm and a 3000 bp 3′ homology arm were amplified by PCR from the Jurkat cell genome and cloned into the pVenus by using a DNA Ligation Kit. Donor DNA was linearized by Eam1105I or KpnI digestion and purified by gel electrophoresis. Targeting vectors with various arm lengths were obtained following the same strategy. Homology arms were amplified by PCR from pVenus-L and cloning was performed with GeneArt Seamless Cloning and Assembly Enzyme Mix (Life Technologies, Carlsbad, CA).
Single-strand annealing (SSA) assay
The SSA assay was performed as previously described24. HEK293T cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) (Hana-Nesco Bio Corp, Tokyo, Japan). Cells (4.4 × 104 were co-transfected with 400 ng of each of the TALEN expression plasmids, 200 ng of the pGL4-SSA reporter plasmid and 40 ng of the pRL-TK reference vector using Polyethylenimine “Max” (Polysciences, Warrington, PA) in a 48-well plate. After 48 h, dual-luciferase assays were carried out using the Dual-Glo luciferase assay system (Promega, Madison, WI) in a ARVO X3 Multilabel Plate Reader (PerkinElmer, Waltham, MA) following the manufacturer's instructions. pGL4-SSA reporter plasmids were constructed as previously described24.
Cell culture and transfection
Jurkat cells were maintained in RPMI-1640 medium (Sigma, St. Louis, MO) supplemented with 10% FBS, 55 μM 2-mercaptoethanol (Life Technologies, Carlsbad, CA), 5 mM HEPES, 1 mM sodium pyruvate (Nacalai Tesque, Kyoto, Japan), 100 U/mL penicillin and 100 μg/mL streptomycin at 37°C with 5% CO2 incubation. Jurkat cells were transfected by electroporation by using a Nepa21 pulse generator (Nepa Gene, Chiba, Japan). Jurkat cells were suspended in OPTI-MEM (Life Technologies) at a density of 2 × 105 cells per electrode chamber with 10 μg each of Left TALEN vector and Right TALEN vector and placed in an electrode chamber. For targeted knock-in through homologous recombination, 2 μg of the targeting vector was added. Two rectangular electric pulses (150 V, 5-ms duration, 50-ms interval, decay constant: 10%), followed by five pulses (20 V, 50-ms duration, 50-ms interval, decay constant: 40%) and another five pulses in the opposite direction of the electric field were delivered. G418 (Nacalai Tesque) at 500 μg/ml was added to the medium 4 d after transfection for selection of targeted knock-in clones. Single-cell clones of genome-modified Jurkat cells and targeted knock-in Jurkat cells were obtained by limiting dilution.
T7 Endonuclease I assay
The T7 Endonuclease I assay was performed as previously described35. Jurkat cells were cultured and transfected as described above. Five days after transfection, genomic DNA was isolated from cells transfected with TALEN-encoding or control plasmids using the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Endogenous loci were amplified by PCR using the following primers: Forward (5′-GGA CCC AGC TCA GGC AGC A-3′) and Reverse (5′-TGG GCA TAG TGG TCA GGA AGA-3′). PCR products were purified with the QIAquick PCR Purification Kit (Qiagen) according to manufacturer's instructions. Purified PCR product (200 ng) was denatured and reannealed in NE Buffer 2 (New England Biolabs, Ipswich, MA) using a thermocycler with the following protocol: 95°C, 5 min; 95–85°C at −2°C/s; 85–25°C at −0.1°C/s; hold at 4°C. Hybridized PCR products were treated with 10 U of T7 Endonuclease I (New England Biolabs) at 37°C for 15 min in a reaction volume of 20 μl. Reactions were stopped by the addition of 2 μl of 0.5 M EDTA and gel electrophoresis of the products was performed.
Sequencing analysis for genome modification
Genomic DNA was extracted using the DNeasy Blood & Tissue Kit (Qiagen) following the manufacturer's protocol. Genomic regions surrounding the TALEN target site were amplified by PCR using the following primers: Forward (5′-CAC CCT CTG TAA AGC CCT GG-3′) and Reverse (5′-CCA GTC CCA GAT TTC CCA CC-3′). PCR products were purified using the QIAquick PCR Purification Kit according to the manufacturer's protocol. DNA sequencing was performed by general Sanger method.
Flow cytometry
APC-conjugated anti-human CD132 (clone: TUGh4) and APC-Rat IgG2b, k Isotype ctrl (clone: RTK4530) (BioLegend, San Diego, CA) antibodies were used for flow cytometry analyses of genome-modified Jurkat cells. Flow cytometry was performed on a FACSCalibur HG flow cytometer with CELL Quest Pro software (Becton Dickinson, Franklin Lakes, NJ). Dead cells were excluded by gating of forward and side scatter.
Jurkat cell stimulation
Jurkat cells (1 × 106 cells/ml) were stimulated with the indicated concentrations of PMA (100 ng/ml; Sigma), ionomycin (1 μg/ml; Sigma) and IL-2 (100 ng/ml; BioLegend) for 48 hr in 24-well plates. We chose five cloned Jurkat cells which had no mutations in IL2RG as wild-type control, at random.
Quantitative PCR
Total cell RNA from Jurkat cells were isolated using ISOGEN (Nippon Gene, Tokyo, Japan) following the manufacturer's protocol. We used 2 μg purified RNA to synthesize the first strand of cDNA with SuperScript II Reverse Transcriptase (Life Technologies) and diluted 1:20 prior to the Quantitative PCR (qPCR) analysis.
qPCR was performed using Power SYBR green PCR master mix (Applied Biosystems Inc. Foster City, CA) and the following primers: BCL2 Forward (5′- GAC TGA GTA CCT GAA CCG GC -3′), BCL2 Reverse (5′-AGT TCC ACA AAG GCA TCC CAG-3′), ACTB Forward (5′-CCC CGC GAG CAC AGA G-3′) and ACTB Reverse (5′-ATC ATC CAT GGT GAG CTG GC-3′). All qPCRs were performed in triplicates. Results were normalized to β-actin and then calculated relative to non-stimulated cells, respectively.
Southern blot analysis
Following electrophoresis, gels were immersed in denaturation buffer (0.5 M NaOH, 1.5 M NaCl) for 30 min, followed by neutralization buffer (0.5 M Tris-HCl [pH 7.5], 1.5 M NaCl) for 10 min. Capillary transfer of DNA from agarose gels to nylon membranes (Amersham Hybond-N+, GE Healthcare, Buckinghamshire, United Kingdom) was conducted overnight with 20× SSC buffer (3 M NaCl, 300 mM sodium citrate, pH 7.0). Transferred DNA was fixed on membranes by UV irradiation and air-dried. Membranes were pre-incubated in hybridization buffer (DIG Easy Hyb; Roche, Basel, Switzerland) at 48°C for 30 min. The solution of DIG-labeled DNA probe was generated with the PCR DIG Probe Synthesis Kit (Roche) following the manufacturer's protocol. To generate the probe, PCR was performed using the following primers: Forward (5′-AGT CAC ACT TCC TCG CCA GT-3′) and Reverse (5′-ACC CAC ACG TTT CCT CTG TC-3′). The probe was heated at 95°C for 5 min and chilled quickly in ice, then diluted 500-fold with hybridization buffer. Membranes were incubated overnight with the diluted probe solution at 48°C. Membranes were washed twice at room temperature for 5 min with 2× SSC and 0.1% SDS and twice at 68°C for 15 min with 0.1× SSC and 0.1% SDS. Subsequently, membranes were washed with washing buffer (0.1 M maleic acid/NaOH [pH 7.5], 0.15 M NaCl and 0.3% Tween 20) for 2 min and incubated with blocking buffer (Blocking Reagent, Roche) for 30 min. Membranes were then incubated with 75 mU/ml Anti-Digoxigenin-AP Fab fragments (Roche) in blocking buffer at room temperature for 30 min and washed twice for 15 min with washing buffer. Finally, membranes were incubated with CDP-Star chemiluminescent substrate (Roche) at room temperature for 5 min, followed by incubation with reaction buffer (0.1 M Tris-HCl, 0.1 M NaCl, pH 9.5) for 3 min. Digital images of membranes were obtained using the ImageQuant LAS 4000 mini imaging system (GE Healthcare).
Statistical analysis
For all statistical analysis, two-tailed Student's t-tests were performed using Excel spreadsheet. p values are given for each individual experiment.
References
Noguchi, M. et al. Interleukin-2 receptor gamma chain mutation results in X-linked severe combined immunodeficiency in humans. Cell 73, 147–157 (1993).
Sugamura, K. et al. The interleukin-2 receptor gamma chain: its role in the multiple cytokine receptor complexes and T cell development in XSCID. Annu. Rev. Immunol. 14, 179–205 (1996).
Cavazzana-Calvo, M. et al. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science 288, 669–672 (2000).
Gaspar, H. B. et al. Gene therapy of X-linked severe combined immunodeficiency by use of a pseudotyped gammaretroviral vector. Lancet 364, 2181–2187 (2004).
Hacein-Bey-Abina, S. et al. Efficacy of gene therapy for X-linked severe combined immunodeficiency. N. Engl. J. Med. 363, 355–364 (2010).
Hacein-Bey-Abina, S. et al. A Serious Adverse Event after Successful Gene Therapy for X-Linked Severe Combined Immunodeficiency. N. Engl. J. Med. 348, 255–256 (2003).
Hacein-Bey-Abina, S. et al. LMO2-Associated Clonal T Cell Proliferation in Two Patients after Gene Therapy for SCID-X1. Science 302, 415–419 (2003).
Hacein-Bey-Abina, S. et al. Insertional oncogenesis in 4 patients after retrovirus-mediated gene therapy of SCID-X1. J. Clin. Invest. 118, 3132–3142 (2008).
Kohn, D. B., Sadelain, M. & Glorioso, J. C. Occurrence of leukaemia following gene therapy of X-linked SCID. Nat. Rev. Cancer. 3, 477–488 (2003).
Bushman, F. D. Retroviral integration and human gene therapy. J. Clin. Invest. 117, 2083–2086 (2007).
Deichmann, A. et al. Vector integration is nonrandom and clustered and influences the fate of lymphopoiesis in SCID-X1 gene therapy. J. Clin. Invest. 117, 2225–2232 (2007).
Schwarzwaelder, K. et al. Gammaretrovirus-mediated correction of SCID-X1 is associated with skewed vector integration site distribution in vivo. J. Clin. Invest. 117, 2241–2249 (2007).
Yáñez, R. J. & Porter, A. C. Therapeutic gene targeting. Gene Ther. 5, 149–159 (1998).
Chapman, J. R., Taylor, M. R. & Boulton, S. J. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell 47, 497–510 (2012).
Kim, Y. G., Cha, J. & Chandrasegaran, S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. U S A. 93, 1156–1160 (1996).
Bibikova, M., Beumer, K., Trautman, J. K. & Carroll, D. Enhancing gene targeting with designed zinc finger nucleases. Science 300, 764 (2003).
Urnov, F. D. et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 435, 646–651 (2005).
Urnov, F. D., Rebar, E. J., Holmes, M. C., Zhang, H. S. & Gregory, P. D. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 11, 636–646 (2010).
Christian, M. et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186, 757–761 (2010).
Miller, J. C. et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 29, 143–148 (2011).
Bedell, V. M. et al. In vivo genome editing using a high-efficiency TALEN system. Nature 491, 114–118 (2012).
Zu, Y. et al. TALEN-mediated precise genome modification by homologous recombination in zebrafish. Nat. Methods 10, 329–331 (2013).
Schneider, U., Schwenk, H. U. & Bornkamm, G. Characterization of EBV-genome negative “null” and “T” cell lines derived from children with acute lymphoblastic leukemia and leukemic transformed non-Hodgkin lymphoma. Int. J. Cancer 19, 621–626 (1977).
Akbar, A. N. et al. Interleukin-2 receptor common gamma-chain signaling cytokines regulate activated T cell apoptosis in response to growth factor withdrawal: selective induction of anti-apoptotic (bcl-2, bcl-xL) but not pro-apoptotic (bax, bcl-xS) gene expression. Eur. J. Immunol. 26, 294–299 (1996).
Ding, Q. et al. Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell 12, 393–394 (2013).
Bousso, P. et al. Diversity, functionality and stability of the T cell repertoire derived in vivo from a single human T cell precursor. Proc. Natl. Acad. Sci. U S A. 97, 274–278 (2000).
Nagai, T. et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 20, 87–90 (2002).
Grez, M., Reichenbach, J., Schwäble, J., Seger, R., Dinauer, M. C. & Thrasher, A. J. Gene therapy of chronic granulomatous disease: the engraftment dilemma. Mol. Ther. 19, 28–35 (2011).
Puck, J. M. et al. Mutation analysis of IL2RG in human X-linked severe combined immunodeficiency. Blood 89, 1968–1977 (1997).
Yusa, K. et al. Targeted gene correction of α1-antitrypsin deficiency in induced pluripotent stem cells. Natur 478, 391–394 (2011).
Dahlem, T. J. et al. Simple methods for generating and detecting locus-specific mutations induced with TALENs in the zebrafish genome. PLoS Genet. 8, e1002861 (2012).
Sander, J. D. et al. ZiFiT (Zinc Finger Targeter): an updated zinc finger engineering tool. Nucleic Acids Res. 38, W462–468 (2010).
Cermak, T. et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39, e82 (2011).
Sakuma, T. et al. Efficient TALEN construction and evaluation methods for human cell and animal applications. Genes Cells 18, 315–326 (2013).
Reyon, D. et al. FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol. 30, 460–465 (2012).
Acknowledgements
This work was supported in part by The Grant of National Center for Child Health and Development, Grant Number 25-1, JSPS KAKENHI Grant Number 24115707, 24659669, 23249071, JST (CREST), NIH grant AR050631, Bristol-Myers RA Research Fund and Mochida Memorial Fund to H.A.
Author information
Authors and Affiliations
Contributions
Y.M. performed the experiments, analyzed the data, drafted and approved the manuscript. T.C. designed the experiments, analyzed the data, drafted and approved the manuscript. K.K., T.M. and S.M. approved the manuscript. S.T. established the protocol for TALEN construction and approved the manuscript. H.A. designed the experiments, drafted and approved the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary info.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. The images in this article are included in the article's Creative Commons license, unless indicated otherwise in the image credit; if the image is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the image. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Matsubara, Y., Chiba, T., Kashimada, K. et al. Transcription activator-like effector nuclease-mediated transduction of exogenous gene into IL2RG locus. Sci Rep 4, 5043 (2014). https://doi.org/10.1038/srep05043
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep05043
This article is cited by
-
Gene correction for SCID-X1 in long-term hematopoietic stem cells
Nature Communications (2019)
-
TALEN-mediated functional correction of human iPSC-derived macrophages in context of hereditary pulmonary alveolar proteinosis
Scientific Reports (2017)
-
Current Progress in Therapeutic Gene Editing for Monogenic Diseases
Molecular Therapy (2016)
-
Newborn Screening for Severe Combined Immunodeficiency
Current Pediatrics Reports (2015)
-
Genome Editing: Potential Treatment for Lysosomal Storage Diseases
Current Stem Cell Reports (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.