Abstract
Mitogen-activated protein kinases (MAPKs) have a number of targets which they regulate at transcriptional and post-translational levels to mediate specific responses. The yeast Hog1 MAPK is essential for cell survival under hyperosmotic conditions and it plays multiple roles in gene expression, metabolic regulation, signal fidelity and cell cycle regulation. Here we describe essential and non-essential roles of Hog1 using engineered yeast cells in which osmoadaptation was reconstituted in a Hog1-independent manner. We rewired Hog1-dependent osmotic stress-induced gene expression under the control of Fus3/Kss1 MAPKs, which are activated upon osmostress via crosstalk in hog1Δ cells. This approach revealed that osmotic up-regulation of only two Hog1-dependent glycerol biosynthesis genes, GPD1 and GPP2, is sufficient for successful osmoadaptation. Moreover, some of the previously described Hog1-dependent mechanisms appeared to be dispensable for osmoadaptation in the engineered cells. These results suggest that the number of essential MAPK functions may be significantly smaller than anticipated and that knockout approaches may lead to over-interpretation of phenotypic data.
Similar content being viewed by others
Introduction
All living cells respond to extracellular stimuli such as hormones, growth factors, cytokines, nutrients and stress. The information is processed by signal transduction systems, which mediate appropriate responses including altered gene expression, metabolism, secretion, proliferation and apoptosis. A conserved family of mitogen-activated protein kinases (MAPKs) serves major roles in intracellular signal transduction from yeasts to mammals1. MAPKs have numerous targets which they regulate at transcriptional and post-translational levels. In the budding yeast Saccharomyces cerevisiae, which has five distinct MAPKs (Hog1, Fus3, Kss1, Slt2/Mpk1 and Smk1)2, the osmoregulatory Hog1 MAPK (a mammalian p38 MAPK homologue) controls gene expression, glycerol accumulation, signal fidelity and cell cycle arrest under hyperosmotic conditions3,4 (Fig. 1a). Consequently, deletion of HOG1 confers osmosensitivity. Several hundred genes are upregulated upon osmotic stress and expression of about 50 of those genes is strongly dependent on Hog15,6. Hog1 affects by phosphorylation the activity of numerous proteins, such as transcription factors7, cell cycle regulators8 and metabolic enzymes9. In this work, we investigated whether Hog1 requires all of those targets for mediating osmoadaptation. For this purpose, we chose a synthetic biological concept, i.e. reconstitution of osmoadaptation in the hog1Δ mutant.
Among Hog1-dependent osmostress-induced genes only GPD1 is essential for osmoadaptation.
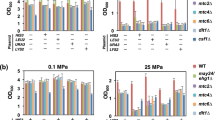
(a) Schematic diagram of the Hog1 MAPK pathway in S. cerevisiae. The HOG pathway consists of two upstream osmosensing branches (Sln1 and Sho1) each with a downstream MAPK cascade (Ssk2/Ssk22 and Ste11 MAPKKKs, Pbs2 MAPKK and Hog1 MAPK). Activation of the HOG pathway leads to rapid translocation of Hog1 into the nucleus, which in turn stimulates expression of osmo-responsive genes via several transcription factors. In addition to gene expression, Hog1 plays roles in glycerol accumulation, control of cell cycle progression, cross pathway inhibition and other aspects of cell physiology. Upstream osmosensing systems are shown in yellow and MAPK cascades in blue. (b) Hog1-dependent osmostress-induced genes identified by microarray analysis6 include genes encoding different stress protective proteins and metabolic enzymes. (c) Deletion of all strongly Hog1-dependent genes except GPD1 does not cause an osmosensitive phenotype. Strains (see further mutants listed in Table S1) were grown on YPD plates with or without 0.8 M KCl for 1–2 days at 30°C.
Yeast is a highly attractive model organism for the study of MAPK signalling systems and for developing and testing synthetic biological approaches10. Engineering signalling pathways has significant potential to provide novel and complementary information that cannot be achieved by traditional genetic approaches such as gene knockout and overexpression11,12. For instance, rewiring signalling components between MAPK pathways13,14, introducing synthetic negative or positive feedback loops15,16, tethering signalling components with specific localization motifs17, assembling or recombining modular signalling domains18,19 and reconstitution of a heterologous MAPK cascade20 are highly informative for understanding the design principles of MAPK pathways and enable generating novel signalling properties. In the present study, we reconstituted osmoadaptation in hog1Δ cells by rewiring osmostress signalling through the MAPK network. This reconstitution approach revealed that osmotic induction of only two Hog1-dependent genes, which encode the enzymes required to produce the osmolyte glycerol, is sufficient for successful osmoadaptation. Moreover, analyses of yeast cells with synthetic osmoadaptation suggest that some of the well-known roles of Hog1 do not seem to be truly essential for osmoadaptation, at least not in the engineered cells. Hence, it appears that the number of MAPK functions essential for specific response may be significantly smaller than anticipated from knockout approaches and genome-wide analyses.
Results
Among Hog1-dependent osmostress-induced genes only GPD1 is essential for osmoadaptation
We and others have previously analyzed the transcriptional response to osmotic shock in Saccharomyces cerevisiae and found that the mRNA level of 200 to 300 genes increased at least 3-fold upon stress5,6. About fifty of those induced genes were highly dependent on the presence of Hog1 and those Hog1-dependent genes encode proteins that presumably contribute to protection against different types of damage or encode enzymes in glycerol, trehalose and glycogen metabolism (Fig. 1b and Table S1). To determine whether those gene products are required for osmoadaptation, we performed growth assay of the corresponding deletion mutants. Growth of the wild-type, hog1Δ and 9 mutant strains chosen as examples (because they have been reported to show osmosensitive phenotype in the Saccharomyces genome database) are shown in Figure 1c. The rest of the mutant strains are shown in Table S1. Only the gpd1Δ strain (GPD1 encodes glycerol-3-phosphate dehydrogenase, the first step in glycerol biosynthesis21,22) showed osmosensitivity similar to the hog1Δ mutant. None of the mutants lacking genes (e.g. ald3Δ, ctt1Δ, hsp12Δ, stl1Δ) whose transcriptional induction is stronger or more dependent on Hog1 than that of GPD1 was osmosensitive. Hence, glycerol biosynthesis appears to be one of the most critical factors for Hog1-dependent osmoadaptation. Moreover, the fact that Hog1 mediates strong upregulation of many genes dispensable for osmoadaptation under laboratory conditions is consistent with a previous report showing no apparent correlation between gene expression and gene dispensability under osmostress conditions23.
Synthetic osmoadaptation in hog1Δ cells using crosstalk between MAPK pathways
It is known that overexpression of GPD1 with a multi copy plasmid partly suppresses the hyper-osmosensitive phenotype of hog1Δ cells22. On the basis of this fact and the result described above, we hypothesized that osmotic up-regulation of GPD1 expression would suppress the phenotype of hog1Δ. We examined this hypothesis by reconstituting osmoadaptation in a Hog1-independent manner. We rewired osmostress signalling to the Fus3/Kss1 MAPKs, which is improperly activated via crosstalk in hog1Δ cells upon osmostress6,24,25. Specifically, we constructed a yeast strain in which GPD1 is expressed under the control of the Fus3/Kss1-dependent FUS1 promoter (Fig. 2a). As shown in Figure 2b, hog1Δ cells carrying a PFUS1-GPD1 gene grew better than control cells on osmotic stress plates containing KCl (or NaCl and sorbitol: data not shown). Growth of the engineered cells (here called synthetic osmoadaptation) was dependent on the presence of Fus3/Kss1. Growth of these cells under osmotic stress (Fig. 2b) correlated well with the ability of those cells to accumulate glycerol (Fig. 2c). However, it should be noted that glycerol accumulation in the hog1Δ cells carrying PFUS1-GPD1 started more slowly than in wild-type cells.
Synthetic osmoadaptation in hog1Δ cells.
(a) Experimental design for synthetic osmoadaptation in hog1Δ using crosstalk and the glycerol biosynthesis pathway. Expression of Hog1-dependent osmostress-induced genes (GPD1 or/and GPP2) was rewired under the control of a Fus3/Kss1 dependent FUS1 promoter. (b) Osmotic induction of GPD1 expression via crosstalk partially suppresses osmosensitivity of hog1Δ in a Fus3/Kss1-dependent manner. Cells of the indicated strains carrying YIp352 or YIp352-PFUS1-GPD1 were grown on YPD plates with or without 0.8 M KCl for 1–2 days at 30°C. (c) Intracellular glycerol accumulation correlates with the cell growth shown in (b). Cells were grown to mid-log phase, subjected to osmotic stress (0.8 M KCl) and intracellular glycerol was monitored at the indicated time points. Values represent the mean and standard deviation of three replicas. (d) Osmotic induction of GPD1 and GPP2 together via crosstalk strongly suppresses osmosensitivity of hog1Δ. Cells of the hog1Δ strains carrying YIp352-PFUS1-GPP2 and/or YIplac128-PFUS1-GPD1 (or GPD14A) were grown as in (b). Cell growth shown in (d) correlates well with the ability of those cells to accumulate glycerol (e) and recover cell volume (f) after osmotic treatment. For cell volume data, line thickness indicates standard deviation of data obtained with approximately 30 cells. See Methods for the details of glycerol assay and cell volume measurement.
To make the hog1Δ cells adapt better to hyperosmotic stress, we examined another strongly Hog1-dependent gene, GPP2, which encodes glycerol-3-phosphate phosphatase26. Gpp2 catalyses the dephosphorylation of glycerol-3-phospate to glycerol, the second and final step in glycerol biosynthesis (Fig. 2a). In contrast to GPD1, osmotic induction of GPP2 alone improved neither growth of hog1Δ cells on high osmolarity plates (Fig. 2d) nor glycerol accumulation (Fig. 2e). These results are consistent with a previous report showing that efficient glycerol production requires accumulation of glycerol-3-phosphate by Gpd127. However, co-expression of GPD1 and GPP2 improved synthetic osmoadaptation. hog1Δ cells carrying both PFUS1-GPP2 and PFUS1-GPD1 genes grew under osmotic stress conditions and accumulated glycerol at a level close to wild type (Fig. 2d, e). Moreover, osmotic induction of an unphosphorylated form of GPD1 (GPD14A), which displays higher enzyme activity28, resulted in even better glycerol accumulation than that of wild-type GPD1 (Fig. 2e). These results indicate that rapid and efficient glycerol accumulation suppresses osmosensitivity of hog1Δ.
Next, we determined whether the synthetic osmoadaptation can restore cell volume after osmotic shock by using a microfluidic device mounted under a fluorescence microscope29. The hog1Δ cells carrying PFUS1-GPD14A/PFUS1-GPP2 recovered cell volume more quickly than the control hog1Δ cells (Fig. 2f). This observation is consistent with recent reports showing that glycerol accumulation is essential for cell volume recovery30,31. Taken together, glycerol accumulation by upregulating expression of two genes, GPD1 and GPP2, is sufficient for synthetic osmoadaptation.
Hog1 is dispensable for regulation of Fps1 gating under hyperosmotic condition
The aquaglyceroporin Fps1 acts as a facilitator for glycerol efflux32,33. Proper Fps1 gating requires its N- and C-terminal regions and unregulated Fps1 causes sensitivity to osmotic34,35, arsenite36 and acetic acid stress37. Fps1 gating upon osmostress appears to be controlled by Hog1 via phosphorylation of Fps136 and its regulator Rgc238 (Fig. 3a). To examine the importance of this step, we introduced a deletion of FPS1 (fps1Δ) or hyperactive Fps1 (N-terminal truncated FPS1-Δ1) into the synthetic osmoadaptation strain (hog1Δ with PFUS1-GPD14A PFUS1-GPP2). The presence or absence of FPS1 did not affect the synthetic osmoadaptation where Fps1 gating cannot be regulated by Hog1, while expression of the unregulated FPS1-Δ1 caused a strong osmosensitive phenotype (Fig. 3b). Moreover, growth of the engineered strains under osmotic condition showed a good correlation with their ability to accumulate glycerol (Fig. 3c) and recover cell volume (Fig. 3d). These results strongly suggest that Hog1-dependent regulation of Fps1 gating is dispensable for synthetic osmoadaptation although control of Fps1 is essential.
Hog1 is dispensable for regulation of Fps1 gating under hyperosmotic condition.
(a) Schematic diagram of Fps1 regulation. Fps1 gating appears to be regulated by phosphorylation at Thr231 and eviction of its positive regulator Rgc2 (not shown), which are mediated by Hog1 and other kinases. Unregulated Fps1 (N-terminal truncated or Thr231 mutants) cannot close the gate and consequently causes osmosensitivity because of constitutive glycerol leakage. (b) The presence or absence of Fps1 does not affect synthetic osmoadaptation, while expression of Fps1-Δ1 prevents it. Cells of the hog1Δ PFUS1-GPP2 PFUS1-GPD14A strains with vector (pRS403), FPS1 deletion, or Fps1-Δ1 (pRS403-FPS1-Δ1) were grown on YPD plates with or without 0.8 M KCl for 1–2 days at 30°C. Cell growth shown in (b) correlates well with the ability of those cells to accumulate glycerol (c) and recover cell volume (d) after osmotic treatment. For cell volume data, line thickness indicates standard deviation of data obtained with approximately 30 cells. See Methods for the details of glycerol assay and cell volume measurement.
Prevention of osmostress-induced abnormal morphology does not affect osmoadaptation
The mechanism how Hog1 prevents crosstalk between the Hog1 and Fus3/Kss1 MAPK pathways, which share several upstream components including Ste20, Ste50 and Ste11, has been extensively studied39. Since previous studies of crosstalk in hog1Δ cells did not consider glycerol accumulation, we examined whether overexpression of GPD14A (PTEF-GPD14A) affects crosstalk. In contrast to sustained activation of Kss1 in hog1Δ cells upon osmotic stress40, we found that activation of Kss1 in hog1Δ cells overexpressing GPD14A was strongly attenuated (Fig. 4a). As expected, the hog1Δ cells overexpressing GPD14A were able to grow well under hyperosmotic condition (Fig. 4b). Importantly, growth supported by overexpressed GPD14A appeared to be better than that of the hog1Δ cells carrying PFUS1-GPD14A/PFUS1-GPP2, which showed strong activation of Kss1 upon osmotic stress (Supplementary Fig. 1). These results suggest that prevention of strong crosstalk does not necessarily require Hog1 itself as long as GPD1 is upregulated before subjection to osmotic stress and that glycerol accumulation and prevention of crosstalk together result in better synthetic osmoadaptation than glycerol accumulation alone.
Prevention of osmostress-induced abnormal morphology does not affect osmoadaptation.
(a) Constitutive expression of GPD14A attenuates crosstalk in hog1Δ. Kss1 phosphorylation upon osmotic stress (0.4 M KCl) was monitored at 0, 30, 60 and 90 minutes by Western blot analysis using an anti-phospho p42/44 antibody. The protein level of Kss1 (detected using anti-Kss1 antibody) served as a loading control. Full-length blots are presented in Supplementary Figure 1. (b, c) Different cell morphology in the presence or absence of crosstalk does not affect synthetic osmoadaptation. Cells of the indicated strains carrying YIplac128-PTEF-GPD14A were grown on YPD plates with or without 0.8 M KCl for 1–2 days at 30°C (b). Cell morphology after one day was observed under the microscope (c). (d) Experimental design for forced osmotic induction of invasive growth. (e) Osmotic induction of filamentous growth (invasive growth) by expressing TEC1T273V or FLO11 under the control of the osmoresponsive STL1 promoter. Cells of different strains in the Σ1278b background were patched on YPD plates with or without 0.8 M KCl, grown for 1–2 days at 30°C (left) and then washed with water (right). (f) Prevention, forced osmotic induction, or constitutive induction of filamentous growth (invasive growth) does not affect osmoadaptation. Cell growth of the indicated Σ1278b strains was examined as in (b).
Crosstalk in the hog1Δ mutant is known to cause a shmoo-like cell morphology and growth arrest24 and a previous study suggested that this abnormal morphology contributes to the osmosensitivity of hog1Δ25. Overexpression of GPD14A partly suppressed the abnormal morphology of hog1Δ (Fig. 4c) and this suppression is probably due to attenuated crosstalk. Although blocking the crosstalk by deletion of both FUS3 and KSS1 genes completely prevented the abnormal morphology, it did not affect growth under hyperosmotic condition in any way (Fig. 4b, c). These results suggest that abnormal morphology itself does not affect osmoadaptation.
To verify that osmoadaptation does not require prevention of osmostress-induced abnormal morphology, we constructed yeast strains in which filamentous growth (invasive growth) is induced upon osmotic stress and examined whether it affects osmoadaptation. We engineered a Σ1278b strain, which is commonly used for the study of filamentous growth41, such that the cells express a constitutively stable TEC1T273 gene (Tec1 is a transcription factor responsible for expression of genes required for invasive growth) under the control of a Hog1-dependent osmoresponsive STL1 promoter (Fig. 4d). As shown in Figure 4e, the wild-type and tec1Δ cells did not invade into the osmotic agar plate, while the cells carrying TEC1T273V (constitutive expression) or PSTL1-TEC1T273V were able to invade into the osmotic stress agar plate. Importantly, all of these strains grew normally under osmotic stress (Fig. 4f). Moreover, yeast cells that upregulated FLO11 (mucin protein responsible for invasive growth) under the control of STL1 promoter showed almost the same growth patterns as cells carrying PSTL1-TEC1T273V (Fig. 4e, f). These results indicate that prevention, forced osmotic induction, or constitutive induction of invasive growth does not affect osmoadaptation. Therefore, prevention of abnormal morphology by Hog1 appears to be dispensable for osmoadaptation.
Discussion
In this report, we reconstituted osmoadaptation in hog1Δ cells by rewiring osmostress signalling through the MAPK network. To our knowledge, this is the first time that yeast osmoadaptation was synthetically mediated through a different MAPK pathway. In addition, our approach made it possible to examine each Hog1-dependent osmostress-induced gene in the hog1Δ mutant background in which all of the Hog1-dependent induction is eliminated. Although our hog1Δ strain showing synthetic osmoadaptation (with PFUS1-GPD1/GPP2) does not have the same capability of establishing osmoresistance as wild type, our data indicate that one of the essential roles of Hog1 for osmoadaptation is osmotic induction of two glycerol biosynthesis genes, GPD1 and GPP2 (Fig. 2). Since Hog1 induces many additional genes, osmotic upregulation of each Hog1-dependent gene may improve the synthetic osmoadaptation. In addition to analysis of systems level properties employing different mutants as system perturbations42,43, such a synthetic approach may also contribute to a quantitative understanding of role of a given Hog1-dependent gene or mechanism in osmoadaptation. However, many of the Hog1-dependent genes do not seem to affect osmoadaptation when deleted individually as suggested by data in Figure 1c, Table S1 and previous reports17,23. Hence, although the physiological reasons why osmotic stress causes upregulation of so many genes remain unclear, this study suggests that the number of essential genes regulated by MAPKs for a specific response or adaptation may be significantly smaller than anticipated.
Nuclear translocation of Hog1 upon osmotic stress44 and the induction of osmoresponsive genes had long been assumed to be necessary for coping with hyperosmotic stress. Contrary to this assumption, Westfall et al. reported that yeast cells lacking Nmd5 (importin-β homologue) required for Hog1 nuclear import or cells in which Hog1 is tethered to the plasma membrane can adapt to hyperosmotic conditions without Hog1-dependent induction of the osmoresponsive genes17. Although our approach is completely different from theirs, these two genetic engineering approaches may suggest that yeast cells can overcome hyperosmotic stress if one or two critical roles of Hog1 are maintained (Hog1 activity and prevention of crosstalk in Westfall's strain; osmotic induction of GPD1 and GPP2 in the strains developed here). This idea is further supported by the fact that overexpression of GPD14A, which causes both crosstalk attenuation and glycerol accumulation, results in improved synthetic osmoadaptation.
Our observation suggests also that Hog1 is dispensable for regulation of Fps1 under hyperosmotic condition although proper Fps1 gating (closing) itself is essential (Fig. 3b–d). This result extends a previous finding that even hog1Δ cells can reduce glycerol transport activity upon hyperosmotic shock33. While this manuscript was in preparation, Levin and coworkers showed that Hog1 closes Fps1 by phosphorylating and displacing the Rgc2 regulator from the C-terminal domain of Fps138. Moreover, they concluded that Hog1 uses the N-terminal domain of Fps1 as a platform to evict Rgc2 from Fps1. However, since deletion of HOG1 does not completely abolish in vivo phosphorylation of Fps136 and Rgc245, other kinases may also be involved in Fps1 gating. At least, we did not observe an osmosensitive phenotype caused by deletion of other MAPK genes (FUS3, KSS1 and SLT2/MPK1) in hog1Δ cells overexpressing GPD14A (Fig. 4b and data not shown), suggesting that Fps1 might be regulated also in a MAPK-independent manner. Changes of turgor pressure or cell volume upon osmotic stress may contribute, probably transiently, to regulation of Fps1 even without Hog1 and/or other MAPKs.
Abnormal cell morphology following osmostress is observed in hog1Δ cells even when GPD14A is overexpressed, while it is completely prevented by deletion of FUS3 and KSS1 (Fig. 4c). Although a previous study showed that prevention of crosstalk by deletion of KSS1 partially suppresses osmosensitivity25, our results indicate that hog1Δ cells with abnormal or normal morphology are capable of acquiring osmoresistance as long as GPD1 is upregulated (Fig. 4b). Therefore, our results suggest that prevention of crosstalk indeed suppresses osmosensitivity, but prevention of osmostress-induced abnormal morphology itself is dispensable. In the Σ1278b strain background, Hog1 acts as a central negative regulator of morphological developments46,47 including fluffy colony morphology, invasive growth and pseudohyphal development, which all are stimulated by the Kss1 MAPK. We demonstrated using a Σ1278b strain background that forced osmotic induction of invasive growth does not impair osmoadaptation (Fig. 4e). Hence, our results strongly suggest that inhibitory regulation of morphological developments is not required for proper osmoadaptation. In addition to investigating how the crosstalk between MAPK pathways is prevented, it would be interesting to understand whether there is a trade-off between osmoadaptation and morphological developments.
Genetic analysis of signal transduction systems by gene deletion or overexpression and phenotypic characterization is well-known to be prone to incorrect interpretation because of compensatory effects, altered protein complex formation and altered pathway crosstalk. Here we present synthetic osmoadaptation in a Hog1-independent manner by using a complementary approach, which provided novel insights into yeast osmoadaptation. Although we focused on glycerol biosynthesis genes, Fps1 regulation and crosstalk inhibition, Hog1 plays many further roles such as controls of cell cycle8 and translation48 and there are many candidate substrates of Hog149. We expect that reconstitution of each role one by one may lead to better understanding of the truly essential Hog1's roles. Moreover, it would be interesting to reconstitute osmoadaptation using orthogonal control systems such as light and hormones. This kind of effort may contribute to creating novel synthetic signalling pathways with predictable behaviours useful for future applications in medicine and biotechnology.
Methods
Yeast media and growth conditions
Standard media, SC (synthetic complete: 2% glucose, 0.67% yeast nitrogen base without amino acids and supplemented with amino acids to satisfy nutritional requirements) and YPD (1% yeast extract, 2% peptone and 2% glucose), were used for yeast cultivation and selection of transformants. For growth assays to examine osmosensitivity, cells were pregrown overnight on YPD plates, resuspended in water to OD600 = 0.1 and 5 μl of a 10-fold dilution series were spotted onto YPD plates with or without KCl. Cell growth or morphology was monitored after 1–2 days culture at 30°C.
Yeast strains and plasmids
Yeast strains, plasmids and primers used in this study are listed in Table S2, Table S3 and Table S4, respectively. Yeast transformation and gene deletion were performed as described previously50. Plasmids for GPD1 and GPP2 expression under the control of FUS1 or TEF promoter were constructed in the YIp352 (URA3 marker) or YIplac128 (LEU2 marker) backbones. A GPD1 fragment was obtained from pCM190HH-GPD1 (Markus Tamás). GPP2, PFUS1 and PTEF fragments were obtained by PCR using yeast genomic DNA or plasmid (pYM-N18 for PTEF) as a template. GPD14A and TEC1T273V mutations were generated by overlap extension PCR-mediated mutagenesis. A plasmid for FPS1-Δ1 expression was constructed by insertion of the FPS1-Δ1 fragment derived from YIpURA3-FPS1-Δ1 (Markus Tamás) into pRS403 (HIS3 marker). PSTL1-TEC1T273V and PSTL1-FLO11 strains were constructed by replacement of their original promoter regions with a LEU2-PSTL1 cassette.
Measurement of intracellular glycerol
Cells were grown to mid-log phase in 30 ml of YPD liquid medium. KCl was added to the medium to a final concentration of 0.8 M and 1 ml aliquots were withdrawn after 0, 2, 4 and 6 hours. Cells were harvested and resuspended in 1 ml of water and boiled at 100°C for 10 min and supernatants were stored at −20°C. OD600 was determined at all-time points. Glycerol concentration was determined using a commercial kit (Roche Applied Science). Reaction was scaled down 12 times to a final reaction volume of 250 μl. Measurements were performed in a 96-well plate using a Polar Star Omega plate reader (BMG Labtech). The mean value of glycerol/OD600 ± S.D. (n = 3) was plotted versus time.
Single cell analysis of cell volume
Single cell analysis of cell volume upon osmotic stress (0.8 M KCl) was performed using a microfluidic system with three inlet channels as described previously29,30. Images of approximately 30 cells were taken sequentially every 30 sec for 5 min, every 1 min for 5 min, every 10 min for 20 min and every 30 min for 90 min, thus yielding a total experiment period of 120 min. The images were analyzed using CellStress software51.
Western blot
Cells were grown to mid-log phase in 30 ml of YPD liquid medium. KCl was added to the medium to a final concentration of 0.4 M and 1 ml aliquots were withdrawn at times 0, 30, 60 and 90 min. Cells were resuspended in standard SDS loading buffer, boiled for 10 min and sedimented at 13,000 × g at 4°C for 10 min to obtain protein extracts. Protein extracts (45 μg) were examined for Western blot analysis using anti-phospho-p44/42 MAPK antibody (Cell Signaling Technology) or anti-Kss1 antibody (y-50, Santa Cruz Biotechnology) as described previously30.
References
Widmann, C., Gibson, S., Jarpe, M. B. & Johnson, G. L. Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol. Rev. 79, 143–180 (1999).
Chen, R. E. & Thorner, J. Function and regulation in MAPK signaling pathways: lessons learned from the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1773, 1311–1340 (2007).
Hohmann, S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 66, 300–372 (2002).
Saito, H. & Posas, F. Response to hyperosmotic stress. Genetics 192, 289–318 (2012).
Posas, F. et al. The transcriptional response of yeast to saline stress. J. Biol. Chem. 275, 17249–17255 (2000).
Rep, M., Krantz, M., Thevelein, J. M. & Hohmann, S. The transcriptional response of Saccharomyces cerevisiae to osmotic shock. Hot1p and Msn2p/Msn4p are required for the induction of subsets of high osmolarity glycerol pathway-dependent genes. J. Biol. Chem. 275, 8290–8300 (2000).
de Nadal, E. & Posas, F. Multilayered control of gene expression by stress-activated protein kinases. EMBO J. 29, 4–13 (2010).
Duch, A., de Nadal, E. & Posas, F. The p38 and Hog1 SAPKs control cell cycle progression in response to environmental stresses. FEBS Lett. 586, 2925–2931 (2012).
Dihazi, H., Kessler, R. & Eschrich, K. High osmolarity glycerol (HOG) pathway-induced phosphorylation and activation of 6-phosphofructo-2-kinase are essential for glycerol accumulation and yeast cell proliferation under hyperosmotic stress. J. Biol. Chem. 279, 23961–23968 (2004).
Furukawa, K. & Hohmann, S. Synthetic biology: lessons from engineering yeast MAPK signalling pathways. Mol. Microbiol. 88, 5–19 (2013).
Kiel, C., Yus, E. & Serrano, L. Engineering signal transduction pathways. Cell 140, 33–47 (2010).
Lim, W. A. Designing customized cell signalling circuits. Nat. Rev. Mol. Cell. Biol. 11, 393–403 (2010).
Park, S. H., Zarrinpar, A. & Lim, W. A. Rewiring MAP kinase pathways using alternative scaffold assembly mechanisms. Science 299, 1061–1064 (2003).
Tatebayashi, K., Takekawa, M. & Saito, H. A docking site determining specificity of Pbs2 MAPKK for Ssk2/Ssk22 MAPKKKs in the yeast HOG pathway. EMBO J. 22, 3624–3634 (2003).
Ingolia, N. T. & Murray, A. W. Positive-feedback loops as a flexible biological module. Curr. Biol. 17, 668–677 (2007).
Bashor, C. J., Helman, N. C., Yan, S. & Lim, W. A. Using engineered scaffold interactions to reshape MAP kinase pathway signaling dynamics. Science 319, 1539–1543 (2008).
Westfall, P. J., Patterson, J. C., Chen, R. E. & Thorner, J. Stress resistance and signal fidelity independent of nuclear MAPK function. Proc. Natl Acad. Sci. USA 105, 12212–12217 (2008).
Mody, A., Weiner, J. & Ramanathan, S. Modularity of MAP kinases allows deformation of their signalling pathways. Nat. Cell Biol. 11, 484–491 (2009).
Peisajovich, S. G., Garbarino, J. E., Wei, P. & Lim, W. A. Rapid diversification of cell signaling phenotypes by modular domain recombination. Science 328, 368–372 (2010).
O'Shaughnessy, E. C., Palani, S., Collins, J. J. & Sarkar, C. A. Tunable signal processing in synthetic MAP kinase cascades. Cell 144, 119–131 (2011).
Larsson, K., Ansell, R., Eriksson, P. & Adler, L. A gene encoding sn-glycerol 3-phosphate dehydrogenase (NAD+) complements an osmosensitive mutant of Saccharomyces cerevisiae. Mol. Microbiol. 10, 1101–1111 (1993).
Albertyn, J., Hohmann, S., Thevelein, J. M. & Prior, B. A. GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae and its expression is regulated by the high-osmolarity glycerol response pathway. Mol. Cell. Biol. 14, 4135–4144 (1994).
Warringer, J., Ericson, E., Fernandez, L., Nerman, O. & Blomberg, A. High-resolution yeast phenomics resolves different physiological features in the saline response. Proc. Natl Acad. Sci. USA 100, 15724–15729 (2003).
O'Rourke, S. M. & Herskowitz, I. The Hog1 MAPK prevents cross talk between the HOG and pheromone response MAPK pathways in Saccharomyces cerevisiae. Genes Dev. 12, 2874–2886 (1998).
Davenport, K. D., Williams, K. E., Ullmann, B. D. & Gustin, M. C. Activation of the Saccharomyces cerevisiae filamentation/invasion pathway by osmotic stress in high-osmolarity glycogen pathway mutants. Genetics 153, 1091–1103 (1999).
Pahlman, A. K., Granath, K., Ansell, R., Hohmann, S. & Adler, L. The yeast glycerol 3-phosphatases Gpp1p and Gpp2p are required for glycerol biosynthesis and differentially involved in the cellular responses to osmotic, anaerobic and oxidative stress. J. Biol. Chem. 276, 3555–3563 (2001).
Remize, F., Barnavon, L. & Dequin, S. Glycerol export and glycerol-3-phosphate dehydrogenase, but not glycerol phosphatase, are rate limiting for glycerol production in Saccharomyces cerevisiae. Metab. Eng. 3, 301–312 (2001).
Oliveira, A. P. et al. Regulation of yeast central metabolism by enzyme phosphorylation. Mol. Syst. Biol. 8, 623 (2012).
Eriksson, E. et al. A microfluidic device for reversible environmental changes around single cells using optical tweezers for cell selection and positioning. Lab Chip 10, 617–625 (2010).
Babazadeh, R. et al. Osmostress-induced cell volume loss delays yeast Hog1 signaling by limiting diffusion processes and by Hog1-specific effects. PLoS One 8, e80901 (2013).
Geijer, C. et al. Initiation of the transcriptional response to hyperosmotic shock correlates with the potential for volume recovery. FEBS J. 280, 3854–3867 (2013).
Luyten, K. et al. Fps1, a yeast member of the MIP family of channel proteins, is a facilitator for glycerol uptake and efflux and is inactive under osmotic stress. EMBO J. 14, 1360–1371 (1995).
Tamás, M. J. et al. Fps1p controls the accumulation and release of the compatible solute glycerol in yeast osmoregulation. Mol. Microbiol. 31, 1087–104 (1999).
Tamás, M. J. et al. A short regulatory domain restricts glycerol transport through yeast Fps1p. J. Biol. Chem. 278, 6337–6345 (2003).
Hedfalk, K. et al. A regulatory domain in the C-terminal extension of the yeast glycerol channel Fps1p. J. Biol. Chem. 279, 14954–14960 (2004).
Thorsen, M. et al. The MAPK Hog1p modulates Fps1p-dependent arsenite uptake and tolerance in yeast. Mol. Biol. Cell 17, 4400–4410 (2006).
Mollapour, M. & Piper, P. W. Hog1 mitogen-activated protein kinase phosphorylation targets the yeast Fps1 aquaglyceroporin for endocytosis, thereby rendering cells resistant to acetic acid. Mol. Cell. Biol. 27, 6446–6456 (2007).
Lee, J. et al. MAPK Hog1 closes the S. cerevisiae glycerol channel Fps1 by phosphorylating and displacing its positive regulators. Genes Dev. 27, 2590–2601 (2013).
Saito, H. Regulation of cross-talk in yeast MAPK signaling pathways. Curr. Opin. Microbiol. 13, 677–683 (2010).
Hao, N., Zeng, Y., Elston, T. C. & Dohlman, H. G. Control of MAPK specificity by feedback phosphorylation of shared adaptor protein Ste50. J. Biol. Chem. 283, 33798–33802 (2008).
Cullen, P. J. & Sprague, G. F., Jr The regulation of filamentous growth in yeast. Genetics 190, 23–49 (2012).
Klipp, E., Nordlander, B., Kruger, R., Gennemark, P. & Hohmann, S. Integrative model of the response of yeast to osmotic shock. Nat. Biotechnol. 23, 975–982 (2005).
Petelenz-Kurdziel, E. et al. Quantitative analysis of glycerol accumulation, glycolysis and growth under hyper osmotic stress. PLoS Comput. Biol. 9, e1003084 (2013).
Ferrigno, P., Posas, F., Koepp, D., Saito, H. & Silver, P. A. Regulated nucleo/cytoplasmic exchange of HOG1 MAPK requires the importin β homologs NMD5 and XPO1. EMBO J. 17, 5606–5614 (1998).
Beese, S. E., Negishi, T. & Levin, D. E. Identification of positive regulators of the yeast fps1 glycerol channel. PLoS Genet. 5, e1000738 (2009).
Furukawa, K., Sidoux-Walter, F. & Hohmann, S. Expression of the yeast aquaporin Aqy2 affects cell surface properties under the control of osmoregulatory and morphogenic signalling pathways. Mol. Microbiol. 74, 1272–1286 (2009).
Furukawa, K., Furukawa, T. & Hohmann, S. Efficient construction of homozygous diploid strains identifies genes required for the hyper-filamentous phenotype in Saccharomyces cerevisiae. PLoS One 6, e26584 (2011).
Warringer, J., Hult, M., Regot, S., Posas, F. & Sunnerhagen, P. The HOG pathway dictates the short-term translational response after hyperosmotic shock. Mol. Biol. Cell 21, 3080–3092 (2010).
Reiter, W. et al. Validation of regulated protein phosphorylation events in yeast by quantitative mass spectrometry analysis of purified proteins. Proteomics 12, 3030–3043 (2012).
Amberg, D. C., Burke, D. J. & Strathern, J. N. Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press (2005).
Frey, S. et al. A mathematical analysis of nuclear intensity dynamics for Mig1-GFP under consideration of bleaching effects and background noise in Saccharomyces cerevisiae. Mol. Biosyst. 7, 215–223 (2011).
Acknowledgements
We thank Markus Tamás (Chemistry and Molecular Biology, University of Gothenburg) for valuable discussion and plasmids as well as Mattias Goksör and Caroline Beck-Adiels (Physics, University of Gothenburg) for letting us use their microfluidic system. This work was supported by the Swedish Research Council (project grant to S.H.), the Olle Engkvist Byggmästare Foundation (to K.F.) and the European Commission via the FP7 project UNICELLSYS (No. 201142 to S.H.).
Author information
Authors and Affiliations
Contributions
K.F. designed and supervised the study; R.B., T.F. and K.F. performed the experimental work; R.B., S.H. and K.F. analysed results and wrote the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Information
Rights and permissions
This work is licensed under a Creative Commons Attribution 3.0 Unported License. The images in this article are included in the article's Creative Commons license, unless indicated otherwise in the image credit; if the image is not included under the Creative Commons license, users will need to obtain permission from the license holder in order to reproduce the image. To view a copy of this license, visit http://creativecommons.org/licenses/by/3.0/
About this article
Cite this article
Babazadeh, R., Furukawa, T., Hohmann, S. et al. Rewiring yeast osmostress signalling through the MAPK network reveals essential and non-essential roles of Hog1 in osmoadaptation. Sci Rep 4, 4697 (2014). https://doi.org/10.1038/srep04697
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep04697
This article is cited by
-
New biomarkers underlying acetic acid tolerance in the probiotic yeast Saccharomyces cerevisiae var. boulardii
Applied Microbiology and Biotechnology (2024)
-
Positive feedback induces switch between distributive and processive phosphorylation of Hog1
Nature Communications (2023)
-
Investigation of osmotic shock effect on pulsed electric field treated S. cerevisiae yeast cells
Scientific Reports (2023)
-
The role of NAD and NAD precursors on longevity and lifespan modulation in the budding yeast, Saccharomyces cerevisiae
Biogerontology (2022)
-
The role of the glycerol transporter channel Fps1p in cellular proteostasis during enhanced proteotoxic stress
Applied Microbiology and Biotechnology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.