Abstract
The disulfide bond is unusual in natural products and critical for thermal stability, cell permeability and bioactivity. DepH from Chromobacterium violaceum No. 968 is an FAD-dependent enzyme responsible for catalyzing the disulfide bond formation of FK228, an anticancer prodrug approved for the treatment of cutaneous T-cell lymphoma. Here we report the crystal structures of DepH and DepH complexed with a substrate analogue S,S'-dimethyl FK228 at 1.82 Å and 2.00 Å, respectively. Structural and biochemical analyses revealed that DepH, in contrast to the well characterized low molecular weight thioredoxin reductases (LMW TrxRs), is an NADP+-independent dithiol oxidase. DepH not only lacks a conserved GGGDXAXE motif necessary for NADP+ binding in the canonical LMW TrxRs, but also contains a 11-residue sequence which physically impedes the binding of NADP+. These observations explain the difference between NADP+-independent small molecule dithiol oxidases and NADP+-dependent thioredoxin reductases and provide insights for understanding the catalytic mechanism of dithiol oxidases involved in natural product biosynthesis.
Similar content being viewed by others
Introduction
The presence of a disulfide bond in natural products is rare but usually critical for their bioactivity, thermal stability and cell permeability. One classic example is the antitumorigen FK228 (also known as FR901228 or depsipeptide, registered as NSC 630176, romidepsin or Istodax; Fig. 1), a selective class I histone deacetylase (HDAC) inhibitor approved for the treatment of cutaneous T-cell lymphoma in 2009 by the US Food and Drug Administration1,2. FK228 is produced by the Gram-negative Chromobacterium violaceum No. 968, with a cage-shaped bicyclic depsipeptide structure and a rare disulfide linkage3,4. The prodrug form of FK228 with a disulfide bond is more stable than its reduced form (red-FK228, Fig. 1) in medium or serum and this disulfide bond facilitates compound diffusion across the cell membrane more efficiently. Once inside the cells, the prodrug FK228 is activated by glutathione reduction so that one of the free thiol groups of red-FK228 interacts strongly with an active-site zinc and thereby prevents substrate binding1. Similar disulfide linkages are also present in a few other antibiotics or antitumor natural products, including holomycin5, gliotoxin6, triostin A and its derivatives7, FR9013758, spiruchostatins9 and thailandepsins10,11,12.
The enzyme responsible for catalyzing the disulfide bond formation in FK228 biosynthesis is DepH13. A BLAST search for proteins homologous to DepH revealed several low molecular weight thioredoxin reductases (LMW TrxRs) from bacteria and fungi and small molecule dithiol oxidoreductases including TdpH from Burkholderia thailandensis E26410 and Ecm17 from Streptomyces lasaliensis7. TrxRs are homodimeric flavoproteins consisting of a cofactor FAD-binding site, an NADP+-binding site and a redox motif CXXC (two cysteine residues separated by two variable amino acids)14,15,16. They catalyze the oxidation or reduction of disulfide bonds through thiol-disulfide exchange between the enzymes and their substrate proteins. Extensive structural studies revealed that the NADP+- and FAD-binding sites of LMW TrxRs are located on opposite faces of the molecules and a conformational change is required for electron transfer from the reduced form of FAD to NADP+17.
Results
Overall structure of DepH
The full-length DepH (residues 1–319) from C. violaceum No. 968 was expressed in E. coli BL21(DE3) as an N-terminal 6 × His-tagged fusion protein and purified to homogeneity13. Decreasing the expression temperature to 16°C and maintaining the purification procedure at 4°C were critical to prevent the recombinant protein from degradation. After extensive trials, the high diffraction-quality crystals were made by the micro-seeding method18.
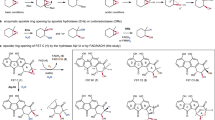
The crystal structure of native DepH was determined by molecular replacement using the structure of a functionally unknown oxidoreductase from Agrobacterium tumefaciens (PDB code: 3FBS, sequence identity 52%, abbreviated as AtuOR in this work) as a search model. The final structure was refined to 1.82 Å resolution with an Rfactor of 16.8% and an Rfree of 20.0% (Table 1). There are two DepH molecules in the asymmetric units related by a non-crystallographic 2-fold axis (Fig. 2A). Similar to the structures of LMW TrxRs13,14,18,19,20,21,22, each DepH polypeptide chain is arranged in two Rossmann-fold domains: an FAD-binding domain (residues 21–138 and 248–319) and a pseudo NADP+-binding domain (residues 139–247) (Fig. 2B). Two antiparallel strands, β7 (residues 135–138) and β16 (residues 248–251), connect the FAD-binding domain with the pseudo NADP+-binding domain. The redox motif 156CXXC159 is located on helix α3 within the pseudo NADP+-binding domain. Under native condition, residues Cys156 and Cys159 are close to each other and form a disulfide bond (2.07 Å) and the Pro157 adopts the trans conformation (Fig. 2C). Similar to LMW TrxRs, the Sγ atom of Cys159 is in the range of electron transfer distance of 3.58 Å with the C4a atom of FAD, while the Sγ atom of Cys156 is away from the FAD (Fig. 2C).
(A) The overall structures of DepH in native form and complexed with dm-FK228. Both structures are homodimers and adopt resembling architectures. Only one monomer of the complexed DepH is shown in gray and superposed with that of the native DepH (green). The other monomer of the native DepH is shown in cyan. The bound FAD and dm-FK228 molecules as well as the redox motif CXXC are shown in stick models in yellow, magenta and orange, respectively. (B) The DepH monomer is consisted of a pseudo NADP+-binding domain and a FAD-binding domain. The secondary structure elements are labelled and the helices are shown as cylinders. (C) A stereo view of the 2Fo-Fc map of the redox motif CPYC and the cofactor FAD (contoured at σ = 1.0). (D, E) Simulated-annealing omit map of the cofactor FAD and the bound dm-FK228 (contoured at σ = 3.5).
It is worth noting that the electron density of FAD is well defined (with B factors near 20 Å2) (Fig. 2D), but no electron density of NADP+ was found in the pseudo NADP+-binding pocket of DepH. The isoalloxazine ring of FAD is planar and stabilized by polar interactions with residues Ser36, Ala38, Ala58, Ala60, Arg62, His70, Val104, Asp290 and Val298, accompanied with non-polar interactions involving residues Ile56, Asp57, Phe133, Trp149, Gly150, Cys159, His160 and Ser252 (Supplementary Fig. 1). The conformation and relative location of FAD are similar to those observed in LMW TrxRs14,15,19,20,21,22,23. The planarity of FAD and the formation of the disulfide bond in the CXXC motif indicate that the native DepH is in the oxidized state.
Molecular basis for FK228 binding
To investigate the molecular basis of substrate binding, we tried to cocrystallize DepH with FK228 in the presence of 5 mM DTT but only got the apo-DepH crystals. Later we obtained and refined the crystal structure of DepH complexed with its substrate analogue dm-FK228 at 2.00 Å resolution. The dm-FK228 molecule contained two methylated thiol groups and was prepared by full methylation of red-FK228 (Fig. 1). The overall complex structure resembles the structure of native DepH (with an r.s.m.d. of 0.7 Å for 294 Cα atoms) (Fig. 2A) and also contains a physiologically related homodimer in an asymmetric unit. Clear and ring-shaped density of dm-FK228 is well defined in the complex structure (Fig. 2E). Due to the crystal packing, one of the two dm-FK228 molecules is buried in the interior of the structures, whose B factor is 39.1 Å2. The other dm-FK228 molecule is exposed to the solvent leading to a higher B factor of 74.0 Å2.
Located in the same line as residues Cys156, Cys159 and the isoalloxazine rings of FAD, dm-FK228 is bound at the other side of the hydrophobic cleft between the FAD-binding domain and the pseudo NADP+-binding domain (Fig. 2A). The dm-FK228 molecule is located in a pocket surrounded by the β8-α3 loop, α4 and the β15–β16 loop from the pseudo NADP+-binding domain, the β17–β18 loop and the η4-α7 loop from the FAD-binding domain and α7* from the other subunit (Fig. 3A). The two aliphatic chains containing sulfur atoms are in the same plane of the peptide ring of dm-FK228 (Fig. 3B). The sulfur atom from the longer aliphatic chain of dm-FK228 is near the CXXC redox motif, with a distance about 3.51 Å from the Sγ atom of the residue Cys156. Although the isoalloxazine ring of FAD is still planar, the distance between the two Sγ atoms of the residue Cys156 and Cys159 is 3.27 Å (Fig. 3C and Supplementary Fig. 2), indicating the breakage of disulfide bond and the existence of the sulfydryl groups24.
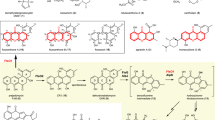
The interactions between DepH and its substrate analog dm-FK228.
(A) The dm-FK228 binding site is located on the dimer interface of DepH. The surfaces of two subunits are shown in gray and pink, respectively. The secondary structures involved in the substrate binding pocket are labeled. (B) Comparison of the structures of the native DepH and its complex with dm-FK228 in the substrate-binding pocket. Cofactor FAD and residues from the native DepH were colored white, while those from complex were depicted cyan and salmon individually. Essential water molecules were depicted as magenta spheres. (C) A close view of the substrate binding pocket. Residues interacting with dm-FK228 are depicted by sticks in, magenta and green, respectively, indicating the residues belong to different monomers. Two water molecules (W174 and W331) mediating the hydrogen bonds between dm-FK228 and DepH are shown by red spheres. (D) Relative enzyme activity of the wild type and mutants of DepH.
A close view of the dm-FK228-binding pocket revealed that non-polar interactions and hydrogen bond interactions are prevalent. The residues involved in hydrophobic interactions are Leu177, Met246 and Pro267 from one subunit and Ile310* and Phe318* from the other subunit. The binding of dm-FK228 triggered a torsion of the side chain in Arg314* to form van der Waals interactions. The dm-FK228 molecule also has a direct hydrogen bond interaction with the carbonyl oxygen atom of Gly295 and water-mediated hydrogen bond interactions with the NH1 atom of Arg294 (through Wat331) and the hydroxyl group of Tyr158 (through Wat174) (Fig. 3C and Supplementary Fig. 3).
To probe the role of these residues in catalysis, we generated DepH mutants by substitution with alanine. Mutations of the two key residues, Tyr158 and Arg294, disrupted the hydrogen bond interactions with the substrate and severely affected the catalytic activity of DepH, with approximate 40% residual activity for Y158A and 20% residual activity for R294A. Mutations of residues Pro267 and Met246 dedicated to hydrophobic interactions had negligible effect on enzyme activity. Although His293 has no direct interaction with the substrate, mutation of this residue to alanine caused a 50% decrease in the activity of DepH, indicating that the π − π stacking between His293 and Arg294 was necessary to maintain the conformation of Arg294. Double mutant H293A/R294A showed no synergistic effects (Fig. 3D).
DepH is an NADP+-independent enzyme
Although the pseudo NADP+-binding domain of DepH is similar to those of LMW TrxRs and excess of NADP+ was added in the protein crystallization solutions, the density of NADP+ was not found in the electron density map of either the native or the complex structure (Fig. 2A). To seek for an interpretation, we compared the structure of DepH with those of LMW TrxRs from Escherichia coli (PDB code: 1TDE), Deinococcus radiodurans (PDB code: 2Q7V), Saccharomyces cerevisiae (PDB code: 3ITJ), Helicobacter pylori (PDB code: 2Q0K), Mycobacterium tuberculosis (PDB code: 2A87), Staphylococcus aureus ssp. (PDB code: 4GCM), Hordeum. vulgare (PDB code: 2WHD) and Arabidopsis thaliana (PDB code: 1VDC) and found that they have an overall resembling architecture and very similar orientations of the loops, α-helices and β-sheets (Supplementary Fig. 4). Superposition of DepH with these structures resulted in r. m. s. d. values of 2.04 Å, 1.95 Å, 2.27 Å, 2.06 Å, 2.04 Å, 1.89 Å, 2.65 Å and 2.15 Å, respectively, for the Cα atoms.
The major differences between DepH and the LMW TrxRs are present in the regions I and II (Fig. 4A and Supplementary Fig. 4). Region I (residues 173–180), located between β9 and α4, consists of a GSGPLSYL motif different from the signature motif GGGDXAXE necessary for NADP+ binding in the LMW TrxRs. Moreover, the residues involved in NADP+ binding are different. There are four basic residues, highly conserved in all LMW TrxRs, anchoring the ribosyl and phosphate groups of the bound NADP+. For example, the four basic residues are Arg127, Arg186, Arg187 and Arg191 in the TrxR (PDB code: 2A87) from M. tuberculosis; in DepH, the corresponding residues, Glu138, Ala197, Ser198 and Asp202, are either negatively charged or non-polar, unfavorable for NADP+ binding (Fig. 4B and Supplementary Fig. 4).
(A) Sequence alignment and structural superimposition of regions I and II of DepH with other homologous proteins, including TdpH from B. thailandensis E264, PA14_09950, CJA_3737, Smed_3568, AtuOR, GliT, HlmI and structure-solved LMW TrxRs from E. coli (1TDE), M. tuberculosis (2A87), D. radiodurans (2Q7V), H. pylori (2Q0K), S. aureus ssp. (4GCM), H. vulgare (2WHD), A. thaliana (1VDC) and S. cerevisiae (3ITJ). Details are described in Supplementary Figure 4. The sequences with known structures were aligned using STRAP43. The sequences lacking of three dimensional structures were aligned using ClustalW44. The alignment results were merged with ESPript45. (B) Comparison of the solvent-accessible surface of the NADP+-binding pocket in TrxR from M. tuberculosis (2A87) (left panel) and that of the pseudo NADP+-binding pocket in the DepH/dm-FK228 complex (right panel). The bound NADP+ in TrxR and dm-FK228 in DepH complex are depicted using stick model and colored in magenta and green, respectively. Four basic residues anchoring the ribosyl and phosphate groups of NADP+ in TrxR were labeled. (C) Superposition of the DepH complex structure (colored in violet) with the structure of the NADP+-dependent LMW TrxR from M. tuberculosis (2A87, colored in teal). The cofactor NADP+ from TrxR and dm-FK228 from DepH are shown in sticks. The presence of NADP+ in DepH would impede substrate binding. (D) Relative activities of DepH in the presence or absence of NAD+ or NADP+. A, no additives; B, with NAD+; C, with NADP+.
Region II (residues 263–272), containing two anti-parallel strands β17 and β18 and a connecting β-turn, is involved in the formation of substrate-binding pocket. The counterparts in LMW TrxRs are three residues shorter and adopt a flexible loop conformation (Fig. 4A). Superimposed with TrxR from M. tuberculosis (PDB code: 2A87, 26% sequence identity with an r. m. s. d. value of 2.04 Å for 273 Cα atoms), we found region II of DepH protrudes into the pseudo NADP+-binding site and actually prevents NADP+ binding in the crystal (Fig. 4C). Furthermore, the residues of Region II have little polar interaction with the residues in the neighborhood, except for a hydrogen bond between main chain atoms between residues Leu268 and Met249 (Supplementary Fig. 5). Removing this special region resulted in insolubility of overexpressed mutant proteins (data not shown). Deletion of residues Glu265 and Gly266 from this region decreased the oxidase activity more than 80% (Fig. 3D), indicating that the importance of the region II to maintain the structural scaffold and catalytic activity of DepH.
The lack of sequence fingerprint necessary for NADP+ binding and the missing of NADP+ in the native and complexed DepH structures suggest that its catalysis might be NAD+/NADP+ independent. To test this hypothesis, we measured the catalytic activity of DepH at various protein concentrations in the absence or presence of NAD+ or NADP+. As shown in Fig. 4D, DepH alone exhibited high catalytic efficiency for converting red-FK228 to FK228, whereas addition of NAD+ or NADP+ actually hampered the catalytic reaction of DepH. All those evidences support DepH as an NADP+-independent enzyme.
Discussion
Several small molecule dithiol oxidases have been identified as the thioredoxin reductases homologues from the biosynthetic gene clusters of disulfide containing natural products. Through structure determination of DepH alone and in complex with the substrate analog dm-FK228, we disclosed the characteristics of DepH as an NADP+-indepdent FAD-binding enzyme. Compared to LMW TrxRs, DepH not only lacks a conserved GGGDXAXE sequence required for NADP+ binding but also contains a connecting β-turn in region II which protrudes into the pseudo NADP+-binding site and prevents NADP+ binding.
We observed that there is a disulfide bond in the native DepH structure whereas the corresponding disulfide bond is broken in the complex structure. As dm-FK228 is a pseudo substrate with two sulfydryl groups protected by methylation, no reaction should have occurred between the enzyme and the substrate analog. The molecular dynamics simulations suggested that there were little conformational changes of the CXXC motif in DepH upon dm-FK228 binding (data not shown). Given the fact that the same level of DTT were applied for protein crystallization and the same beamline was used for data collection, we speculated that unusual breakage of disulfide bond in the complex structure might have been the effect of radiation damages or other unexpected factors.
Recently, two FAD-dependent, homodimeric enzymes, GliT from Aspergillus fumigatus and HlmI from Streptomyces clavuligerus, were identified as dithiol oxidases and to catalyze the oxidation of intramolecular dithiol groups to disulfide during the biosynthesis of gliotoxin6 and holomycin5. Although GliT and HlmI are analogous to the LMW TrxRs superfamily, the electron transfer during the dithiol-disulfide exchange involving the CXXC motif is independent of NADP+ and utilizes molecular oxygen as the terminal electron acceptor. To compare DepH with other small molecule dithiol oxidoreductases, we conducted a sequence alignment among GliT, HlmI and DepH as well as other structurally unknown but functionally homologous enzyme, such as TdpH, PA14_09950, CJA_3737 and Smed_3568. As shown in Fig. 4A and Supplementary Fig. 4, all of these enzymes have almost the same redox motif CP/LY/FC, which is totally different from those of the canonical LMW TrxRs, whose redox motifs are CAT/VC. Moreover, these enzymes involved in the oxidation of dithiols into disulfide lack the signature sequence GGGDXAXE necessary for NADP+ binding. In addition, the aforementioned four highly conserved basic residues absent in DepH are accordingly missing in these homologous proteins as well as GliT and HlmI. Based on the sequence alignment and the information that NADP+ was not necessary for the catalytic activity of DepH, we concluded that DepH is a dithiol oxidase which uses molecular oxygen as the terminal electron acceptor to catalyze intramolecular disulfide formation in FK228.
The crystal structures of DepH also shed new light on understanding the catalytic mechanism of dithiol oxidases. We found that there might be an oxygen channel of DepH to the vicinity of the N5 nitrogen of FAD. This channel is mainly formed by residues Arg62, His155 and Cys156, which are strictly conserved in GliT, HlmI and other dithiol oxidases (Supplementary Fig. 6 and Supplementary Fig. 4). This channel is similar to the putative oxygen access channel of yeast Erv225 and might be suitable for FAD to transfer electrons to molecular oxygen. Interestingly, this channel is blocked by the binding of substrate analogue dm-FK228 in the complex structure (Supplementary Fig. 6). The catalytic cycle of a flavo-oxidase typically comprises two half-reactions26: in the reductive half-reaction, the substrate is oxidized by the flavin; in the oxidative half-reaction, an electron acceptor re-oxidizes the reduced flavin and the product is released. Based on the crystal structural information in this work, we proposed that the oxidative half-reaction of DepH is implemented by the ping-pong mechanism (Supplementary Fig. 7). The whole catalytic procedure starts when substrate red-FK228 first binds to the active pocket with the free thiol group of the longer aliphatic chain close to the catalytic motif CXXC. The shorter aliphatic chain containing the other thiol group is exposed to the solvent and locates in the same plane as the longer one Like GliT and HlmI, the reductive half-reaction is initiated by the attack of the thiol group of the longer aliphatic chain of red-FK228 on residue Cys156 in the redox motif to yield a covalent disulfide adduct with the enzyme. Intra-molecular attack of the second thiol of the shorter aliphatic chain upon proton abstraction by an active site base will lead to formation of the FK228 and generation of the reduced form of DepH. After product leaving, the FADox generates the CXXC disulfide in the DepH to form FADH2. Finally, the molecular oxygen accesses the active site and accepts the electron from FADH2 to release hydrogen peroxide and regenerate of FADox.
Methods
Preparation of reduced FK228 (red-FK228) and dimethylated FK228 (dm-FK228)
FK228 was routinely prepared from the fermentation broth of C. violaceum No. 968 in one of our laboratories. Preparation of red-FK228 was accomplished by using tris(carboxylethyl)phosphine hydrochloride (TCEP, Sigma-Aldrich) as the reducing agent. The reaction was performed at room temperature by mixing 20 mg (36 μmol) of FK228 and 100 μmol of TCEP in 1 ml of acetonitrile for 20 min. The red-FK228 was purified by preparative HPLC, confirmed by liquid chromatography mass spectrometry (LC-MS; [M + H]+ = 542 m/z) and lyophilized. To prepare dm-FK228, 10 mg red-FK228 (18 μmol) was dissolved in 1 ml of 50% acetonitrile and 3 μl of CH3I (50 μmol) was added. Then 14 mg of K2CO3 (100 μmol) was added into the reaction mixture and the reaction was performed at 60°C for 15 min. The final product dm-FK228 was purified by preparative HPLC, confirmed by LC-MS ([M + H]+ = 571 m/z) and lyophilized.
Expression and purification of DepH proteins
The full-length DepH (residues 1–319) from C. violaceum No. 968 was expressed in E. coli BL21(DE3) as an N-terminal 6 × His-tagged fusion protein and purified to homogeneity as previously reported13. Decreasing the expression temperature to 16°C and maintaining the purification procedure at 4°C were critical to prevent the recombinant protein from degradation. The DepH protein was further purified by gel filtration chromatography using a Superdex 75 10/30 column (GE Healthcare), which showed that it exists as a homodimer in solution. Peak fractions were dialyzed against buffer containing 50 mM Tris, 50 mM NaCl, 1 mM DTT, pH 8.0 and then concentrated in an Amicon-10 (10 K MWCO) spin concentrator to approximately 9.7 mg/ml.
Mutations were introduced into the pET28a-depH template DNA by site-directed mutagenesis using the QuickChange kit (Strategene) and the primers listed in Supplementary Table 1. The sequence of each clone was verified by DNA sequencing. All DepH mutants were expressed and purified similarly as the wild-type protein.
Crystallization and data collection
Initial crystallization trials were performed using the sitting-drop vapor-diffusion method at 20°C with commercially available kits. Micro-crystals of DepH were produced in 2 μl drops containing an 1:1 mixture of the protein solution and a reservoir solution consisting of 15% PEG4000, 0.2 M (NH4)2SO4, 0.1 M citric acid, pH 5.0. The best diffraction-quality crystals were obtained by the micro-seeding method27 at an optimized reservoir condition with 500 μl of well solution (11.75% PEG4000, 0.2 M (NH4)2SO4, 0.1 M citric acid, pH 5.0) and 100 μl paraffin oil. We did not try the cocrystallization of DepH with the substrate red-FK228 because red-FK228 is sensitive to oxidation and will be catalyzed to form FK228 by DepH during the cocrystallization process. Cocrystallization of DepH with FK228 was performed using the same method in the presence of 5 mM DTT but only yielded the apo-DepH crystals. Fortunately, cocrystallization of DepH with dm-FK228 turned out to be successful. The protein drops in which seeds were transferred contained 1.8 μl of the protein solution and 0.2 μl of 10 mM dm-FK228 dissolved in a mixture of dimethyl sulfoxide and PEG 200 (1:1 ratio). The molar ratio of protein to ligand was close to 1:4. We attempted to cocrystallize DepH with NADP+ and to get the tertiary structure of DepH/dm-FK228/NADP+ by controlling the ratio of protein to NADP+ from 1:4 to 1:10, but failed to find any NADP+ densities in the obtained crystal structures.
All crystals were mounted in nylon loops and flash-frozen in liquid nitrogen. The cryoprotectant for both native and complex crystals consisted of the reservoir solution plus 15% ethylene glycol. Diffraction data were collected on the SSRF BL17U1 beamline (Shanghai, China). Intensity data were integrated and scaled by using the program HKL200028. The statistics for data collection are listed in Table 1.
Structure determination and refinement
The structure of DepH was determined by molecular replacement method using the program PHASER29 in the CCP4 suite30 and the monomer structure of an oxidoreductase from A. tumefaciens (PDB code: 3FBS) as a search model. The initial phase was improved using OASIS31 and PARROT32 and then used to rebuild the main chain of DepH by ARP/wARP33. Refinement was performed with PHENIX34 and a random selection of 5% reflections was set aside for cross-validation. The model was extended and rebuilt manually with Coot35.
The structure of the DepH/dm-FK228 complex was obtained and refined by molecular replacement using the program PHASER29 and the DepH coordinates as the search model. The atomic model was refined using the same protocol aforementioned. The model of dm-FK228 was generated by the PRODRG36 server. MolProbity37 and PROCHECK38 were used to assess the overall quality of the structural models. The dimer interface of DepH was analyzed using the PISA server39. All graphic presentations of structures were generated using PyMOL40 and CCP4MG41. All simulated-annealing omit maps were calculated using CNS42.
Enzymatic activity assay
Dithiobis(5-nitropyridine) (DTNP, Sigma-Aldrich) was dissolved in acetonitrile (4 mM) and used as a color development substrate. The reaction buffer was 20 mM Tris-HCl, pH7.0 and 100 mM NaCl and 96-well cell culture plate (Corning Incorporated) were used for the reactions. The measurements were performed on BioTek HT microplate reader (Winooski, VT05404, USA) and the detecting wavelength was set at 385 nm.
Each reaction included 2.5 μl of 1 mM red-FK228 (final concentration 100 μM), variable volume of 0.1 mg/ml protein sample, with or without NAD+ or NADP+ and a variable volume of the reaction buffer up to total volume of 25 μl. The reaction was carried out at room temperature for 30 min and stopped by incubating in boiling water for 1 min. After cooling, the reaction mixture was spun down and 20 μl of the supernatant was transferred into 96-well plate, mixed with 20 μl 50% acetonitrile and 20 μl 4 mM DTNP. The final 60 μl reaction system was shaken for 5 min at room temperature to develop the color reaction and then read by the plate reader at 385 nm.
To examine the NAD+ or NADP+ effect on enzyme catalysis, 1 μl of 5 mM NAD+ or 1 μl of 5 mM NADP+ (final concentration 200 μM) was mixed with 0.1, 0.2 or 0.4 μM DepH. In the control reactions, no NAD+ or NADP+ was added. For the routine activity assay of DepH mutants, a final concentration of 0.2 μM enzymes without NAD+ or NADP+ was used in the reactions and the wild type DepH was used as the control.
Additional Information
Accession Coordinates. Atomic coordinates and structure factors for the apo- and complexed DepH structures have been deposited in the Protein Data Base with the codes 4JN9 and 4JNA.
References
Furumai, R. et al. FK228 (depsipeptide) as a natural prodrug that inhibits class I histone deacetylases. Cancer Res 62, 4916–4921 (2002).
VanderMolen, K. M., McCulloch, W., Pearce, C. J. & Oberlies, N. H. Romidepsin (Istodax, NSC 630176, FR901228, FK228, depsipeptide): a natural product recently approved for cutaneous T-cell lymphoma. J Antibiot 64, 525–531 (2011).
Shigematsu, N. et al. FR901228, a novel antitumor bicyclic depsipeptide produced by Chromobacterium violaceum No. 968. II. Structure determination. J Antibiot 47, 311–4 (1994).
Cheng, Y. Q., Yang, M. & Matter, A. M. Characterization of a gene cluster responsible for the biosynthesis of anticancer agent FK228 in Chromobacterium violaceum No. 968. Appl Environ Microbiol 73, 3460–3469 (2007).
Li, B. & Walsh, C. T. Streptomyces clavuligerus HlmI is an intramolecular disulfide-forming dithiol oxidase in holomycin biosynthesis. Biochemistry 50, 4615 (2011).
Scharf, D. H. et al. Transannular disulfide formation in gliotoxin biosynthesis and its role in self-resistance of the human pathogen Aspergillus fumigatus. J Am Chem Soc 132, 10136–10141 (2010).
Watanabe, K. et al. Total biosynthesis of antitumor nonribosomal peptides in Escherichia coli. Nat Chem Biol 2, 423–8 (2006).
Bok, J. W. et al. Chromatin-level regulation of biosynthetic gene clusters. Nat Chem Biol 5, 462–4 (2009).
Masuoka, Y. et al. Spiruchostatins A and B, novel gene expression-enhancing substances produced by Pseudomonas sp. Tetrahedron Lett 42, 41–44 (2001).
Wang, C., Flemming, C. J. & Cheng, Y. Q. Discovery and activity profiling of thailandepsins A through F, potent histone deacetylase inhibitors, from Burkholderia thailandensis E264. MedChemCommun 3, 976–981 (2012).
Wang, C. et al. Thailandepsins: bacterial products with potent histone deacetylase inhibitory activities and broad-spectrum antiproliferative activities. J Nat Prod 74, 2031–8 (2011).
Biggins, J. B., Gleber, C. D. & Brady, S. F. Acyldepsipeptide HDAC inhibitor production induced in Burkholderia thailandensis. Org Lett 13, 1536–9 (2011).
Wang, C., Wesener, S. R., Zhang, H. & Cheng, Y. Q. An FAD-dependent pyridine nucleotide-disulfide oxidoreductase is involved in disulfide bond formation in FK228 anticancer depsipeptide. Chem Biol 16, 585–593 (2009).
Akif, M., Suhre, K., Verma, C. & Mande, S. C. Conformational flexibility of Mycobacterium tuberculosis thioredoxin reductase: crystal structure and normal-mode analysis. Acta Crystallogr D Biol Crystallogr 61, 1603–1611 (2005).
Obiero, J., Pittet, V., Bonderoff, S. A. & Sanders, D. A. R. Thioredoxin system from Deinococcus radiodurans. J Bacteriol 192, 494–501 (2010).
Hernandez, H. H., Jaquez, O. A., Hamill, M. J., Elliott, S. J. & Drennan, C. L. Thioredoxin reductase from Thermoplasma acidophilum: a new twist on redox regulation. Biochemistry 47, 9728–37 (2008).
Oliveira, M. A. et al. Insights into the Specificity of Thioredoxin Reductase− Thioredoxin Interactions. A Structural and Functional Investigation of the Yeast Thioredoxin System. Biochemistry 49, 3317–3326 (2010).
D'Arcy, A., Villard, F. & Marsh, M. An automated microseed matrix-screening method for protein crystallization. Acta Crystallogr D Biol Crystallogr 63, 550–554 (2007).
Waksman, G., Krishna, T. S. R., Williams Jr, C. H. & Kuriyan, J. Crystal Structure of Escherichia coli Thioredoxin Reductase Refined at 2 A Resolution: Implication for a Large Conformational Change during Catalysis. J Mol Biol 236, 800–816 (1994).
Kirkensgaard, K. G., Hagglund, P., Finnie, C., Svensson, B. & Henriksen, A. Structure of Hordeum vulgare NADPH-dependent thioredoxin reductase 2. Unwinding the reaction mechanism. Acta Crystallogr D Biol Crystallogr 65, 932–941 (2009).
Gustafsson, T. N., Sandalova, T., Lu, J., Holmgren, A. & Schneider, G. High-resolution structures of oxidized and reduced thioredoxin reductase from Helicobacter pylori. Acta Crystallogr D Biol Crystallogr 63, 833–843 (2007).
Dai, S. et al. Crystal Structure of Arabidopsis thaliana NADPH Dependent Thioredoxin Reductase at 2.5 Å Resolution. J Mol Biol 264, 1044–1057 (1996).
Zhang, Z. et al. Crystal structure of Saccharomyces cerevisiae cytoplasmic thioredoxin reductase Trr1 reveals the structural basis for species-specific recognition of thioredoxin. BBA- Proteins Proteom 1794, 124–128 (2009).
Lennon, B. W., Williams Jr, C. H. & Ludwig, M. L. Crystal structure of reduced thioredoxin reductase from Escherichia coli: structural flexibility in the isoalloxazine ring of the flavin adenine dinucleotide cofactor. Protein Sci 8, 2366–2379 (1999).
Vitu, E., Bentzur, M., Lisowsky, T., Kaiser, C. A. & Fass, D. Gain of function in an ERV/ALR sulfhydryl oxidase by molecular engineering of the shuttle disulfide. J Mol Biol 362, 89–101 (2006).
Mattevi, A. To be or not to be an oxidase: challenging the oxygen reactivity of flavoenzymes. Trends Biochem Sci 31, 276–283 (2006).
Ducruix, A. & Giegé, R. Methods of crystallization. Crystallization of nucleic acids and proteins: A practical approach. Ducruix, A. & Giegé, R. (Eds.) 73–98. (IRL Press, Oxford, 1992).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276, 307–326 (1997).
Mccoy, A. et al. Phaser crystallographic software. J Appl Crystallogr 40, 658–674 (2007).
Collaborative Computational Project, N. The CCP4 Suite: Programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50, 760–763 (1994).
He, Y. et al. OASIS and molecular-replacement model completion. Acta Crystallogr D Biol Crystallogr 63, 793–799 (2007).
Cowtan, K. Recent developments in classical density modification. Acta Crystallogr D Biol Crystallogr 66, 470–478 (2010).
Langer, G., Cohen, S. X., Lamzin, V. S. & Perrakis, A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc 3, 1171–1179 (2008).
Adams, P. D. et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221 (2010).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501 (2010).
Schuttelkopf, A. W. & van Aalten, D. M. F. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr 60, 1355–1363 (2004).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66, 12–21 (2009).
Morris, A. L., MacArthur, M. W., Hutchinson, E. G. & Thornton, J. M. Stereochemical quality of protein structure coordinates. Proteins 12, 345–64 (1992).
Krissinel, E. & Henrick, K. Inference of macromolecular assemblies from crystalline state. J Mol Biol 372, 774–797 (2007).
Schrodinger, L. L. C. The PyMOL Molecular Graphics System, Version 1.3r1. (2010).
McNicholas, S., Potterton, E., Wilson, K. S. & Noble, M. E. M. Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr D Biol Crystallogr 67, 386–394 (2011).
Brunger, A. T. et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54, 905–921 (1998).
Gille, C. & Frömmel, C. STRAP: editor for STRuctural Alignments of Proteins. Bioinformatics 17, 377–378 (2001).
Chenna, R. et al. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31, 3497–3500 (2003).
Gouet, P., Robert, X. & Courcelle, E. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res 31, 3320–3323 (2003).
Acknowledgements
We thank J. He and S. Huang for data collection at SSRF BL17U1 beamline, L. Han and R. Wang for docking and molecular dynamics calculations, Z.-X. Xia for critical reading the manuscript. This work was supported by grants from China MOST and NSFC (2009CB918600, 2011CB710800 and 20111ZX09506-001 to J.Z. and 2011CB966304 and 31000329 to J.L.) and US NIH/NCI (R01CA152212 to Y.-Q.C.).
Author information
Authors and Affiliations
Contributions
C.W. purified the protein samples and performed activity assays, J.L. obtained the crystals and structures, Z.-M.Z. assisted in protein purification and crystallization, Y.-Q.C. and J.Z. designed and supervised the project, J.L., C.W., Y.-Q.C. and J.Z. analyzed the data and wrote the manuscript. All authors provided feedback on the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Info
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Li, J., Wang, C., Zhang, ZM. et al. The structural basis of an NADP+-independent dithiol oxidase in FK228 biosynthesis. Sci Rep 4, 4145 (2014). https://doi.org/10.1038/srep04145
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep04145
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.