Abstract
Methamphetamine (METH) abuse is a worldwide threat, without any FDA approved medications. Anti-METH IgGs and single chain fragments (scFvs) have shown efficacy in preclinical studies. Here we report affinity enhancement of an anti-METH scFv for METH and its active metabolite amphetamine (AMP), through the introduction of point mutations, rationally designed to optimize the shape and hydrophobicity of the antibody binding pocket. The binding affinity was measured using saturation binding technique. The mutant scFv-S93T showed 3.1 fold enhancement in affinity for METH and 26 fold for AMP. The scFv-I37M and scFv-Y34M mutants showed enhancement of 94 and 8 fold for AMP, respectively. Structural analysis of scFv-S93T:METH revealed that the substitution of Ser residue by Thr caused the expulsion of a water molecule from the cavity, creating a more hydrophobic environment for the binding that dramatically increases the affinities for METH and AMP.
Similar content being viewed by others
Introduction
Methamphetamine (METH) (Figure 1 a) and related stimulants are one of the most severe drug threats worldwide. (+)-Methamphetamine alone contributed to an estimated annual economic burden of $23.4 billion in the United States1. However, there are no FDA approved medications to treat METH addiction and thus effective treatments are greatly needed. Current METH addiction treatments mainly comprise behavior modifications or palliative interventions which only relieve some organ-based symptoms. These approaches neither remove nor block METH from its sites of action, rendering them ineffective in reducing METH-related medical complications and formation of its toxic metabolites. Moreover, the effectiveness of stand-alone cognitive-behavioral therapy for METH addiction is challenging, since METH addiction has the highest relapse rates of any illicit drug addiction2,3.
Chemical structures of stimulants and scFv6H4:METH binding.
(a) Chemical structure of (+)-amphetamine and (+)-methamphetamine. (b) A stereo representation of scFv6H4:METH complex. The variable light chain (VL) is shown in cyan, variable heavy chain (VH) in pink and METH in yellow. The CDR loops are labeled as L1, L2, L3 for the light chain (green) and as H1, H2, H3 for the heavy chain (red). The aromatic ring of METH is surrounded with seven aromatic amino acid residues. The cationic nitrogen (blue) of METH forms hydrogen bonds with GluH101(red) and HisL89(green). There are two water molecules Ow5 and Ow6 (brick red) also present at the side wall of binding cavity. They make hydrogen bond (forest green) with each other and with the side chains of SerH35 and SerH93 (red).
In pursuit of effective treatments for METH abuse, that could be used in conjunction with behavioral therapies, anti-METH monoclonal antibodies (mAbs) have been developed4,5. The first generation mAbs have been well characterized in vivo and in vitro6,7,8 and have shown significant preclinical efficacy. In METH abuse rodent models, anti-METH mAbs significantly reduce METH brain concentrations and relapse to METH self-administration and shorten the duration of METH-induced locomotor activity9,10,11. A chimeric anti-METH mAb, Ch-mAb7F9, developed from these endeavors is now in Phase 1 clinical trials.
These anti-METH mAbs act as pharmacokinetic antagonists by retaining METH in the vasculature and thus sequestering it from its sites of action in the brain and heart. The variable domains of the heavy and light chains form METH binding cavities in these IgG molecules. Since only these variable regions are involved in direct interactions with the ligand, we have constructed an anti-METH single-chain variable-fragment (ScFv) from one of our most potent mAb, mAb6H4 (IgG, κ light chain, KD = 4 nM, measured using radioimmunoassay). The constructed scFv6H4 showed no change in affinity for METH with mAb6H412. This scFv format has some clear advantages over the mAb format. In particular the scFv can be produced more economically and since it is only one-sixth of the size of the original antibody, the protein dosage required to deliver the same number of binding fragments is one-third that of the mAb (IgG).
Despite of their promise the current anti-METH scFv, as well as the available mAbs are challenged by the fact that they have very low affinity for amphetamine (AMP) (Figure 1 a). AMP is pharmacologically active metabolite (Figure 1 a), produced by demethylation in the body. Although AMP is less potent stimulant than METH, an immunotherapy capable of removing both of these drugs from their site of action would be highly desirable. Therefore we sought to generate mutants of scFv6H4 with enhanced binding affinity for both METH and AMP improving its therapeutic potential. Unfortunately, hapten design studies suggest that we might have reached the limit of generating very high affinity antibodies by traditional immunization methods12. Indeed, screening over 30,000 cell lines from mice immunized with METH-conjugate vaccines, failed to produce a mAb with high affinity for both METH and AMP13.
Designing antibodies that bind with high affinity and specificity to small molecule drugs of abuse is extremely challenging. METH has a calculated surface area of 206 Å2 with only one charged atom available for hydrogen bonding at physiological pH (~7.4). Thus, in order to engineer scFvs with high METH and AMP binding affinities, we turned to X-ray crystallography data of scFv6H4 in complex with METH.
The crystal structure of the scFv6H4:METH complex shows METH sitting in a deep pocket with the cationic amino tail pointing towards the bottom of the pocket (Figure 1b)14. Aromatic residues from the complementarity determining region (CDR) loops of the light and heavy chains form a hydrophobic barrel around the phenyl ring of METH covering 75% of its total surface area14 (Figure 1 b). The secondary amine of METH forms a hydrogen bond with the imidazole ring of His89 of the light chain and another hydrogen bond with the carboxylate of Glu101 from the heavy chain. In addition to METH, the pocket accommodates two water molecules which form hydrogen bonds with the two Ser residues SerH35 and SerH93.
We hypothesized that the affinity of an anti-METH scFv6H4 to METH and AMP could be enhanced using structure-driven mutations aimed to optimize the shape and the hydrophobicity of the binding pocket. Due to the small size of METH (149.23 Da) and the very limited number of critical binding interactions with the antibody, the precise crystallographic delineation of these interactions were vital in our efforts to enhance binding affinity. From these efforts we developed scFv-S93T, an antibody that shows highly enhanced affinity for both METH and AMP. In this report, we also present the high-resolution crystal structure of scFv-S93T in complex with METH. Clinically, this antibody, could be an ideal candidate for treating METH overdose, or could be serve as the basis of further customization.
In addition to scFv-S93T, we designed and constructed four additional scFv6H4 mutants: scFv-I37M, scFv-Y34W, scFv-Y34M and scFv-H89E. We found that three of the mutants (scFv-S93T, scFv-I37M and scFv-Y34M) exhibited dramatically increased affinity to either METH or AMP, or both. Two of the mutants (scFv-Y34W and scFv-H89E) resulted in substantially lower affinity or lacked binding activity altogether, underscoring the fragile balance of complex interactions between METH and the binding pocket.
Results
We designed the following mutations (shown in Figure 2 a and b) with the corresponding rationale for each mutation. S93T Mutation: As METH and AMP have only one charged atom each (Figure 1 a) and as both have a predominant hydrophobic nature, we sought to increase the hydrophobic character of the binding pocket by forcing a water molecule out, while preserving the side chain charges for the most part. The Ser H93 residue located on the sidewall of the cavity interacts with a water molecule and was replaced with a slightly larger residue threonine. I37M Mutation: We substituted an isoleucine residue situated at the bottom of the pocket with a longer residue methionine (IleH37Met). The binding pocket of I37M has potential for tighter packing environment for METH and AMP. H89E Mutation: The cationic nitrogen of METH forms two hydrogen bonds of different strength with HisL89 and GluH101. The hydrogen bond with GluH101 is stronger compared to HisL89. We substituted histidine with glutamic acid to make both interactions equally strong. Y34M and Y34W Mutations: In order to generate a mutant of scFv that binds to both METH and AMP with high affinity, we substituted the aromatic residue tyrosine with a methionine (Y34M) and tryptophan (Y34W) residue. The bulkier side chain of tryptophan could compliment the loss of the methyl group in AMP (Figure 1 a), provided it adopted a favorable conformation. The methionine side chain is more flexible and therefore, we anticipated that it could adopt a suitable conformation to optimize packing for AMP or METH.
Point mutation sites on scFv6H4 structure and sequence.
(a) The original amino acid residues of the native structure are shown in grey and the mutations are shown in red. Portions of the light chain are shown in light green and parts of the heavy chain in pink. The point mutations SerH93Thr (S93T) and IleH37Met (I37M) are on the VH chain while TyrL34Trp (Y34W), TyrL34Met (Y34M) and HisL89Glu (H89E) are on VL chain. (b) Amino acid sequence of scFv6H4 with Kabat numbering. Framework regions and CDR regions from VH (red and pink) and VL (blue and light blue) are designated above the sequence. The linker region is represented in grey. Amino acids selected for point mutations are highlighted in yellow. Amino acids after point mutations are represented in red beneath the wild type amino acids. Point mutations from VH chain (S93T, I37M) are in the framework regions, the point mutations Y34M, Y34W are in the CDR L1 loop and H89E in the CDR L3 loop of the VL chain.
In vitro affinity for METH
All the newly produced scFv6H4 mutants scFv-S93T, scFv-I37M, scFv-Y34M, scFv-Y34W and svFv-H89E were tested for METH affinity. The dissociation constants (KD) were measured using saturation binding equilibrium dialysis experiments between scFvs and METH. The KD (±SEM) for wild type scFv6H4 is 0.79 (±0.19) nM. The KDs of the scFv-S93T, scFv-I37M and scFv-Y34M were 0.25 (±0.014), 0.61 (±0.005) and 5.0 (±0.87) nM, respectively (Figure 3 a). ScFv-S93T mutant showed statistically significant 3.1 fold improvement in affinity for METH (p < 0.05) with respect to wild type scFv6H4 (Figure 3c).
Affinity determination of scFv mutants with METH and AMP.
(a) In Vitro saturation binding analysis to determine the specific affinity of scFv mutants with METH. Representative specific binding curves are shown with mean and ±SEM of each triplicate data points for each scFv. The KDs of the scFv6H4, scFv-S93T, scFv-I37M and scFv-Y34M were 0.79, 0.25, 0.61 and 5.0 nM, respectively. (b) In vitro competition binding analysis to determine the affinity of scFv mutants for AMP. Representative percent binding curves are shown for each scFv with mean and ±SEM obtained from triplicate data points. The IC50s of the scFv6H4, scFv-S93T, scFv-I37M and scFv-Y34M were 54.6, 2.12, 0.58 and 6.0 μM, respectively. (c) Histogram summarizing fold change in METH affinity KD of scFv mutants with respect to wild type scFv6H4. The wild type scFv6H4 is set at a value of 1. The fold change increase in scFv-S93T, scFv-I37M was 3.13 and 1.3, respectively. The fold change decrease in scFv-Y34M was observed as 0.16. (d) Histogram summarizing fold change in AMP affinity IC50 of scFv mutants with respect to wild type scFv6H4. The wild type scFv6H4 is set at a value of 1. The fold change increase for AMP affinity in scFv-S93T, scFv-I37M and scFv-Y34M was observed as 26, 94 and 8, respectively.
In vitro affinity for AMP
All the mutants were evaluated for AMP affinity by a competition based equilibrium dialysis method, in which unlabeled AMP competes with 3H-AMP to determine binding affinity. This method gives us the half minimal inhibitory concentration (IC50), which in this case is a good approximation to KD12. The IC50 (±SEM) for wild type scFv6H4 was 54.6 (±7.6) μM. The IC50s of the scFv-S93T, scFv-I37M and scFv-Y34M were 2.12 (±0.20), 0.58 (±0.25) and 6.0 (±0.91) μM, respectively (Figure 3 b). The scFv-Y34W and svFv-H89E affinity for AMP were below our threshold of 50 μM affinity. The mutants scFv-S93T, scFv-I37M and scFv-Y34M showed statistically significant improvement in affinity for AMP with respect to wild type scFv6H4 (p < 0.01) by 26, 94 and 8 fold, respectively (Figure 3 d).
Deleterious mutations
Ligand binding is a delicate balance involving a multitude of complex interactions and some changes in the binding pocket could have negative effects. Two of the mutants (H89E and the Y34W) did not show any measurable affinity for METH or AMP (data not shown). HisL89 forms a hydrogen bond with the cationic nitrogen and the replacement of His with a negatively charged residue appeared to have caused an unfavorable electrostatic environment for binding. The replacement of Tyr with a larger residue Trp (Y34W) was intended to create a tighter environment for AMP, but the mutation appears to have caused disruption of the pocket and nearly complete loss of binding. However, replacement of Tyr by a more flexible Met (Y34M) enhanced the AMP binding significantly and reduced binding slightly for METH.
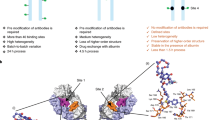
Crystal structure of scFv-S93T:METH
In order to examine the structural basis for the enhancement of affinity, we analyzed the crystal structure of the scFv-S93T mutant in complex with METH at 2.0 Å resolution. The structure was refined to Rfactor and Rfree of 19.4 and 25.4% respectively (Table 1). The overall structure of the S93T is similar to the native structure14; however, the mutation alters the nature of the binding site by expelling the water molecule interacting with Ser93 from the binding pocket (Figure 4 a and b) leaving only one water molecule (Ow5) in the binding pocket. The hydrogen bonding interactions of this water molecule are similar to those in the native structure, except that it interacts with the side chain γ-oxygen of the mutated Thr residue in the place of the second water molecule. The hydrogen bonds with the side chain of SerH35, the main chain carbonyls of TyrH33 and Thr93 are retained. The secondary amine of METH forms the hydrogen bonds with the carboxyl oxygen of GluH101 of the heavy chain (N···O distance 2.6 Å) and HisL89 of the light chain (N···N distance 3.0 Å).
Crystal structure of scFvS93T in complex with METH.
(a) A stereo view of the omit map displaying the electron density of METH (yellow), the water molecule Ow5 (red) and two residues in the cavity (ThrH93 and Ser H35) are depicted. The map is computed at 2 Å resolution and contours are drawn at 1.5 σ level. (b) Binding pocket interactions of scFv-S93T:METH. The VH is represented in pink and VL is represented in cyan. The figure depicts an electrostatic surface of the cross section of the scFv-S93T:METH complex passing through the binding pocket. The electronegative regions are shown in red and positive areas are shown in blue. The opening of the binding pocket is composed of non-polar aromatic amino acid residues; but the lower part of the cavity around the cationic nitrogen is electronegative in nature. There is one water molecule located at the lower right hand side of the pocket. Electrostatic potential was calculated using PyMol vacuum electrostatistics.
Role of water molecule interactions with hydrocarbon atoms in METH binding
As shown in Figure 5, the replacement of SerH93 by a ThrH93 resulted in the expulsion of a water molecule from the binding pocket, improving the affinity for both METH and AMP. In the case of anti-METH scFv6H4, the binding pocket has considerable hydrophobic characteristics, yet has two water molecules buried deep inside. These waters do not form any hydrogen bonds with the amide nitrogen of METH, but participate in hydrogen bonding interactions with backbone carbonyl atoms and side chain hydroxyl atoms of Ser residues (Figure 5 a). The water molecule Ow5 participates altogether in four and Ow6 in three hydrogen bond interactions. Ow6 however has two contacts with hydrophobic hydrocarbon atoms of METH molecule with contact distances of 4.0 and 4.2 Å (Figure 5 a), but the Ser Oγ atom is farther away (6.5 Å) from these hydrocarbon atoms.
Comparisons of the binding pocket molecular interactions of wild-type scFv6H4:METH and the mutant scFv-S93T:METH.
(a) Binding pocket interactions in the scFV6H4:METH structure. Ow6 of scFv6H4 has 3 hydrogen bond interactions and has two contacts with hydrophobic hydrocarbon atoms of METH molecule with contact distances of 4.0 and 4.2 Å. (b) Binding pocket interactions in the scFv-S93T and METH interaction. The side chain γ-oxygen atom of ThrH93 positions itself to form a hydrogen bond with water molecule Ow5, but it is farther away from the METH molecule compared to Ow6 with the contact distances to 6.0 and 6.1 Å from 4.0 and 4.2 Å.
Compared to the native structure, the scFv-S93T mutant is devoid of the water molecule Ow6, but has an extra methyl group by virtue of the mutation. Therefore the mutation increases the hydrophobic character of the pocket and alters some interactions in the pocket. The side chain of Thr residue is oriented to form a hydrogen bond between its γ-oxygen atom and Ow5. At this position, the γ-oxygen atom has moved slightly towards the METH compared to the Ser residue in the native structure, but still keeping distances of 6.1 and 6.0 Å from the hydrocarbon atoms of METH (Figure 5 b). Therefore, the mutation increases the separation between the hydrophilic oxygen and the hydrocarbon atoms.
Discussion
The purpose of the study was to design higher affinity antibodies against METH and AMP. The affinity of an antibody for its ligand (expressed in terms of KD) indicates the degree of binding site occupancy at a given concentration. In other words, a higher affinity antibody will bind and neutralize more METH or AMP at a given drug concentration than a lower affinity antibody. Thus, to ensure the highest efficacy based on a wide range of potential serum concentrations of METH, antibody affinity should be maximized. Indeed it has been shown in animal models that the higher affinity anti-METH mAbs are significantly more effective than low affinity mAbs4,6.
The present study shows that the structure-based engineering has promise as a technique for producing higher affinity antibodies as therapeutic agents for small molecules such as drugs of abuse. In vitro techniques that are commonly employed to improve affinity of antibodies to their antigens include phage display, yeast display and ribosomal display15,16,17,18. These methods introduce mutations randomly throughout the entire sequence and use a variety of selection processes to screen for high affinity antibodies and have been successful for a variety of antibody targets. METH, however, poses a special challenge due to its smaller size and the limited number of residues that directly interact with it. Therefore, we employed a structure-based method to improve the affinity by remodeling the METH binding pocket. Through this approach, we were able to increase the affinity to METH and its active metabolite AMP by designing and testing a small number of mutants based on crystallographic information. The usefulness of structure-based drug design is well documented19,20. For example, using structure-based computational techniques, Clark et al improved the affinity of an antibody fragment against the I-domain of integrin VLA120. In that case however, the interacting surface was more extensive than in the METH antibody reported here. There were a large number of residues involved in the molecular interactions, resulting in many more parameters available for manipulation. The METH binding, on the contrary, presents an extreme situation with a limited number of parameters.
This study also underscores the role of water molecules in ligand binding as demonstrated by the high affinity mutant scFv-S93T. Since hydrophobic interactions were first described by Walter Kauzmann more than five decades ago21, their crucial role in protein folding and ligand binding has become evident. Structured water molecules in the binding pocket play an important role in ligand binding. Ordered water molecules can modify the environment of the binding pocket to optimize binding through favorable interactions. In certain cases, however, the presence of a water molecule may generate less favorable contacts and removal of those water molecules could promote binding. In the present study, the introduction of a methyl group through the substitution of Ser → Thr, a water molecule could be expelled increasing the affinities for both METH and AMP.
There are other examples of water molecules altering the affinity of proteins to ligands and other proteins. An illustration of water molecules amending the specificity and affinity favorably to accommodate variations in the ligands is provided by the crystal structures of L-arabinose-binding protein in complex with L-arabinose, D-fucose and D-galactose22. These structures show that the water molecules can adjust their position or even move out of the cavity to accommodate size, shape or hydrophobicity variations of the ligand. An example illustrating the crucial role of water molecules in protein-protein interactions is the crystal structure of the Ile → Val mutant of von Willebrand factor A1 domain. The mutation led to incorporation of a water molecule into the protein structure increasing its affinity for the receptor glycoprotein23. The scFv-S93T crystal structure presents a different scenario where a water molecule moves out in response to the increased hydrophobicity and the shrinkage of the pocket size favorably for METH and AMP binding.
The overall purpose of this study was to utilize the anti-METH scFv6H4 crystal structure to design rational point mutations to identify high affinity mutants. Three of the five mutants that we generated showed significant improvement in binding to either METH or AMP, or both. We determined the crystal structure of the highest affinity mutant, scFv-S93T, revealing the important loss of a water from the binding pocket. We utilized the scFv platform for affinity improvement due to its ease of production, structural studies and sequence manipulation24. Clinically, with its short half-life, scFv is an ideal candidate for treating METH overdose and could be adapted for longer-term clinical use through molecular customization such as conjugation to polyethylene glycol or potentially even nanoparticles25. These mutants could be further tailored to chimeric or humanized mAbs for preclinical and clinical testing.
Methods
Cloning of anti-METH scFv6H4 mutants
Point mutations were created in scFv6H4 (wild type) sequence to make scFv-S93T, scFv-Y34M scFv-Y34W, scFv-I37M and scFv-H89E following the procedures described in Molecular Cloning: A Laboratory Manual26. Mutants were engineered with EcoRI restriction site on N-terminus and on the carboxy terminus with His-tag and XbaI restriction site. ScFv6H4 mutants were synthesized using Genscript services and were received cloned in pUC57 vector. For maintenance, all the mutants were transformed into E. coli, DH5α (Invitrogen) cells. The transformed colonies were selected through ampicillin resistance. The mutants (scFv-S93T, scFv-Y34M, scFv-Y34W, scFv-I37M and scFv-H89E) cDNA were extracted from vector pUC57 using EcoRI and XbaI (NEB) restriction sites. DNA was then gel purified and ligated to the vector pPiczα-A (Invitrogen) using the same restriction sites. The plasmids were transformed into E. coli, DH5α (Invitrogen) cells for amplification on LB Agar plates and selected through ampicillin resistance. Plasmids were isolated and tested for mutation and in-frame cloning by PCR amplification, restriction digestion and DNA sequencing (University of Arkansas for Medical Sciences DNA sequencing core facility). Plasmids were linearized using SacI (NEB) and transformed into Pichia pastoris, X33 cells using electroporation. Transformed colonies containing cDNA of scFv6H4 mutants were selected using Zeocin (Invitrogen) resistance on YPD (Yeast Extract Peptone Dextrose) agar plates.
Expression of anti-METH scFv6H4 mutants
Selected transformed colonies with strong Zeocin resistance were tested for the expression of scFv-S93T, scFv-Y34M scFv-Y34W, scFv-I37M and scFv-H89E. For expression, selected colonies were inoculated in BMGY (Buffered Glycerol-complex Medium) for amplification. For scFv6H4 mutants expression, cells were harvested and re-suspended into BMMY (Buffered Glycerol-complex Medium). Protein expression was induced by feeding to a final v/v concentration of 0.5% of methanol at 24, 48, 72 and 96 hr. ScFv6H4 mutant proteins containing His-tag in its carboxy terminus were secreted into the media. The protein sample in media was collected after centrifugation at 15000 g for 15 min. The supernatant was tested for expression using SDS-PAGE.
Purification of anti-METH scFv6H4 mutants
The expressed scFv6H4 mutants were purified using nickel sepharose immobilized metal affinity chromatography, HiPrep FF 16/10 column (GE healthcare). ScFv6H4 mutants were purified in one-step elution using elution buffer (20 mM sodium phosphate buffer, 500 mM NaCl and 500 mM imidazole, pH 7.4). Collected fractions were pooled, buffer exchanged and concentrated to 1.4 mg/ml in scFv administration buffer (15 mM sodium phosphate buffer and 150 mM NaCl, pH 7.4) for binding studies.
Saturation equilibrium dialysis to determine KD for METH affinity
Binding of scFv6H4, scFv-S93T, scFv-Y34M and scFv-I34M with METH was tested using Rapid Equilibrium Dialysis devices (RED, Thermo Scientific) with inserts of 6–8 kDa MWCO membranes. Saturation binding assay was performed as described in Nanaware-Kharade et al25. The scFv concentration used in all saturation binding experiments was optimized to maximize the signal to noise ratio as well as minimize errors resulting from possible ligand depletion. The saturation binding experiment involves two parts: determination of total and non-specific binding. Total binding was determined by saturating scFv with the increasing concentration of 3H-METH in range 0.01 to 40 nM. To determine the non-specific binding, we added 10 μM of unlabeled METH as a competitor of 3H-METH. Data points were tested in triplicates and at least three independent experiments were performed for each scFv.
Inhibition equilibrium dialysis to determine IC50 values for AMP affinity
Binding affinity of scFv6H4 and mutants to AMP was determined using the RED devices described above. Titration experiments were used to determine the concentration of scFv6H4, scFv-S93T, scFv-Y34M, scFv-I34M, scFv-H89E and scFv-Y34W that will bind to 20% of a 3H-AMP solution of 50,000 dpm and used for the competition binding experiment. In this experiment a constant concentration of 3H-AMP 50,000 dpm was allowed to compete with various concentrations of AMP (0.01–1000 μM). Protein was placed on the sample side and on the buffer side constant 3H-AMP and increasing concentrations of AMP were placed. Each concentration of AMP was tested in triplicate. Plates were sealed and gently shaken for 18 hr at room temperature to achieve equilibrium between sample side (red) and buffer side (white). To determine binding, 50 μl aliquots were taken from the sample and the buffer sides separately and quantified using liquid scintillation spectrophotometry.
Data analysis for comparison of affinities
For evaluating METH binding we measured the KD, while for AMP we measured the IC50. At least three independent experiments were performed with each scFv to test the METH and AMP affinities. KD for METH saturation binding was determined using nonlinear curve fitting accounting for specific and nonspecific binding. IC50 for AMP inhibition binding was determined using the percent bound vs. AMP concentration curve27. Unpaired, one-tailed Student's t-test was performed to obtain the statistical significance of the differences in binding affinities. KDs of scFv mutants observed from three independent experiments were compared with KDs of scFv6H4 and statistical significance was calculated for the difference in affinities. The same analysis was repeated for the comparison of AMP affinities using IC50 values from three independent experiments. Fold changes were calculated using the mean of the respective affinity values (KD or IC50) observed for scFv mutants and compared with mean of KD or IC50 observed for the wild type scFv6H4 for the respective analysis. Saturation and inhibition binding figures were generated using Graphpad Prism 5.0d software.
Crystallization and data collection of scFv-S93T in complex with METH
For crystallization, the protein sample was concentrated to 12.3 mg/ml in a buffer containing 5 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 150 mM sodium chloride and 5 mM octyl b-D-glucopyranoside detergent at pH 8.3 using the 20 ml Pierce concentrators with 9 kDa molecular weight cut off (MWCO) membrane. METH was added to the protein solution to a final concentration of 5 mM. All the crystals scFV-S93T:METH were grown using the vapor diffusion, hanging drop method at 287 K. Initial screening of crystallization was designed based on the crystal condition of scFv6H414. For scFv-S93Tcrystallization, 500 μl of reservoir solution contained 0.917 M sodium citrate pH 8.2, 0.324 M Imidazole-malate at pH 8.3. Hanging drops were made using 1 μl protein and 1 μl reservoir solution. The crystals grew in 3–4 weeks at 287 K. Using the reservoir solution as a cryo-protectant, the crystals were flash cooled in liquid nitrogen. The diffraction data were collected at Stanford Synchrotron Radiation Laboratory (SSRL), Palo Alto, CA.
Structure determination and refinement of scFv-S93T:METH
Diffraction data were indexed and scaled using the software package HKL200028. The structure solution was carried out using the scFv6H4:METH structure as the starting model14. The model building was carried out using the program COOT29 and the refinement of the structure was carried out using the program Refmac530 in the CCP4 suite of programs31 with 2.0 Å diffraction data. Table 1 provides data collection and refinement statistics. Ray-traced graphics for structure figures were produced using The PyMOL Molecular Graphics System, Version 1.5.0.4 (Schrödinger, LLC).
Additional information
Accession numbers: The atomic coordinates and structure factors for scFv-S93T:METH has been deposited in the Research Collaborator for Structural Bioinformatics Protein Data Bank, Rutgers University, New Brunswick, NJ (http://www.rcsb.org). PDB ID code 4GQP.
References
Nicosia, N., Pacula, R. L., Kilmer, B. & L. R. a. C. J. The Economic Cost of Methamphetamine Use in the United States, 2005. Drug Policy Research Center. RAND Corporation Monograph Series. www.rand.org/pubs/monographs/2009/RAND_MG829.pdf 2009).
Galloway, G. P. et al. An examination of drug craving over time in abstinent methamphetamine users. Am. J. Addict. 19, 510–514 (2010).
Hillhouse, M. P., Marinelli-Casey, P., Gonzales, R., Ang, A. & Rawson, R. A. Predicting in-treatment performance and post-treatment outcomes in methamphetamine users. Addiction 102 Suppl 1, 84–95 (2007).
Owens, S. M., Atchley, W. T., Hambuchen, M. D., Peterson, E. C. & Gentry, W. B. Monoclonal antibodies as pharmacokinetic antagonists for the treatment of (+)-methamphetamine addiction. CNS. Neurol. Disord. Drug Targets. 10, 892–898 (2011).
Peterson, E. C. & Owens, S. M. Designing immunotherapies to thwart drug abuse. Mol. Interv. 9, 119–124 (2009).
Laurenzana, E. M. et al. Use of anti-(+)-methamphetamine monoclonal antibody to significantly alter (+)-methamphetamine and (+)-amphetamine disposition in rats. Drug Metab Dispos. 31, 1320–1326 (2003).
Byrnes-Blake, K. A., Laurenzana, E. M., Landes, R. D., Gentry, W. B. & Owens, S. M. Monoclonal IgG affinity and treatment time alters antagonism of (+)-methamphetamine effects in rats. Eur. J. Pharmacol. 521, 86–94 (2005).
Gentry, W. B. et al. Safety and efficiency of an anti-(+)-methamphetamine monoclonal antibody in the protection against cardiovascular and central nervous system effects of (+)-methamphetamine in rats. Int. Immunopharmacol. 6, 968–977 (2006).
Byrnes-Blake, K. A. et al. Pharmacodynamic mechanisms of monoclonal antibody-based antagonism of (+)-methamphetamine in rats. Eur. J. Pharmacol. 461, 119–128 (2003).
Milesi-Halle, A., Hendrickson, H. P., Laurenzana, E. M., Gentry, W. B. & Owens, S. M. Sex- and dose-dependency in the pharmacokinetics and pharmacodynamics of (+)-methamphetamine and its metabolite (+)-amphetamine in rats. Toxicol. Appl. Pharmacol. 209, 203–213 (2005).
McMillan, D. E. et al. Effects of murine-derived anti-methamphetamine monoclonal antibodies on (+)-methamphetamine self-administration in the rat. J. Pharmacol. Exp. Ther. 309, 1248–1255 (2004).
Peterson, E. C., Laurenzana, E. M., Atchley, W. T., Hendrickson, H. P. & Owens, S. M. Development and preclinical testing of a high-affinity single-chain antibody against (+)-methamphetamine. J. Pharmacol. Exp. Ther. 325, 124–133 (2008).
Peterson, E. C. et al. Using hapten design to discover therapeutic monoclonal antibodies for treating methamphetamine abuse. J. Pharmacol. Exp. Ther. 322, 30–39 (2007).
Celikel, R., Peterson, E. C., Owens, S. M. & Varughese, K. I. Crystal structures of a therapeutic single chain antibody in complex with two drugs of abuse-Methamphetamine and 3,4-methylenedioxymethamphetamine. Protein Sci. 18, 2336–2345 (2009).
Hanes, J., Schaffitzel, C., Knappik, A. & Pluckthun, A. Picomolar affinity antibodies from a fully synthetic naive library selected and evolved by ribosome display. Nat. Biotechnol. 18, 1287–1292 (2000).
Wu, H. et al. Stepwise in vitro affinity maturation of Vitaxin, an alphav beta3-specific humanized mAb. Proc. Natl. Acad. Sci. U. S. A 95, 6037–6042 (1998).
Kurtzman, A. L. et al. Advances in directed protein evolution by recursive genetic recombination: applications to therapeutic proteins. Curr. Opin. Biotechnol. 12, 361–370 (2001).
Boder, E. T., Midelfort, K. S. & Wittrup, K. D. Directed evolution of antibody fragments with monovalent femtomolar antigen-binding affinity. Proc. Natl. Acad. Sci. U. S. A. 97, 10701–10705 (2000).
Jorgensen, W. L. The many roles of computation in drug discovery. Science 303, 1813–1818 (2004).
Clark, L. A. et al. Affinity enhancement of an in vivo matured therapeutic antibody using structure-based computational design. Protein Sci 15, 949–960 (2006).
Kauzmann, W. Some factors in the interpretation of protein denaturation. Adv. Protein Chem. 14, 1–63 (1959).
Quiocho, F. A., Wilson, D. K. & Vyas, N. K. Substrate specificity and affinity of a protein modulated by bound water molecules. Nature 340, 404–407 (1989).
Celikel, R., Ruggeri, Z. M. & Varughese, K. I. von Willebrand factor conformation and adhesive function is modulated by an internalized water molecule. Nat. Struct. Biol. 7, 881 (2000).
Holliger, P. & Hudson, P. J. Engineered antibody fragments and the rise of single domains. Nat. Biotechnol. 23, 1126–1136 (2005).
Nanaware-Kharade, N., Gonzalez, G. A., III, Lay, J. O., Jr, Hendrickson, H. P. & Peterson, E. C. Therapeutic anti-methamphetamine antibody fragment-nanoparticle conjugates: synthesis and in vitro characterization. Bioconjug. Chem. 23, 1864–1872 (2012).
Sambrook, J. & Russel, D. W. Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press; 2001).
Akera, T. & Cheng, V. K. A simple method for the determination of affinity and binding site concentration in receptor binding studies. Biochim. Biophys. Acta 470, 412–423 (1977).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 (2004).
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. Biol. Crystallogr. 53, 240–255 (1997).
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol Crystallogr. 67, 235–242 (2011).
Acknowledgements
This work is supported in part by a grant from the Arkansas Biosciences Institute (K.I.V.) and a grant from National Institutes of Health DA018039 (E.C.P.) and by grants from NCRR (5P20RR016460-11) and NIGMS (8 P20 GM103429-11) at NIH. We would like to thank Guillermo A. Gonzalez III, Brandon Linz and Jaya Nair for their valuable technical contributions to this work. Shraddha Thakkar is a graduate student in the UALR/UAMS joint Bioinformatics program.
Author information
Authors and Affiliations
Contributions
E.C.P. and K.I.V. conceived, designed and supervised the project. K.I.V. designed the mutants. S.T. and N.N.K. expressed the protein, carried out binding affinity studies and analyzed the data. S.T. and R.C. crystallized the protein. S.T. solved the structure. S.T., N.N.K., E.C.P. and K.I.V. wrote the paper. S.T. prepared most of the figures. All authors reviewed the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Thakkar, S., Nanaware-Kharade, N., Celikel, R. et al. Affinity improvement of a therapeutic antibody to methamphetamine and amphetamine through structure-based antibody engineering. Sci Rep 4, 3673 (2014). https://doi.org/10.1038/srep03673
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep03673
This article is cited by
-
A Highly Sensitive Detection System based on Proximity-dependent Hybridization with Computer-aided Affinity Maturation of a scFv Antibody
Scientific Reports (2018)
-
PEGylation of a High-Affinity Anti-(+)Methamphetamine Single Chain Antibody Fragment Extends Functional Half-Life by Reducing Clearance
Pharmaceutical Research (2016)
-
A Nanotechnology-Based Platform for Extending the Pharmacokinetic and Binding Properties of Anti-methamphetamine Antibody Fragments
Scientific Reports (2015)
-
Contrasting effects of ammonium and nitrate inputs on soil CO2 emission in a subtropical coniferous plantation of southern China
Biology and Fertility of Soils (2015)
-
A tool kit for rapid cloning and expression of recombinant antibodies
Scientific Reports (2014)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.