Abstract
eIF4E is frequently over-expressed in different cancers and causes increased translation of oncogenic proteins via deregulated cap-dependent translation. Inhibitors of the eIF4E:eIF4G interactions represent an approach that would normalize cap-dependent translation. Stapled peptides represent an emerging class of therapeutics that can target protein: protein interactions. We present here molecular dynamics simulations for a set of rationally designed stapled peptides in solution and in complex with eIF4E, supported with biophysical and crystallographic data. Clustering of the simulated structures revealed the favoured conformational states of the stapled peptides in their bound or free forms in solution. Identifying these populations has allowed us to design peptides with improved affinities by introducing mutations into the peptide sequence to alter their conformational distributions. These studies emphasise the effects that engineered mutations have on the conformations of free and bound peptides and illustrate that both states must be considered in efforts to attain high affinity binding.
Similar content being viewed by others
Introduction
mRNAs are hypothesized to compete with one another for binding to the eIF4F protein complex and in turn their subsequent translation. The eIF4F complex consists of eIF4E, eIF4A and eIF4G proteins. eIF4E initiates cap-dependent translation by binding to the cap structure (m7GTP) found at the 5′ end of mRNA. eIF4F forms a complex with the 40S ribosomal subunit and eIF3. This complex shuttles along the 5′-untranslated region (5′-UTR) of the mRNA until it arrives at the AUG initiation codon1. The short, unstructured 5′-UTRs of most cellular mRNAs enable the eIF4E containing complex to scan efficiently for the translation initiation codon (AUG). In comparison, the lengthy, G + C-rich, highly structured 5′-UTRs typical of proto-oncogenic mRNAs (e.g. cyclin D1, VEGF) hinder recognition of the AUG start codon by the initiation complex, leading to mRNAs being poorly translated2. eIF4E contributes to malignancy by increasing the translation of mRNAs with structured 5′UTRs, either when over-expressed or when the eIF4F complex is not regulated correctly2,3,4. Failure to regulate the eIF4F complex frequently occurs in cancers when the 4E-binding proteins (4E-BPs), which regulate eIF4F activity by displacing eIF4G from eIF4E, are hyperphosphorylated by mTOR5.
Inhibiting the protein interface between eIF4E and eIF4G is attractive for the design of anti-cancer therapeutics. Peptides derived from eIF4G1 and 4E-BP1 containing the residues responsible for their interactions with eIF4E (YXXXXLΦ motif, where Φ signifies any hydrophobic residue), are structural mimics of each other6. The tyrosine (Y4) (Figure 1) is engaged in multiple van der Waal contacts with eIF4E and an h-bond between its side chain hydroxyl and the carbonyl backbone of P38 in eIF4E. The leucine (L9) exploits a shallow cavity on the surface of eIF4E and interacts with W73 of eIF4E via a hydrogen bond between its backbone and the indole of the tryptophan. The conserved hydrophobic residue (L10) packs against L131 and L135 of eIF4E. Crystal structures of both peptides complexed to eIF4E are approximately 50% α-helical; however they contain negligible helicity in solution6. The binding regions of both eIF4G and 4E-BP1 become structured upon binding eIF4E7,8. The peptide binding interface of eIF4E is very planar and lacks any distinctive cleft to recognise either 4E-BP1 or eIF4G1, in contrast to the deep peptide binding grooves in Mdm2:p53 or Bcl-xl:BH3 complexes9,10. The lack of any major cavity in the eIF4E peptide binding site makes this a challenging target for small molecules (figure 1).
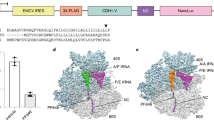
Crystal strucure of the eIF4GD5S peptide in complex with eIF4E.
(PDB ID: 4AZA). eIF4E is shown with surface representation and the peptide with cartoon representation. All residues from the peptide are represented in stick representation and labelled. Hydrogen bonds between Y4:P38 and L9:W73 and those formed by S5, in order to cap the helix, are shown.
We have therefore embarked on a program to inhibit the eIF4E and eIF4G interaction by designing novel stapled peptide inhibitors. Stapling peptides entails the introduction of an all-hydrocarbon linkage between turns of a helix to stabilize their secondary structures11,12,13. This enables: 1) high affinity by reducing the entropic cost of binding, 2) increased in vivo half-life by improving their proteolytic stability, 3) potential enhancement of their cellular uptake and intra-cellular activity14,15. However, stabilization of α-helices in peptides results in the removal of conformational freedom from systems that are usually highly plastic. In addition, the plastic protein-peptide interfaces also frequently modulate each other16. Understanding these effects and any interactions that the staple may engage in with the target surface are crucial in the development of strategies for the incorporation of macrocyclic links into peptides12,17,18,19.
In this work we analyse the effect different hydrocarbon stapling strategies have on the conformational populations of eIF4E interacting peptides in their free and complexed states using molecular dynamics (MD) simulations. We use our understanding to design mutations to overcome any deleterious effects induced by the hydrocarbon staples in the conformations of the peptides, before or after binding, thus increasing their affinities. The findings are supported by biophysical and crystallographic analyses.
Results
Peptide template selection
The template chosen for the design of stapled peptides against eIF4E was 1KKRYSREFLLGF12 (eIF4GD5S, table 1) and was derived from the eIF4G1 epitope that interacts with eIF4E. D5 was mutated to S to optimize the N-capping motif formed when the peptide is bound to eIF4E20. S5 forms a hydrogen bond with the backbone amide of R6, E7 and F8 (figure 1). To initiate the design process, MD simulations of eIF4GD5S bound to eIF4E coupled with replica exchange molecular dynamics (REMD) simulations of the unbound peptide were performed in order to understand the conformational space sampled by the peptide in both states.
MD simulations of the free and bound forms of the eIF4GD5S peptide
Multiple-copy MD simulations, initiated from the crystal structure of eIF4GD5S peptide in complex with eIF4E, were performed to generate an ensemble of structures. These were then clustered to identify favourable conformations of the bound peptide (table 2). The clustering identified one conformation (Cluster 1) as dominant, with a representative structure showing high similarity to the crystal structure (figure 1 and 2a). Comparison of the crystal structure with representative structures from the other clusters (which totalled ~14% of the total population sampled), showed that sites of structural variation were mainly localized at the C-terminus of the bound peptide (supplementary figure S1a).
The representative structure of each cluster is the peptide conformation with the least distance from the centroid of its population.
(a) Superimposition of representative structures from the largest cluster of the eIF4E:eIF4ED5S complex (Cluster 1, Yellow) and the largest cluster of eIF4ED5S in the solution state (Cluster1, Blue). The superimposition was performed by taking the heavy atoms of the peptide without the first three residues “KKR”. The first two residues “KK” from the peptide are not shown for clarity. (b) Scatter plot of the centre-of-mass distance (Comd, Å) measured between the side-chains of residue pairs Y4:F8 and F8:F12 in the eIF4E:eIF4ED5S complex simulations. (c) Root mean square deviation (RMSD) of the heavy atoms of residues in the peptide without the first three residues “KKR”. This analysis was performed for clusters with the largest population in the complex (com) and solution (sol) state simulations by comparing structures within (Inset: Com, Sol) and between (Inset: Com and Sol) these states. (d) Superimposition of representative structures from major clusters 1, 4 and 5 of the eIF4E:sTIP-01 complex simulations. F12 shows a stable orientation, while F8 either points away (cyan) from or towards (magenta) the protein surface. Y4 shows significant differences between the structures. (e) Same plot as in (b) for eIF4E:sTIP-01 complex simulation (f) Superimposition of representative structures from different clusters of the solution state simulations of sTIP-01 depicting the “in” (magenta), “out” (Cyan) and “intermediate” (Blue) pose of F8.
Residues Y4, R6, L9, L10 and F12 all make significant energetic contributions (table 3) to the eIF4E:eIF4D5S interaction. Y4 is involved in hydrogen-bond and van der Waals interactions with eIF4E, R6 forms a salt-bridge with E132, whilst L9, L10 and L12 all form hydrophobic interactions with eIF4E. The conserved hydrogen-bond between Y4 and P38 is maintained in approximately half the population of the largest cluster (table 2, cluster 1). F8 forms few direct contacts with eIF4E and this correlates to its low energetic contribution (table 3). However, Y4, F8 and F12 show minimal structural variations with respect to the representative structures of the other clusters, which suggests that the spatial arrangement of the three aromatic residues is important in anchoring the peptide to the protein surface and in maintaining favourable packing interactions. Pair-wise distance measurements between the centres of mass of Y4, F8 and F12 sidechains, show a clustered population suggesting minimal conformational variation (figure 2b). This indicates that F8 forms π-π interactions with Y4 and F12 to facilitate optimal interactions with eIF4E.
REMD simulations of the unbound eIF4GD5S peptide were initiated from the bound-state experimental conformation. The ensemble of structures was then clustered and yielded a single dominant population (table 2). The structure, associated with this major cluster, orientates R6, L9 and L10 into spatial positions that are similar to their bound orientations (figure 2a). In contrast, the three aromatic residues (Y4, F8 and F12) show a large variation in their orientations between the two states (figure 2c). This suggests that the eIF4GD5S peptide undergoes a significant reorganization of these residues during the process of binding, to attain the most favourable native bound-state. The representative structures from the other clusters show low conformational similarity to the bound peptide (supplementary figure S1b). It therefore seems likely that this major cluster represents a metastable population of peptides that are predisposed to binding to eIF4E. However, to attain the bound conformation of the eIF4ED5S peptide, the residues Y4, F8 and F12 must undergo a conformational transition. Simulations of apo eIF4E show the presence of a small shallow pocket, formed by residues V69, F72, W73, L135 and I138 (supplementary figure S2), which may act as an initial docking point to recognise L9 in populations of peptide that are pre-predisposed to interact with eIF4E.
Initial design and characterization of I, I + 4 hydrocarbon stapled eIF4E binding peptides
Two I, I + 4 staples were inserted at positions 7 and 11, and, 8 and 12, to generate the sTIP-01 and sTIP-02 peptides, respectively. For sTIP-01, the two solvent exposed residues E7 and G11 were replaced to ensure that the peptide maintained its optimal interactions with eIF4E, whilst in sTIP-02, the hydrophobic staple was designed to replace F8 and the interactions made by F12 with eIF4E. The Kd of sTIP-01 shows an ~4.5 fold increase in Kd over eIF4GD5S, whilst sTIP-02 shows no improvement in binding (table 1). CD spectra (figure S3 and table 1) also revealed that the staple induces a significant increase in helicity for both sTIP-01 and sTIP-02 compared to the unstapled eIF4GD5S. To elucidate the reasons for the relative lack of improvement in the Kd of both stapled peptides, further MD simulations were performed to analyse the structural effects of the alternative hydrocarbon staples on the bound and free states of the peptide.
sTIP-01
MD simulations of the sTIP-01:eIF4E complex were subjected to cluster analysis, which isolated three distinct conformational populations (table 2). An alignment of the representative structure from each cluster showed fluctuations in the structure of the N-terminal of the peptide, whilst the section of the peptide (residues 7 to 11) enclosed by the I, I + 4 staple exhibited a stable α-helical fold (figure 2d). The residues Y4 and F8 both exhibited conformational plasticity. In contrast, F12 is restrained with minimal variation across the representative structures. The h-bond between Y4 and P38 is only observed in clusters 4 and 5 when F8 is projecting away from eIF4E. This enables Y4 to orient itself to form the hydrogen bond. In the representative structure for cluster 1, F8 packs against eIF4E, sterically preventing Y4 from interacting with P38 and reducing its energetic contribution to the peptide: protein interaction (table 3). The incorporation of the staple in the eIF4GD5S peptide changes the conformational dynamics of the bound peptide, impeding F8 and F12 from binding to eIF4E in the orientations observed in the linear peptide. This disruption to the side-chain packing causes a scattered distribution of the distances between the three residues (Y4, F8 and F12), when compared to the linear eIF4GD5S peptide (figure 2b and 2e).
REMD simulations of the sTIP-01 peptide in solution revealed no single dominant conformation (table 2). Representative structures for each cluster showed conservation of the α-helix within the peptide, whilst the unrestrained N-terminal region of the peptide, which contains Y4, exhibited structural plasticity (figure 2f). Interestingly, the F8 side chain samples 3 distinct conformations, two of which correspond to the “in” and “out” conformations of the peptide in its bound states (figures 2d and 2f). The third position corresponds to an intermediate state between these orientations, which is similar to the conformation of F8 seen in the linear bound peptide. However, F12 has a strictly conserved orientation in all the clusters. These observations infer several possibilities: 1) sTIP-01 interacts initially with F8 in the “in” orientation and prevents Y4 forming a hydrogen bond with P38, 2) sTIP-01 interacts initially with F8 in the “out” conformation and allows hydrogen bond formation between Y4 and P38 and 3) sTIP-01 binds in the intermediate state and transitions to the “in” or “out” conformations. An additional possibility is that F8 may undergo transitions between each state when bound to eIF4E. The poor binding of sTIP-01 to eIF4E results from the staple creating a geometric restraint on the peptide, which adversely affects its conformation in the complex and also prevents it from attaining a stable solution state.
sTIP-02
Clustering of the sTIP-02 conformations when bound to eIF4E reveals one major population (cluster 2) and three evenly distributed smaller subpopulations (clusters 1, 4 and 5). Analysis revealed that differences in the bound representative structures are mainly located in the extended N-terminal beyond Y4 and that there is little variation in the helix, which is enclosed and stabilized by the I, I + 4 staple (figure 3a). The hydrogen bond between Y4 and P38 is stable in all the derived clusters (table 2), whilst the staple that replaces F8 and F12 interacts, via hydrophobic interactions, with eIF4E (table 3 and figure 3a). In its unbound state, sTIP-02 exists in one major cluster (cluster 1) with a conformation very different from that in the bound state (figure 3a). This is also apparent in the differences observed between the representative structures of the bound and unbound clusters (figure 3b). The clusters with structures most similar to the bound state are clusters 3 and 4; however they constitute only about 13% of the total population of sTIP-02 in solution (supplementary figure S4a). The majority of the solution states sampled by sTIP-02 are therefore significantly different from those sampled by the bound peptide and hence would require a significant conformational reorganization to bind to eIF4E. The incorporation of a hydrocarbon linkage at positions 8 and 12 therefore fails to stabilise the unbound states of the peptide sufficiently enough into conformations that would improve the potency of sTIP-02 in comparison to the linear eIF4GD5S.
The representative structure of each cluster is the peptide conformation with the least distance from the centroid of its population.
(a) Representative structures from major clusters 1, 2, 4 and 5 of the eIF4E:sTIP-02 complex (yellow) simulation were superimposed with the representative structure of the largest cluster isolated in the sTIP-02 solution simulation (blue). (b) The large conformational difference between sTIP-02 in the bound and unbound sates is quantitatively represented by comparing the RMSD between the largest clusters derived from either state. (c) Overlay of the representative structures from the largest cluster of the eIF4E:sTIP-03 MD simulation (yellow) and the largest cluster from the sTIP-03 solution simulation (blue). Both representative structures have very similar conformations except for the orientation of Y4. (d) Quantitative representation of the similarity between the bound and unbound conformation of the peptide for clusters with the largest population in the eIF4E:sTIP-03 complex and the sTIP-03 solution state simulations.
Initial design and characterization of I, I + 3 hydrocarbon stapled eIF4E binding peptides
sTIP-03
The I, I + 3 stapling system21 was also used to stabilize the helix of the eIF4GD5S peptide and was inserted at positions 8 and 11 (table 1), leading to the loss of the F8 sidechain. However F8 only makes a small contribution to the peptide:protein interaction and the staple should compensate for the role of F8 in stabilising the interactions of Y4 and F12 with eIF4E (table 3). The I, I + 3 staple resulted in a peptide with greater helicity than to sTIP-01/02 (table 1) and showed a 17-fold improvement in Kd compared to eIF4GD5S. Clustering of the simulated structures showed that the bound-state of the peptide primarily adopts one favourable conformation (table 2 and figure 3c). However, only about 33% of the population in this cluster shows the presence of the hydrogen-bond between Y4 and P38, which is primarily due to a subtle change in the orientation of Y4 (figure 3c). Both the staple and F12 maintain a stable orientation and interact with the protein (table 3). REMD simulations of unbound sTIP-03 reveal one major subpopulation and 3 other significant clusters (table 2). In the major cluster, the peptide samples conformations close to the complexed state (figure 3d), except for the orientation of Y4 (figure 3c). The populations in clusters 2 and 4 are also similar to the major cluster, but the orientation of the hydrocarbon linker in cluster 2 is different (supplementary figure S4b). Cluster 3 has a significantly different solution state conformation. Thus bound sTIP-03 has Y4 and the staple optimally arranged to interact with eIF4e, whilst F12 shows an increase in its free energy of binding to eIF4E. This improved packing of F12 is caused by H37 of eIF4E interacting with the I, I + 3 staple. This, together with the observation that its solution state conformation is close to its bound conformation, accounts for the high binding affinity observed.
Optimization of I, I + 4 hydrocarbon stapled eIF4E binding peptides
The I, I + 4 hydrocarbon linkage between positions 7 and 11 (sTIP-01) creates a geometric restraint on the peptide, resulting in inefficient packing and attenuation of peptide binding to eIF4E. However, it still remains a highly attractive stapling position, as it minimizes perturbation of the key interactions with eIF4E. Comparison of the sTIP-01 and sTIP-03 systems revealed that in addition to the difference in the stapling systems, factors that likely contribute to the improved binding observed for sTIP-03 are the more efficient packing of F12 and the absence of a phenyl sidechain at position 8. Therefore substitution of F8 or F12 could lead to more optimal packing and elimination of the deleterious effects caused by the staple in sTIP-01. The following point mutations (F8A and F12&, & = 2-aminobutyric acid = 2AB) were introduced into sTIP-01. Both these derivative peptides (sTIP-01F8A and sTIP-01F12&) showed improved Kd values and helicity when compared to sTIP-01 (table 1).
sTIP-01F8A
The simulations of sTIP-01F8A in complex with eIF4E yield four clusters of conformations (table 2). The structural variation between the representative structures for each cluster localizes mostly to the N-terminal residues, including Y4 and to subtle changes in the orientation of the side-chain of F12 (figure 4a). The change in the orientation of Y4 in clusters 1, 2 and 3, causes a loss in the Y4:P38 hydrogen-bond. The increased conformational freedom of the F12 side chain here contrasts with that in sTIP-01, where it is highly inflexible and this is directly related to the presence of F8. Comparison of the energetic contributions towards the sTIP-01/01F8A interactions reveals that the F12 residue makes no improved contribution (table 3). This shows that despite the increased flexibility of the residue, there is no enhanced packing of the peptide with eIF4E. Examination of the REMD simulations of sTIP-01F8A in solution reveals one major cluster and two other distinct populations (table 2). As expected the representative structures of these peptide groupings reveal a high degree of structural variation in the N-terminus (figure 4b). However the orientation of F12 is remarkably similar to its packing arrangements observed in the bound conformations of the peptide, as are other key features in the helical portion of the peptide. Therefore when compared to sTIP-01, where there is a much greater requirement of conformational rearrangements of the peptide in its bound and free states, the degrees of freedom open to sTIP-01-F8A are reduced, thus making it a more potent binder of eIF4E. However, although sTIP-01F8A was able to rescue the peptide from the detrimental effects observed for sTIP-01, the binding affinity shows no increase over the linear peptide eIF4GD5S.
The representative structure of each cluster is the peptide conformation with the least distance from the centroid of its population.
(a) Superimposition of representative structures from major clusters 1, 2, 4 and 5 of the eIF4E:sTIP-01F8A complex simulation, comparing the structural variations of the peptide in different sub-populations. (b) Superimposition of the major clusters 1, 2 and 3 from the sTIP-01F8A solution state simulations showing the structural variations of the peptide in different sub-populations. (c) The representative structures from the largest cluster of the eIF4E:sTIP-01F12&complex simulation (yellow) and the largest cluster of the sTIP-01F12&solution simulation (blue) overlaid with each other, showing that both structures are similar, except for subtle variations observed in the orientation of the side-chains. & = 2-aminobutyric acid = 2AB. (d) The low conformational difference between sTIP-01F12& in the bound and unbound sates is quantitatively represented by comparing the RMSD between the largest clusters derived from either state.
sTIP-01F12&
Clustering of the bound sTIP-01F12& conformations reveals one significant population as opposed to the dispersed populations observed in sTIP-01 and sTIP-01F8A (table 2). Unlike in sTIP-01, the F8 sidechain does not adopt multiple conformational states (figure 4c) and the hydrogen bond between Y4 and P38 is present in 60% of the structures in this cluster. More importantly, the side-chains of Y4, F8 and the 2AB residue (used to replace F12) pack efficiently against the protein (figure 4c). REMD simulations of sTIP-01F12& in solution and subsequent clustering shows 4 distinct groups with the largest constituting 54% of the sampled frames (table 2). Comparison of these different clusters shows that the peptide conformations are largely similar with major differences observed for Y4 and F8 (Supplementary figure S5a). In addition, the unbound peptides have conformations which are largely similar to their bound-state conformations when in complex, except for subtle changes in the side-chain orientations (figures 4c and 4d). The alteration observed in the dynamics of the residue side chains induced by the hydrocarbon staple in sTIP-01 is overcome with the substitution of F12 by 2AB, which stabilizes the conformations of the peptide.
sTIP-04
Point mutations at positions F8 and F12 of sTIP-01 generated analogues with improved helicity and potency, but with no improvement in Kd over the linear eIF4GD5S peptide (table 1). In prior work we identified the mutations F8Q and F12L, which in conjunction, improved the binding of the eIF4G1 peptide to eIF4E20. These mutations were incorporated into a I, I + 4 stapled peptide termed sTIP-04. The resulting peptide had a significant increase in helicity and ~25-fold improvement in Kd compared to eIF4GD5S (table 1). Cluster analysis of the simulations of sTIP-04 in complex with eIF4E revealed a single cluster that is significantly more favoured (~80% of sampled frames) than the other clusters (table 2). The representative structure of this major cluster showed that the F12L mutation facilitated packing of sTIP-04 peptide against eIF4E (figure 5a).
The representative structure of each cluster is the peptide conformation with the least distance from the centroid of its population.
(a) Representative structures from the largest clusters of the eIF4E:sTIP-04 complex MD simulations and the sTIP-04 solution simulation were overlaid. Both the bound and unbound structures of sTIP-04 show high structural similarity. (b) The similarity in the conformations is shown by a large population showing small a small deviation of the RMSD values between the two clusters. (c) Crystal structure of sTIP-04 peptide in complex with eIF4E (4BEA). The 2Fo-Fc electron density map (blue) for sTIP-04 is shown at 1.5 sigma. (d) The N-terminal of the eIF4GD5S peptide (yellow) has a very high structural similarity to sTIP-04 (blue), but its conformation begins to diverge from that of the stapled peptide at position L10. This results in displacement of residues G11 and F12 with respect to their sTIP-04 counterparts. This displacement occurs due to unwinding of the C-terminal helical turn, which occurs in order to accommodate the interactions of F8 and F12 with each other and with the surface of eIF4E. sTIP-04 residues are labelled in italics whilst eIF4GD5S are not.
However energy decomposition analysis revealed that despite the increased contribution of L12 to the free energy of binding compared to the sTIP-01 analogues, it is not as energetically significant as the contribution made by F12 in sTIP-03 (table 3). The F8Q substitution makes a negligible contribution to the binding energy of the interaction and in this respect is similar to the F8A substitution in sTIP-01F8A. However, the side-chain and backbone amide of Q8 is involved in forming an interaction-network with the side-chain of S5, which also involves the backbone amides of R6 and X7 (linker) (figures 5a and S6). This network makes a significant contribution towards stabilizing the bound form of sTIP-4. Analysis of the REMD simulations of sTIP-04 reveals two significant clusters in solution (table 2), whose representative structures are very similar except for structural variations in the N-terminus (supplementary figure S5b). The simulated conformations of the peptide, when in solution, are also very similar to the bound-state structure, as shown by the low deviation between the two states (figure 5b) and the similarity between the representative structures from the largest clusters of the two simulations (figure 5a). Residues S5 and Q8 of unbound sTIP-04 also form an interaction network with each other and the backbone amides of R6 and the linker at position 7 (figures 5a and supplementary S6). This ensures that the structure of the unbound peptide, in addition to the helical stabilization induced by the staple, is pre-structured further for favourable binding to eIF4E. The combination of improved packing of sTIP-04 with eIF4E, improved stabilization of the bound form, a single distinct structural cluster when bound and partial restraint on the N-terminal structural fluctuations in the bound and unbound forms, results in an improved interaction when compared to the other stapled peptides (except sTIP-03).
Crystallographic analysis of the eIF4E:sTIP-04 complex and validation of MD simulations
Crystallization of the eIF4E:sTIP04 complex showed that the conserved residues required for the peptide to interact with the protein (Y4, L9 and L10) are all orientated in the “correct” position, confirming the findings of the MD simulations (figure 5a and 5c). When compared to the eIF4GD5S peptide in the 4AZA crystal structure, the C-terminal turn of the helix stabilised by the hydrocarbon staple in sTIP-04 possesses greater helicity. The N-terminus of the eIF4GD5S peptide has a very high structural similarity to sTIP-04 but its conformation begins to diverge from that of the stapled peptide at position L10. This results in residues G11 and F12 being significantly displaced with respect to their sTIP-04 counterparts (figure 5d). The displacement of G11 and F12 in eIF4GD5S is caused by the unwinding of the C-terminal helical turn, which occurs in order to accommodate the interactions of F8 and F12 with each other and with eIF4E. As shown by the MD simulations of sTIP-01 bound to eIF4E, the presence of the hydrocarbon staple prevents the F8 and F12 residues from interacting with eIF4E in the orientations observed in the crystal structure, due to stabilisation of the C-terminal end of the helix. The presence of F8Q and F12L in sTIP-04 promotes stabilization of the peptide interactions, with no detrimental effect on any of the other interactions made with eIF4E. However Q8, as also shown and supported by MD simulations, forms no direct interactions with eIF4E (figure 5d). This enables optimal packing of L12 and the formation of 2 hydrogen bonds to the side chain and backbone amide of S5, respectively. These interactions further stabilise the bound form of sTIP-04 and add to the stabilization brought about by the hydrogen bond interaction network between the hydroxyl side chain of S5 and the backbone amides of R6, Q8 and X7 (initiating residue of the hydrocarbon staple), which caps the exposed free amides of the first turn of the peptide helix. The crystallisation of eIF4E:sTIP-04 highlights the versatility of MD simulations in quickly assessing the effects of stapling upon linear peptides.
Discussion
Restraining the conformational freedom of peptides via hydrocarbon linkages can have very dramatic effects upon the structural dynamics of the peptide, when either bound or free in solution and as a result have severe consequences on the avidity of the peptide:protein interface. An example of this is sTIP-01, where the incorporation of an I, I + 4 staple has the potential not to perturb any of the key interactions with eIF4E, whilst stabilising the helix. However, the outcome of this modification was to impose several barriers to engaging high affinity interactions. These barriers include the stabilisation of the unbound peptide into several metastable conformations that require a reorganizational penalty to bind to eIF4E, as well as effecting the stacking interactions between Y4, F8 and F12, which prevents them from attaining the orientation required for efficient packing as observed in the linear peptide. However, to attain the optimum mode of interaction, it is clear the peptides need to undergo subtle rearrangements, with the amount of reorganization required varying with sequence.
sTIP-02 and sTIP-03 also provide insights on manipulating the interactions between the staple and the surface of eIF4E. In the case of sTIP-02, the staple replaces the interactions of F12 with eIF4E. In sTIP-03, an additional interaction is formed between the staple and eIF4E, which cooperatively leads to improved packing for the terminal F12 side chain. However, the I, I + 4 stapling of sTIP-02, at positions 8 and 12 of the peptide leads to the detrimental stabilisation of the free peptide in conformations that are non-conducive for a high affinity interaction. Interestingly, sTIP-02 is the only stapled peptide where we isolated a metastable conformation in solution where the helix is not stabilised by the staple. This observation supports the findings of Kutchukian et al.22, who also observed metastable non-helical conformations of stapled peptides. In contrast, in sTIP-03, the I, I + 3 staple induces populations of peptides with conformations similar to those that are adopted in its bound state. In the case of the point mutant analogues of sTIP-01 (sTIP-01F8A, sTIP-01F12&), both variants bound with improved affinities by preventing the propagation of conformations in solution that require significant conformational changes to bind to eIF4E. However neither analogue showed improved potency compared to the linear peptide, mainly due to the lack of features which improved the stability of the bound complex.
Our current studies demonstrate that alternative helical stabilization strategies give rise to diverse molecular mechanisms for binding and that those improvements in affinity result from compensatory interactions. Staples predominately increase the helicity of the peptide in solution, but this can result in populations of peptides with conformations that are not ideal for binding, require significant reorganizational energies to bind and may be compromised by non optimal interactions at the peptide-protein interface. In the 2 high affinity peptides designed here (sTIP-04 and sTIP-03), such limitations have been overcome, by optimising packing effects at the interface, stabilising the bound complex and inducing greater helical stabilization in solution of conformations that are predisposed to interact with eIF4E. Both sTIP-03 and 04 possess a large degree of helicity in solution (45% and 63% respectively) and, interestingly, neither readily stabilises the Y4:P38 hydrogen bond when bound to eIF4E. However, they stabilise their bound conformations via two very different mechanisms: sTIP-03 optimises the intermolecular interactions with eIF4E, whilst sTIP-04 forms an intricate hydrogen bond network to stabilise the bound form of the peptide. This hydrogen bond network also stabilises the conformations of the free peptide in states that are conducive to binding eIF4E.
In this work we utilised two paradigms to improve the potency of stapled peptides, both of which involved optimising the unbound forms of the peptide. In addition, the affinity for eIF4E is also increased either by stabilising the bound conformation of the peptide further or by utilizing extra interactions to improve binding with eIF4E. However, there remains one avenue that can still potentially be manipulated, wherein an unfavourable conformation, identified by the MD simulations, is taken and optimised further in search for a new and more potent interaction. The observations made in this work will further the design of peptidomimetics for potential lead compounds against eIF4E and lead to greater understanding of the effects of incorporating staples into helices targeting binding interfaces.
Methods
Peptide reagents
eIF4GD5S as well as the FAM labeled tracer peptide (KKRYSRDFLLALQK-(FAM)) were ordered from and synthesized by Mimotopes, Clayton, Australia. Stapled peptides (sTIP) were synthesized by Anaspec (San Diego, Ca). Stapled peptides (sTIP) with an (i, i + 4) hydrocarbon linkage were generated by replacing the respective amino acids with the olefin-bearing unnatural amino acids (S)-2-(4′pentenyl) alanine and (S)-2-(4′-pentenyl) alanine and stapled, via olefin metathesis, using the Grubbs catalyst. Stapled peptides (sTIP) with an (i, i + 3) hydrocarbon linkage were generated by replacing the respective amino acids with the olefin-bearing unnatural amino acids (R)-2-(4′pentenyl) alanine and (S)-2-(4′-pentenyl) alanine and stapled, via olefin metathesis, using the Grubbs catalyst. The stapled peptides were purified using HPLC to >90% purity. All peptides were amidated at their C-terminus and acetylated at their N-terminus. Peptides were purified using HPLC to >90% purity.
Protein expression and purification
Full-length human eIF4E was expressed and purified as described previously23,24,25.
Surface plasmon resonance
SPR experiments were carried out as described previously20.
Competitive fluorescence anisotropy assay
See Supplementary Information.
Circular dichroism measurements
See Supplementary Information.
Crystallization and data collection
See Supplementary Information.
Computer simulations
Starting structures and system setup
Chains A and B from the crystal structure of eIF4E in complex with the eIF4GD5S peptide (PDB ID: 4AZA) were used to construct the different complexes used in the MD studies. Unresolved loops in chains A and B were built using the eIF4E:eIF4G1 peptide structure (PDB ID: 2W97) as a template. The N-terminus of eIF4E (1–30) was not modelled or used in the MD simulations. The sequence of the peptide eIF4GD5S is “KKRYSERFLLGF” and is sequentially numbered from 1–12 in the main text. The hydrocarbon linker (staple) in the various designed stapled peptides and other non-natural amino acids (e.g. 2 Amino Butyric acid) were modelled using the XLEAP module of AMBER. The RESP (Restrained Electrostatic Potential) based atomic charges for the hydrocarbon linker were derived through the R.E.D. server using the RESP-A1A (HF/6-31G*) charge model and Guassian_2009_C.01 quantum mechanics program26. Other force field parameters were derived from all-atom ff99SB27 force field in AMBER1128,29. In-silico mutations on the peptide were performed using PyMOL molecular visualization software30. The N-terminal of the protein and peptide was acetylated (ACE), whilst the C-terminal was methylated (NME) for the protein and amidated (NHE) for the peptide, respectively. The starting structures were placed in a cuboid water box such that the minimum distance from the edge of the box was at-least 12Å. TIP3P water model was used for solvation31. The solvated systems were neutralized by adding appropriate numbers of chloride ions using the TLEAP module of AMBER. A total of seven systems were prepared (table 1) and the total number of atoms ranged from 37,401 to 38,199.
General molecular dynamics simulations
Multi-copy Molecular Dynamics Simulations were performed using the PMEMD module of AMBER1128,29, employing the all-atom ff99SB force field parameters27. The solvated systems were initially relieved of any unfavourable interactions by subjecting them to 1000 steps of energy minimization, which involved using 500 steps each of steepest descent and then followed by conjugate gradient algorithms. The conjugate gradient algorithms were used as follows: 1) Cartesian restraints were imposed on the solute while the solvent molecules were then allowed to relax around it, 2) The solvent was then restrained and the solute was energy minimized, 3) The final stage involved no restraints and the whole system was minimized. Force constant of 500 kcal mol−1 Å2 was used during the restraining steps. The systems were then gradually heated to 300 K over a period of 30 ps using the NVT ensemble. Following this, each system was equilibrated under NPT conditions for 500 ps. They were then subjected to the production phase of molecular dynamics simulation using the NPT ensemble for a period of 50 ns each. Three replicates of each of the seven systems were performed in order to improve the sampling and remove the biasness in our analysis data set. This amounts to a total of 21 simulations with a combined simulation period of 1.05 microseconds. The simulation temperature of 300 K was set using langevin dynamics32,33, with a collision frequency of 1.0 ps−1. The pressure was maintained at 1 atm using weak-coupling34 with a pressure relaxation time of 1 ps. Periodic boundary conditions in x, y and z directions were appropriately applied. Particle Mesh Ewald method (PME) was used for treating the long range electrostatic interaction35. All bonds involving hydrogen atoms were constrained using the SHAKE algorithm36. A time step of 2 fs was used and the coordinates were saved every 1 ps. The first 20 ns of the production run were discarded in the analysis of the trajectories to reduce the biasness caused by the similarity in the starting structures of the different systems.
Explicit solvent replica exchange molecular dynamics simulations (REMD)
REMD is an advanced molecular dynamics method37,38. In this method, different replicas of the system are coupled to different temperatures, over a temperature range and an exchange is made between the replicas at fixed intervals, using a Metropolis-type criterion. This method has been widely used to study the conformational properties of biomolecules and is known to improve the sampling efficiency39,40. REMD was apllied, as implemented in AMBER11, to sample the conformational space of the free peptides in solution using explicit representation of the solvent molecules. The starting structures of the different peptides were derived from their corresponding states in complex with eIF4E. The peptides were solvated with TIP3P water in a truncated octahedron box with at-least 10 Å buffer. The total number of atoms for the different systems ranged from 8475 to 9362. A total of 24 replicas were selected over a temperature range of 300–400 K and the temperatures selected in this range were 300.0, 302.1, 304.9, 307.7, 310.6, 313.6, 316.8, 320.2, 323.7, 327.4, 331.2, 335.3, 339.4, 343.7, 348.2, 352.9, 357.7, 362.8, 368.1, 373.7, 379.7, 386.0, 392.8 and 400.0 K. The solvated peptides were energy minimized, following the same protocol as mentioned for the complex system. The non-coupled 24 replicas were then heated to their respective temperatures and equilibrated using standard molecular dynamics simulations individually, under NPT condition for a period of 200 ps, before subjecting them to production dynamics for a period of 20 ns, using REMD under the NVT ensemble. Exchange between replicas were attempted every 1 ps and the minimum acceptable exchange ratio was optimized to be around 20%. A total of seven different simulations for each of the seven peptides were performed with a combined simulation period of 140 ns. The first 10 ns of the production run from each system was discarded during the analysis to account for the similarity in the initial starting structure.
Clustering analysis
The ensemble of structures, generated from traditional MD simulation of the protein-peptide complex systems and the REMD for the free peptides, were clustered into distinct sets based on the conformation of the peptide. Clustering was performed using the average-linkage algorithm41 with the desired cluster number set to five. Pairwise RMSD between the structures was used as a distance matrix. The clustering method was applied on the heavy atoms of the peptide excluding the first three residues “KKR”. A total of 3000 structures for the complex and 1000 structures for the free peptides were used for clustering. The structures were extracted at equal intervals over the entire range of the simulation trajectories considered for analysis.
Energy decomposition and hydrogen bond analysis
Molecular free energy decomposition based on the MM/GBSA (Molecular Mechanics / Generalized Born Surface Area)42 analysis was performed on the simulated trajectories in order to obtain a quantitative description of the energetic contribution for the peptide: protein interaction. This was computed using the MMPBSA.py script in AMBER43 using over 1000 structures as input. Water molecules and Cl− ions were stripped from the extracted structures and the solvent effect was represented using a Generalized Born Solvation Model44. A salt concentration of 0.15 mM was used. The surface area was calculated by employing a recursive method, which approximates a sphere around an atom beginning from an icosahedra shape. Hydrogen bond analysis was performed using the PTRAJ module in AMBER. A total of 10000 structures were analysed, using a distance and angle cut-off of 3.5Å and 120°, respectively.
References
Topisirovic, I., Svitkin, Y. V., Sonenberg, N. & Shatkin, A. J. Cap and cap-binding proteins in the control of gene expression. Wiley Interdiscip Rev RNA 2, 277–298 (2011).
Montanaro, L. & Pandolfi, P. P. Initiation of mRNA translation in oncogenesis: the role of eIF4E. Cell Cycle 3, 1387–1389 (2004).
Culjkovic, B., Topisirovic, I. & Borden, K. L. Controlling gene expression through RNA regulons: the role of the eukaryotic translation initiation factor eIF4E. Cell Cycle 6, 65–69 (2007).
De Benedetti, A. & Graff, J. R. eIF-4E expression and its role in malignancies and metastases. Oncogene 23, 3189–3199 (2004).
Gingras, A. C. et al. Regulation of 4E-BP1 phosphorylation: a novel two-step mechanism. Genes Dev 13, 1422–1437 (1999).
Marcotrigiano, J., Gingras, A. C., Sonenberg, N. & Burley, S. K. Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G. Mol Cell 3, 707–716 (1999).
Fletcher, C. M. et al. 4E binding proteins inhibit the translation factor eIF4E without folded structure. Biochemistry 37, 9–15 (1998).
Gross, J. D. et al. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell 115, 739–750 (2003).
Vassilev, L. T. et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303, 844–848 (2004).
Sattler, M. et al. Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science 275, 983–986 (1997).
Kim, Y. W., Grossmann, T. N. & Verdine, G. L. Synthesis of all-hydrocarbon stapled alpha-helical peptides by ring-closing olefin metathesis. Nat Protoc 6, 761–771 (2011).
Baek, S. et al. Structure of the stapled p53 peptide bound to Mdm2. J Am Chem Soc 134, 103–106 (2012).
Walensky, L. D. et al. Activation of apoptosis in vivo by a hydrocarbon-stapled BH3 helix. Science 305, 1466–1470 (2004).
Brown, C. J. et al. Stapled Peptides with Improved Potency and Specificity That Activate p53. ACS Chem Biol, 8, 506–12 (2012).
Verdine, G. L. & Hilinski, G. J. Stapled peptides for intracellular drug targets. Methods Enzymol 503, 3–33 (2012).
Dastidar, S. G., Lane, D. P. & Verma, C. S. Multiple peptide conformations give rise to similar binding affinities: molecular simulations of p53-MDM2. J Am Chem Soc 130, 13514–13515 (2008).
Joseph, T. L., Lane, D. & Verma, C. S. Stapled peptides in the p53 pathway: computer simulations reveal novel interactions of the staples with the target protein. Cell Cycle 9, 4560–4568 (2010).
Phillips, C. et al. Design and structure of stapled peptides binding to estrogen receptors. J Am Chem Soc 133, 9696–9699 (2011).
Stewart, M. L., Fire, E., Keating, A. E. & Walensky, L. D. The MCL-1 BH3 helix is an exclusive MCL-1 inhibitor and apoptosis sensitizer. Nat Chem Biol 6, 595–601 (2010).
Zhou, W. et al. Improved eIF4E binding peptides by phage display guided design: plasticity of interacting surfaces yield collective effects. PLoS One 7, e47235 (2012).
Kim, Y. W., Kutchukian, P. S. & Verdine, G. L. Introduction of all-hydrocarbon i, i + 3 staples into alpha-helices via ring-closing olefin metathesis. Org Lett 12, 3046–3049 (2010).
Kutchukian, P. S., Yang, J. S., Verdine, G. L. & Shakhnovich, E. I. All-atom model for stabilization of alpha-helical structure in peptides by hydrocarbon staples. J Am Chem Soc 131, 4622–4627 (2009).
Brown, C. J. et al. Stabilizing the eIF4G1 alpha-helix increases its binding affinity with eIF4E: implications for peptidomimetic design strategies. J Mol Biol 405, 736–753 (2011).
Brown, C. J., Verma, C. S., Walkinshaw, M. D. & Lane, D. P. Crystallization of eIF4E complexed with eIF4GI peptide and glycerol reveals distinct structural differences around the cap-binding site. Cell Cycle 8, 1905–1911 (2009).
Brown, C. J., McNae, I., Fischer, P. M. & Walkinshaw, M. D. Crystallographic and mass spectrometric characterisation of eIF4E with N7-alkylated cap derivatives. J Mol Biol 372, 7–15 (2007).
Vanquelef, E. et al. R. E. D. Server: a web service for deriving RESP and ESP charges and building force field libraries for new molecules and molecular fragments. Nucleic Acids Res 39, W511–517 (2011).
Hornak, V. et al. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins 65, 712–725 (2006).
Case, D. A. et al. The Amber biomolecular simulation programs. J Comput Chem 26, 1668–1688 (2005).
Ponder, J. W. & Case, D. A. Force fields for protein simulations. Adv Protein Chem 66, 27–85 (2003).
Schrodinger, L. L. C. The PyMOL Molecular Graphics System, Version 1.3r1 (2010).
Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys 79, 926–935 (1983).
Pastor, R. W., Brooks, B. R. & Szabo, A. An analysis of the accuracy of Langevin and molecular dynamics algorithms. Mol. Phys. 65, 1409–1419 (1988).
Loncharich, R. J., Brooks, B. R. & Pastor, R. W. Langevin dynamics of peptides: the frictional dependence of isomerization rates of N-acetylalanyl-N′-methylamide. Biopolymers 32, 523–535 (1992).
Berendsen, H. J. C., Postma, J. P. M., van Gunsteren, W. F., DiNola, A. & Haak, J. R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690 (1984).
Darden, T., York, D. & Pedersen, L. Particle mesh Ewald: An N [center-dot] log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993).
Ryckaert, J.-P., Ciccotti, G. & Berendsen, H. J. C. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 23, 327–341 (1977).
Nymeyer, H., Gnanakaran, S. & Garcia, A. E. Atomic simulations of protein folding, using the replica exchange algorithm. Methods Enzymol 383, 119–149 (2004).
Sugita, Y. & Okamoto, Y. Molecular mechanism for stabilizing a short helical peptide studied by generalized-ensemble simulations with explicit solvent. Biophys J 88, 3180–3190 (2005).
Gnanakaran, S., Nymeyer, H., Portman, J., Sanbonmatsu, K. Y. & Garcia, A. E. Peptide folding simulations. Curr Opin Struct Biol 13, 168–174 (2003).
Sanbonmatsu, K. Y. & Garcia, A. E. Structure of Met-enkephalin in explicit aqueous solution using replica exchange molecular dynamics. Proteins 46, 225–234 (2002).
Shao, J., Tanner, S. W., Thompson, N. & Cheatham, T. E. Clustering Molecular Dynamics Trajectories: 1. Characterizing the Performance of Different Clustering Algorithms. J. Chem. Theory Comput. 3, 2312–2334 (2007).
Bashford, D. & Case, D. A. Generalized born models of macromolecular solvation effects. Annu Rev Phys Chem 51, 129–152 (2000).
Miller, B. R. et al. MMPBSA.py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput. 8, 3314–3321 (2012).
Onufriev, A., Bashford, D. & Case, D. A. Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins 55, 383–394 (2004).
Author information
Authors and Affiliations
Contributions
S.T.Q., D.L. and R.L. performed the experiments. C.J.B. and D.L. planned the experiments. C.J.B., D.L. and C.S.V. wrote the paper. C.J.B., C.V., R.W.B. and D.P.L. designed and directed the research. All authors read the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Lama, D., Quah, S., Verma, C. et al. Rational Optimization of Conformational Effects Induced By Hydrocarbon Staples in Peptides and their Binding Interfaces. Sci Rep 3, 3451 (2013). https://doi.org/10.1038/srep03451
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep03451
This article is cited by
-
Engineering an autonomous VH domain to modulate intracellular pathways and to interrogate the eIF4F complex
Nature Communications (2022)
-
Control of the eIF4E activity: structural insights and pharmacological implications
Cellular and Molecular Life Sciences (2021)
-
Peptidomimetics: A Synthetic Tool for Inhibiting Protein–Protein Interactions in Cancer
International Journal of Peptide Research and Therapeutics (2020)
-
Monitoring flux in signalling pathways through measurements of 4EBP1-mediated eIF4F complex assembly
BMC Biology (2019)
-
Therapeutic anti-cancer activity of antibodies targeting sulfhydryl bond constrained epitopes on unglycosylated RON receptor tyrosine kinase
Oncogene (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.