Abstract
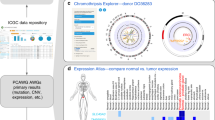
The UCSC Cancer Genomics Browser (https://genome-cancer.ucsc.edu) offers interactive visualization and exploration of TCGA genomic, phenotypic and clinical data, as produced by the Cancer Genome Atlas Research Network. Researchers can explore the impact of genomic alterations on phenotypes by visualizing gene and protein expression, copy number, DNA methylation, somatic mutation and pathway inference data alongside clinical features, Pan-Cancer subtype classifications and genomic biomarkers. Integrated Kaplan–Meier survival analysis helps investigators to assess survival stratification by any of the information.
Similar content being viewed by others
Introduction
Cancer is largely a genomic disease, with each cancer arising from a signature set of somatic alterations. These alterations are observed directly as point mutations, copy number and structural variants in tumor DNA and indirectly as changes in transcription, protein activity and epigenetic states.
The Cancer Genome Atlas (TCGA) project is a comprehensive and coordinated effort to accelerate our understanding of the molecular basis of cancer through the application of genomic technologies. TCGA Pan-Cancer analysis looks to identify genomic similarities between tumors regardless of tissue or organ of origin. Existing knowledge for one group of tumors might help demystify another group with a similar genomic subtype and treatments that are established for the first group might prove effective for the second. In order for this to happen, researchers must be able to evaluate the genomic data for groups of samples, observe patterns shared by samples of a given subtype and determine if the same pattern is present in a related subtype in a different cancer. Further, researchers should be able to interrelate genomic subtypes with clinical features and evaluate how these new genomic subtypes affect clinically relevant phenotypes such as survival.
The UCSC Cancer Genomics Browser (https://genome-cancer.ucsc.edu)1,2 supports these efforts with flexible visualization options combined with a comprehensive, up-to-date and curated TCGA data collection. It allows researchers to identify and assess genomic signatures in cancer subtypes, to compare and contrast subtypes defined using different data or methods and to assess their impact on survival independently or in conjunction with clinical data.
Many tools offer heatmap visualization, from the Eisen Cluster/TreeView software (http://rana.lbl.gov/EisenSoftware.htm) to the R statistical package (http://www.r-project.org/). Most of these tools generate static images, with fixed sort order and no option to select which data is displayed. One exception is Next-Generation Clustered Heatmaps (http://bioinformatics.mdanderson.org/main/NG-CHM:Overview), which allows users to zoom into the heatmap and explore selected regions interactively. However, cancer genomics analysis demands visualization of multi-omic, clinical and subtype data together and frequently exceeds the capability of any single heatmap tool.
There are many choices for cancer genomics visualization tools, as reviewed3. They fall into three general categories: genome-based, gene-based and pathway-based. Genome-based methods such as the UCSC Cancer Genomics Browser and IGV (http://www.broadinstitute.org/igv/) are well suited for exploring alterations that follow genomic coordinates, such as copy number variations or DNA methylation profile. However, because genome-based visualizations only allow the user to see one or two genomic regions at once, they are generally not as effective for exploring possible connections between alterations in multiple different genomic regions. Gene-based visualizations are offered by GiTools (http://www.gitools.org/), cBio Portal (http://www.cbioportal.org/public-portal/), IntOGen (http://www.intogen.org/home), IGV and the UCSC Cancer Genomics Browser. These tools are useful for exploring alterations in multiple genes at once, especially when the set of genes has known functional significance such as known marker genes or genes in a common pathway. The UCSC Cancer Genomics Browser further allows probe-based visualization in its gene-based viewing mode, which is particularly effective for assessing exon expression or DNA methylation data in genes of interest. However, these heatmap based-methods do not generally indicate how genes interact in a pathway. Pathway-based methods such as Cytoscape (http://cytoscape.org/) and cBio Portal allow the user to see known functional connections between genes, but are limited to known pathways (which are limited in coverage) or predicted pathways (which tend to have high error rates). In short, no tool meets all purposes and all tools have scenarios in which they are most effective. The UCSC Cancer Genomics Browser is effective for genome-based or gene-based analysis, scales well to larger cohorts and offers additional features for cancer analysis such as integrated Kaplan-Meier plots.
Results
The power of this approach is illustrated in Figure 1, which shows a heatmap of the somatic mutation profile of the significantly mutated genes in the TCGA acute myeloid leukemia (AML) cohort, as well as the corresponding AML subtype designations for these samples. Compared to the other cancers within TCGA, AML is a distinct, relatively homogenous group, yet it can be further divided into subtypes by clustering genomic data such as miRNA expression4 or DNA methylation (Peter Laird, Personal communication) (Figure 1b). miRNA expression and DNA methylation subtypes show significant concordance, in particular, between miRNA cluster 3 and DNA methylation cluster 3 (the orange subtypes) and also between the miRNA cluster 5 and methylation cluster 5 (the green subtypes). The double cluster 3 is strongly enriched for mutations of NPM1, a nucleoplasmin involved in the regulation of TP53; this pathway connection is reflected in the mutually exclusive pattern of TP53 and NPM1 mutations (Figure 1a). The double cluster 5 shows relatively few mutations and is strongly enriched for samples with favorable cytogenetic risk4. The UCSC Cancer Genomics Browser's dynamic Kaplan-Meier plot confirms this by demonstrating that the samples in miRNA cluster 5 show a substantial advantage in overall survival (Figure 1c). Altogether, this analysis connects cancer subtypes, their underlying genomic data and their clinical outcome. This analysis is done in seconds and interactively.
Using the UCSC Cancer Genomics Browser to explore relationships between somatic mutation profiles, genomic subtypes and survival.
(a) Somatic mutations for the most-significantly mutated genes in TCGA AML tumor samples3. Samples are arranged in rows and genes in columns. Red indicates that the tumor sample harbors non-synonymous coding mutations in the corresponding gene while white indicated that such mutations were not detected. (b) Column 1 represents the miRNA expression clusters3, Column 2 represents the DNA methylation clusters (Peter Laird, Personal communication) and Column 3 represents cytogenetic risk category for the AML cohort. For each column, each cluster or category was assigned a distinct color from the D3 color map (https://github.com/mbostock/d3/wiki/Ordinal-Scales), with five clusters for miRNA expression (cluster 1–5) and nine for DNA methylation (cluster 1–9) and three for cytogenetic risk category (favorable, intermediate, poor). A strong concordance is observed between miRNA cluster 3 (orange), DNA methylation cluster 3 (also orange) and intermediate cytogenetic risk (light blue); and between miRNA cluster 5 (green), DNA methylation cluster 5 (also green) and favorable cytogenetic risk (dark blue). (c) The integrated Kaplan-Meier plot confirms that miRNA cluster 5 (green line) has a more favorable overall survival profile. The colors of the lines correspond to the colors of the miRNA clusters. See https://genome-cancer.ucsc.edu/proj/site/hgHeatmap/#?bookmark=sr1.
The TCGA Pan-Cancer analysis revealed that tumor samples cluster largely by their tissue of origin5. For example, lung squamous cell carcinoma (LUSC) clusters with other squamous cell carcinomas such as head and neck (HNSC) and some bladder urothelial carcinoma (BLCA) samples, while lung adenocarcinoma (LUAD) falls into a distinct cluster. The contrast between LUSC and LUAD is illustrated in Figure 2a, a gene expression heatmap for a set of 600 genes with at least three-fold differential expression between LUSC and LUAD. The associated clinical heatmap (Figure 2b) shows that, in lung cancer, the subtypes defined by Pan-Cancer whole-genome RNAseq analysis (predominantly LUAD-like and squamous-like subtypes) are largely consistent with the cancer types defined by pathology (LUAD and LUSC cohorts), but there are exceptions.
Using the UCSC Cancer Genomics Browser to compare lung cancer subtypes defined by pathology or Pan-Cancer RNAseq analysis and identify a subtype of discordant samples with poor recurrence free survival prognosis.
(a) The heatmap shows gene expression for TCGA LUAD and LUSC samples in a combined lung cohort, over a set of genes differentially expressed (> 3 fold) between the two cohorts. (b) Column 1 is user custom annotation that partitions the samples into three groups: LUAD, concordant LUSC (where the cohort and the RNA subtype are consistent) and discordant LUSC (where the cohort and RNA subtype are inconsistent). Column 2 is tumor's Pan-Cancer RNA subtype designation. Red: LUAD-like. Light blue: squamous-like. Column 3 is tumor type by pathology, which is equivalent to the TCGA cohort. Dark blue: LUAD. Light blue: LUSC. Column 4 is the Shedden gene expression signature where higher signature score (red) predicts poor overall survival in LUAD8. The discordant LUSC samples have a gene expression pattern weakly resembles LUAD. (c) The Kaplan-Meier plot indicates that the discordant LUSC samples (bottom, yellow) have a worse recurrence free survival profile than the concordant LUSC samples (top, brown). See https://genome-cancer.ucsc.edu/proj/site/hgHeatmap/#?bookmark=sr2.
Most LUSC samples are designated as squamous-like by RNAseq analysis. However, 19 out of 347 LUSC samples fall into the LUAD-like RNA subtype. This is not too surprising, since LUAD and LUSC are difficult to distinguish histologically, with as much as 40% disagreement between pathologists6. Misdiagnosis is potentially fatal: Bevacizumab (Avastin) is the recommended chemotherapy for LUAD, but can lead to a life-threatening hemorrhage in LUSC patients7. A user-defined custom annotation column divides the samples into three categories: LUAD, discordant LUSC (those for which the RNA subtype is inconsistent with the cohort) and concordant LUSC (for which the RNA subtype and cohort are consistent). In Figure 2a, we observe that the discordant LUSC samples have gene expression levels that are more similar to LUAD than LUSC, although their similarity to LUAD is modest. This suggests that these samples may be near the borderline for subtype classification, for example, tumors that are less differentiated, therefore difficult to classify by pathology. Many of these discordant LUSC samples lack the 3q26 amplification commonly seen in LUSC and other squamous cell carcinomas, where PIK3CA and ECT2 are located8 (Figure 3).
The UCSC Cancer Genomics Browser shows that many of the discordant LUSC samples (LUSC by pathology, LUAD-like by Pan-Cancer RNAseq analysis) lack the 3q26 amplification commonly seen in LUSC and other squamous cell carcinomas, where PIK3CA and ECT2 are located.
LUSC: lung squamous cell carcinoma. LUAD: lung adenocarcinoma. See https://genome-cancer.ucsc.edu/proj/site/hgHeatmap/#?bookmark=sr3.
Using the UCSC Cancer Genomics Browser's genomic signature function, users can easily define gene expression signatures. We further analyzed the discordant samples against the Shedden gene expression signature where a high signature value predicts poor overall survival in LUAD9. We are able to confirm the Shedden signature in TCGA LUAD data in terms of both overall and recurrence free survival (Figure 4). The Shedden signature value is high for the discordant LUSC samples, suggesting that if they are misclassified by pathology and truly LUAD samples, then their prognosis is likely to be poor (Figure 2b). Consistent with this, the browser's Kaplan-Meier plot suggests that the patients of the discordant LUSC group have worse recurrence free survival prognosis than either the LUAD group or the concordant LUSC group (Figure 2c). This demonstrates how the UCSC Cancer Genomics Browser, in combination with the Pan-Cancer subtypes, can lead to testable hypotheses with a direct impact on patient care.
Confirms the Shedden gene expression signature in TCGA lung adenocarcinoma (LUAD) data in terms of both overall and recurrence free survival.
(a) UCSC Cancer Genomics Browser heatmap view of the gene expression data over the set of genes that used by Shedden et. al. to predict overall survival in LUAD. When these genes are up-regulated, their expression profile predicts poor overall survival in LUAD1. (b) TCGA LUAD sample annotation shown in the UCSC Cancer Genomics Browser clinical heatmap as four columns. Column 1 is the user-defined two subgroups of samples. Red group: samples with high Shedden signature value. Green group: samples with low signature value. Column 2 is the Shedden signature score built using the browser's genomic signature functionality. Red to green color represents higher to lower signature scores computed dynamically. Column 3 is the tumor sample type. Blue: primary tumor. Green: recurrent tumor. Column 4 is LUAD gene expression subtype. Dark blue: Bronchioid. Light Blue: Magnoid. Orange: Squamoid. (c) Shedden signature high score subgroup (red line) has a worse overall survival compared to the low score subgroup (green line). See https://genome-cancer.ucsc.edu/proj/site/hgHeatmap/#?bookmark=sr4.
Interactive online exploration is easy using the UCSC Cancer Genomics Browser in part because it hosts a comprehensive collection of TCGA data. There are currently 376 genomic datasets from 27 TCGA cancer types, including data on gene and exon expression, total protein and phosphoprotein expression, segmented and gene-level copy number variation, DNA methylation, somatic mutation and pathway inference, as well as all the associated public-tier TCGA patient and tumor sample clinical data (Supplementary Table 1). TCGA data is automatically updated on the browser on a monthly basis. We further curate the overall and recurrence-free survival information based on patient follow-up data, which are used in the integrated survival analysis. Pan-Cancer subtypes are defined by the Pan-Cancer working group using gene expression, copy number, DNA methylation, protein expression, or somatic mutation data individually, or integratively by iCluster10, PARADIGM pathway inference11, Cluster of Clusters and SuperCluster method5 are available.
Pan-Cancer subtypes defined by RNA (k = 16 subtypes) or protein expression (k = 8) show strong overall concordance with the tumor's tissue of origin (Supplementary Figure 3). Both subtype designations use tissue dependent data as input that reflect both tumors' somatic changes as well as innate genomic profile of the tissue of origin. Pan-Cancer subtypes defined using only copy number alterations or mutation reflect somatic changes in tumor DNA and show less concordance with the tissue of origin (Supplementary Figure 4). DNA methylation data subtypes are defined using a selected set of loci unmethylated in almost all the matched normals. The subtypes that reflect tumor-specific hyper-methylation show a unique profile according to tumor's tissue of origin, with each tissue often further breaking down into several subtypes (Peter Laird, Personal communication). Investigators can upload custom sample annotations to the browser, such as their own subtype classifications, to view with TCGA genomic and clinical data, compare against Pan-Cancer subtypes and evaluate using online survival analysis. Researchers can also save browser views as bookmarks to share interesting findings with collaborators or for later revisits. In addition to data visualization, TCGA data are available at the browser as bulk downloads and selected data slices.
Discussion
TCGA holds promise for a comprehensive understanding of human cancer. The project has produced the most uniform and comprehensive catalog of cancer-specific genomic aberrations to date. The TCGA Pan-Cancer project furthers this effort by identifying patterns of aberrations that are shared by different cancers regardless of their tissue of origin. But to further this effort and to derive most value from the large amounts of TCGA data, there must be powerful, flexible tools to allow researchers and clinicians to connect genomic aberrations, cancer subtypes and clinical features such as survival and response to therapy.
The UCSC Cancer Genomics Browser presents the TCGA data and Pan-Cancer subtypes in a coherent, integrated system for both TCGA researchers and the scientific community at large. It provides direct access to and visualization of data at specific genes or genomic regions on samples of interest. It displays genomic aberrations alongside clinical and annotation features in a flexible, dynamic display, sorted by the features of the user's selection. Integrated Kaplan-Meier plots allows researchers to assess when change of cancer subtypes coincide with changes in survival. We have shown examples of this in acute myeloid leukemia and in adenocarcinoma and squamous lung cancer. We will continue to develop the UCSC Cancer Genomics Browser with mechanisms for custom input of genomic data, new genesets from the literature and queries and profiles that select across genomic data types.
Methods
The UCSC Cancer Genomcs Browser can be accessed by navigating to https://genome-cancer.ucsc.edu/ and following the central hyperlink. Public datasets include TCGA, Cancer Cell Line Encyclopedia12 and the Stand Up To Cancer (SU2C) Breast Cancer Dream Team13 as well as other datasets from the literature. For each dataset displayed, a “Dataset details” link opens a pop-up window that details the data provenance. For TCGA datasets, the data was downloaded from the TCGA Data Coordination Center (DCC) or from the results of the TCGA Firehose pipeline at the Broad Institute (http://gdac.broadinstitute.org/).
Genomic data and views
The UCSC Cancer Genomcs Browser displays genomic, clinical and annotation data in multiple views: Heatmap, Proportions and Boxplot. The default Heatmap view allows the visualization of each sample in the cohort separately, with rows representing samples and columns representing genes, genomic regions, clinical or phenotypic parameters. Hovering over a data point will display its sample, data type and value in a separate tooltip box. Proportion and Boxplot viewing modes summarize genomic data at each gene or genomic location, with Proportions view showing the data in rank order and Boxplot view illustrating the data distribution at each position.
Genomic data can be visualized along genomic coordinates or according to a set of genes using the Chromosome or Genes viewing mode respectively. Chromosome mode displays the genomic data by position, from whole-genome scale (the default mode) to single base-pair resolution. Genes mode displays genomic data for selected sets of genes where users can select an existing gene set from the browser's database or to construct their own. In either mode, users can dynamically zoom to any scale of interest.
Annotation data
Each genomic dataset has a number of annotation fields associated with it, including Pan-Cancer subtypes and clinical features. Certain annotation fields are displayed by default; others can be added to the display by clicking the plus (+) icon. In Heatmap view, genomic and annotation data is sorted first by the values in the leftmost column and then by each column after that. By clicking on a column, users can hide or reposition an annotation, dynamically changing the sort order.
Each annotation column is color-coded by its contents. Continuous data are rendered as a color continuum, while discrete, categorical data are rendered as a set of multi-colored blocks. As with the genomic data, hovering over an annotation column will display the sample and value in a separate tooltip box.
For annotation fields that span multiple cohorts, such as a Pan-Cancer subtype classification, coloring is consistent across cohorts. This allows the user to quickly determine when samples from different cohorts belong to the same subtype.
Controlling the display
Users can zoom into the display either horizontally or vertically. By clicking and dragging horizontally across the genomic data, users can zoom into selected genes or genomic regions. By clicking and dragging the mouse vertically across the annotation data, users can zoom into selected samples. Controls alongside the genomic and annotation data allow the user to zoom out horizontally and vertically. In addition to zooming, the user can resize the display of any dataset by dragging the size control at the lower right of the dataset display.
Subgroups and online statistics
Subgroups allow users to group samples according to an arbitrary combination of annotation data values. Statistical analysis results from comparing two subgroups of samples are displayed at the bottom of the genomic map, indicating whether values vary significantly by subgroup. For example, it can be used to indicate if mutation frequencies or gene expression levels vary significantly by user-defined groupings.
Genomic signatures
Genomic signatures are an algebraic expression over a set of genes. The resulting signature values form a new annotation column, which can be used to sort the samples according to genomics data. A signature can be as simple as a single gene, enabling users to sort the genomic data by TP53 expression or ERG copy number changes. Users can also make more complex signatures such as the Shedden gene expression signature that predicts overall survival in lung adenocarcinoma, or one that predicts drug response.
Annotation upload and download
Investigators can upload their own sample annotations as custom data. They can also download annotation data currently displayed at the browser, further analyze it using other tools such as Microsoft Excel or R and upload the results back into the browser. These functions together allow users to perform advanced analysis on the browser.
Kalpan-Meier plots
The UCSC Cancer Genomcs Browser can dynamically generate Kalpan-Meier (KM) plots for datasets with curated survival data, which includes all TCGA cohorts. Groups of samples are colored and labeled according to the leftmost annotation column so that users can quickly assess if any annotation field has an impact on survival. Advanced options in KM plots allow investigators to select other survival parameters. For example, in TCGA datasets, users can choose to assess either recurrence free survival or overall survival.
Bookmarks
Bookmarks provide users with hyperlinks to save the state of the browser and share analysis insights with others. When a user enters a bookmark URL in a web browser, a new UCSC Cancer Genomics Browser session will open with the same saved view and the user can then proceed interactively, perhaps trying something different the second time. User accounts allow users to save named bookmarks under their account.
References
Goldman, M. et al. The UCSC Cancer Genomics Browser: update 2013. Nucleic Acids Res. 41, D949–954 (2013).
Zhu, J. et al. The UCSC Cancer Genomics Browser. Nat Methods 6, 239–240 (2009).
Schroeder, M. P., Gonzalez-Perez, A. & Lopez-Bigas, N. Visualizing multidimensional cancer genomics data. Genome Med. 5, 9 (2013).
Cancer Genome Atlas Research Network. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 368, 2059–2074 (2013).
Stuart, M. J. et al. The cancer genome atlas pan-cancer analysis project. Nat. Genet. 45, 1113–1120 (2013).
Stang, A., Pohlabeln, H., Müller, K. M., Jahn, I., Giersiepen, K. & Jöckel, K. H. Diagnostic agreement in the histopathological evaluation of lung cancer tissue in a population-based case-control study. Lung Cancer 52, 29–36 (2006).
Stinchcombe, T. E. & Socinski, M. A. Current treatments for advanced stage non-small cell lung cancer. Proc Am Thorac Soc. 6, 233–41 (2009).
Fields, A. & Justilien, V. The guanine nucleotide exchange factor (GEF) Ect2 is an oncogene in human cancer. Adv Enzyme Regul. 50, 190–200 (2010).
Director's Challenge Consortium for the Molecular Classification of Lung Adenocarcinoma. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nat Med. 14, 822–827 (2008).
Shen, R., Olshen, A. B. & Ladanyi, M. Integrative clustering of multiple genomic data types using a joint latent variable model with application to breast and lung cancer subtype analysis. Bioinformatics 25, 2906–2912 (2009).
Vaske, C. J. et al. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics 26, i237–i245 (2010).
Barretina, J. et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 483, 603–607 (2012).
Heiser, L. M. et al. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc Natl Acad Sci U S A. 109, 2724–2729 (2012).
Acknowledgements
This work was supported by National Cancer Institute (NCI) [5U24CA143858]; American Association for Cancer Research (AACR) Stand Up To Cancer (SU2C) program [SU2C-AACR-DT0409]; and UCSF Helen Diller Family Comprehensive Cancer Center. We gratefully acknowledge the contributions from the TCGA Research Network and its TCGA Pan-Cancer Analysis Working Group (contributing consortium members are listed in Supplementary Note 1).
Author information
Authors and Affiliations
Contributions
The main manuscript was written by M.S.C. and J.Z. and reviewed by all authors. B.C., T.S., M.G. and S.M. are all central to the development of the UCSC Cancer Genomics Browser, under the direction of D.H. and J.Z.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Note 1
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Cline, M., Craft, B., Swatloski, T. et al. Exploring TCGA Pan-Cancer Data at the UCSC Cancer Genomics Browser. Sci Rep 3, 2652 (2013). https://doi.org/10.1038/srep02652
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep02652
This article is cited by
-
Identification of miR-143-3p as a diagnostic biomarker in gastric cancer
BMC Medical Genomics (2023)
-
Bufalin targeting BFAR inhibits the occurrence and metastasis of gastric cancer through PI3K/AKT/mTOR signal pathway
Apoptosis (2023)
-
High Level of Ubiquitin Conjugate Enzyme E2O Indicates Poor Prognosis of Patients with Hepatocellular Carcinoma
Current Medical Science (2023)
-
CD97 serves as a novel biomarker of immune cell infiltration in hepatocellular carcinoma
World Journal of Surgical Oncology (2022)
-
PCGEM1 promotes proliferation, migration and invasion in prostate cancer by sponging miR-506 to upregulate TRIAP1
BMC Urology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.