Abstract
The transcriptional regulatory protein Fnr, acts as an intracellular redox sensor regulating a wide range of genes in response to changes in oxygen levels. Genome sequencing of Herbaspirillum seropedicae SmR1 revealed the presence of three fnr-like genes. In this study we have constructed single, double and triple fnr deletion mutant strains of H. seropedicae. Transcriptional profiling in combination with expression data from reporter fusions, together with spectroscopic analysis, demonstrates that the Fnr1 and Fnr3 proteins not only regulate expression of the cbb3-type respiratory oxidase, but also control the cytochrome content and other component complexes required for the cytochrome c-based electron transport pathway. Accordingly, in the absence of the three Fnr paralogs, growth is restricted at low oxygen tensions and nitrogenase activity is impaired. Our results suggest that the H. seropedicae Fnr proteins are major players in regulating the composition of the electron transport chain in response to prevailing oxygen concentrations.
Similar content being viewed by others
Introduction
H . seropedicae is an endophytic diazotroph belonging to the Betaproteobacteria that can fix nitrogen under micro-oxic and nitrogen limiting conditions. It is found in association with different crops such as rice, maize, sugar cane and sorghum1,2,3. H. seropedicae SmR1 is an aerobic bacterium which has a branched respiratory chain comprising different types of terminal oxidases, which potentially could allow the bacteria to exploit respiratory flexibility and survive under microaerobic conditions4. Genome sequencing revealed genes coding for three Fnr-like proteins in H. seropedicae SmR14. In many organisms the Fnr protein, which belongs to the CRP-FNR family of transcriptional regulators5, acts as a positive or negative regulator of genes required for the metabolic switch in response to O2 levels6. The Fnr protein of Escherichia coli contains an N-terminal sensory domain, which binds an oxygen-labile [4Fe–4S]2+ cluster under oxygen limiting conditions7 and a C-terminal DNA-binding domain, which recognizes a partially palindromic sequence called the Fnr-box or anaerobox, TTGAT-N4-ATCAA5,8.
Various Fnr-related transcriptional regulators of the CRP-FNR family have been reported to be involved in biological nitrogen fixation. The Fnr protein from Klebsiella pneumoniae is required to relieve inhibition of NifA activity by its partner regulatory protein NifL under anaerobic conditions9. In Rhizobium leguminosarium UPM791 FnrN is responsible for the expression of the high affinity oxidase encoded by fixNOQP that supports growth under microaerobic conditions and is essential for nitrogen fixation10. Similarly the FixK2 protein is also essential for nitrogen fixation in Bradyrhizobium japonicum and Sinorhizobium meliloti11,12,13. Representatives of the CRP-FNR family are also known to act negatively in repressing genes related to nitrogen fixation, such as the FixK1 protein from B. japonicum12.
As the H. seropedicae genome encodes three Fnr-like proteins, we were interested to determine the potential involvement of these three Fnr homologs in nitrogen fixation and in the control of gene expression in response to oxygen limitation. Several representatives of the Betaproteobacteria encode more than one Fnr-like protein in their genome. For example, Burkholderia pseudomalei 1710b and Herminiimonas arsenicoxydans have genes coding for two Fnr-like proteins, whereas Cupriavidus metalidurans CH34 and Ralstonia eutropha H16 encode three and five Fnr-like proteins, respectively. However, to date the functions of these Fnr-like paralogs have not been determined.
In the current study, we sought to attribute function to the three Fnr-like proteins found in H. seropedicae and in particular to examine their role in the regulation of electron transport chain composition. We demonstrate that deletion of all three fnr alleles results in a growth phenotype under microaerobic conditions, implying that Fnr proteins may be involved in controlling the expression of respiratory oxidases. By comparing the transcription profiles of the wild-type and triple fnr mutant strains and performing further gene expression and biochemical analyses, we observe that the Fnr proteins not only activate genes required for expression and activity of the high affinity cbb3-type oxidase, but also have a major influence on the regulation of the cytochrome bc1 complex and on cytochrome c biogenesis. This suggests that the H. seropedicae Fnr proteins facilitate distribution of the electron flux through cytochrome carriers and the heme-copper oxidase branch of the respiratory chain to increase coupling efficiency under oxygen-limiting conditions.
Results
H. seropedicae encodes three proteins that share homology with E.coli Fnr
The H. seropedicae SmR1 genome4 contains three genes encoding homologs of the Fnr protein, which we designate as fnr1, (Locus Tag: Hsero_3197; Ref seq: YP_003776587.1), fnr2 (Locus Tag: Hsero_2381; Ref Seq: YP_003775788.1) and fnr3 (Locus Tag: Hsero_2538; Ref Seq: YP_003775945.1) The H. seropedicae Fnr1, Fnr2 and Fnr3 proteins share 38.4%, 37.5% and 26.9% identity respectively with E. coli Fnr. As shown in Figure 1, the Fnr1 and Fnr3 proteins are more similar to each other than to Fnr2. When compared with E. coli Fnr, the three Fnr paralogs have characteristic sequence features that are hallmarks of Fnr proteins, including three conserved N-terminal cysteines plus a central cysteine that are thought to co-ordinate the oxygen-labile [4Fe–4S]2+ cluster5. In addition all three deduced proteins contain the predicted dimerization helix sequence located at the beginning of the C-terminal domain and a helix-turn-helix DNA binding motif characteristic of members of the CRP-FNR family.
Alignment between H. seropedicae Fnr1, Fnr2 and Fnr3 proteins and E. coli Fnr.
Identical amino acids are indicated by asterisks ( * ), high similarity amino acids are indicated by colons ( : ) and low similarity amino acids by dots ( . ). Conserved cysteines required for binding of the [4Fe-4S]+ are shown in bold and indicated by thick arrows. The double underlined sequence represents the region of the N-terminal sensory domain that comprises the eight-stranded β-roll. The α-helix required for dimerization is boxed. Highlighted in light-grey is the DNA-binding domain with residues that are important for Fnr-box recognition indicated by thin arrows.
A phylogenetic affiliation for the CRP-FNR superfamily of transcriptional regulators has been proposed5,14. Proteins from the Fnr and FnrN groups sense oxygen directly via an [4Fe–4S]2+ cluster, whereas the FixK members, which lack the conserved cysteine residues required for ligating the cluster, either sense oxygen indirectly, or simply relay the O2 signal, via the two-component regulatory system FixL-FixJ13. In order to classify the H. seropedicae Fnr proteins, we carried out a phylogenetic reconstruction and observed that all three are in a clade together with other members of the Fnr group of CRP-FNR superfamily from Betaproteobacteria (Supplementary Fig. S1). This finding is in agreement with the cysteine motif arrangement found in the H. seropedicae Fnr proteins, which is characteristic of the Fnr group and clearly divergent from the FnrN and FixK groups commonly represented in the Alphaproteobacteria5. Among the Betaproteobacteria, H. seropedicae Fnr1, Fnr2 and Fnr3 branched into a group with the Fnr proteins from Janthinobacterium sp. Marseille (Minibacterium massiliensis) and H. arsenicoxydans. Within this group H. seropedicae Fnr2 is more divergent from Fnr1 and Fnr3 and also from the J. sp. Marseille and H. arsenicoxydans Fnr proteins.
Construction of H. seropedicae Δfnr mutant strains
To study the role of the three Fnr proteins, single, double and triple fnr deletion mutant derivatives of H. seropedicae strain SmR1 were constructed using a sacRB::Km cartridge as described in the Methods section. This strategy allowed the construction of 7 unmarked deletion strains in which all possible combinations of fnr genetic backgrounds are available (Supplementary Fig. S2). The promoters and non-coding regions of fnr were retained in these ORF deletions, enabling transcriptomic analysis of fnr mutant strains. Deletion mutants were given the prefix MB, with a number indicating which fnr deletion is present (for example MB1 lacks fnr1, whereas MB13 lacks fnr1 and fnr3 respectively).
Influence of fnr on growth at low oxygen concentrations
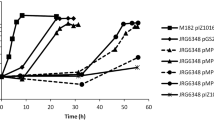
As Fnr proteins are known to sense oxygen and have an important role as transcriptional regulators during the switch from aerobic to oxygen-limiting conditions, we were interested to determine if deletion of the three H. seropedicae fnr genes would influence growth under hypoxic conditions. Accordingly, we compared the growth curves of wild-type strain SmR1 with that of the triple deletion fnr strain MB231, when grown in malate minimal medium with an initial oxygen concentration in the gas phase of 5% and supplemented with 2 or 20 mM ammonium chloride. Oxygen consumption during growth of the cultures was monitored using a gas chromatograph equipped with a molecular sieve column and a TCD detector. Under these conditions, the rate of oxygen depletion in the gas phase was similar for both the wild-type and MB231 strains (Figure 2). However, after 4 hours when the oxygen concentration had decreased to approximately half of the initial concentration, the growth rate of the triple fnr deletion strain was clearly slower than that of the wild-type. Moreover, the optical density (O.D600) of the triple mutant strain reached only 0.5 and 0.6 after 10 hours growth in the presence of 2 and 20 mM NH4Cl, respectively, compared with an O.D600 of 0.8 reached by the wild type strain in both ammonium chloride concentrations (Supplementary Fig. S3). In contrast we observed no difference in growth rate, when both strains were grown under aerobic conditions with 20.8% oxygen in the gas phase (Supplementary Fig. S4). These results imply that the absence of fnr imposes a growth rate penalty under oxygen-limiting conditions, which may suggest the involvement of at least one of the three Fnr proteins in the regulation of terminal oxidases in response to oxygen, as observed in other bacteria.
Influence of fnr genes on growth during oxygen limitation.
The growth of H. seropedicae SmR1 (black squares) and MB231 (grey squares) strains were assayed in NFbHP-Malate minimal media supplemented with high ammonium concentration (20 mM NH4Cl) under 5% initial oxygen concentration. The oxygen depletion in the gas phase was monitored for SmR1 (black circles) and MB231 (grey circles). Every two hours 0.5 mL samples from the flask gas phase were analysed by gas chromatography. The data represents the mean of three independent assays performed in duplicate. Error bars indicate standard deviations. In some case these are not visible as they are smaller than the graph points.
Transcriptional profiling of wild-type and fnr strains using RNA-seq
The influence of the Fnr proteins on global gene expression under microaerobic conditions was assessed using RNA sequencing. To avoid problems associated with growth rate differences, we grew the wild-type and the fnr ablated strain, MB231 under aerobic conditions (20.8% oxygen) to an optical density of 0.4 and then switched the cultures to microaerobic conditions (initial oxygen concentration of 2%) for 1.5 hours prior to RNA extraction. Comparison of global gene expression patterns revealed that 187 H. seropedicae genes were differentially expressed by more than 3-fold, with p values <0.05. In some cases, depending on the genomic context, genes with p-values slightly higher than 0.05 were also considered as being differentially expressed. Of these, 143 were down-regulated in the fnr triple mutant strain, indicating that these genes are activated either directly or indirectly by Fnr under oxygen-limiting conditions. 44 genes were up-regulated in the fnr ablated strain, implying that they are targets for Fnr-mediated repression. A complete listing of differentially expressed genes is provided in Supplementary Dataset 1.
Of the 187 regulated genes, 70 (37.43%) are classified in the cellular process category and 58 genes (31.02%) belong to the metabolism category according to the Clusters of Orthologous Genes (COG) functional classification15 (Supplementary Fig. S5a). In the cellular process category, 29 genes are related to signal transduction mechanisms, whereas in the metabolism category, 30 genes are related to energy production and conversion (Supplementary Fig. S5b). Most of the genes from the energy and production subcategory, encode important proteins required for synthesis and activity of many of the respiratory electron transport chain components (Table 1 and Supplementary Dataset 1). These findings suggest that the H. seropedicae Fnr proteins may facilitate efficient adaptation to the variable oxygen concentrations found in different environments.
H. seropedicae possesses a branched aerobic respiratory chain comprising four different types of terminal oxidases4. These are represented by the aa3-type (cox) and cbb3-type (fix) oxidases in the heme copper oxidase branch and the bd-type and bo3-type oxidases representing the ubiquinol oxidase branch (Figure 3a). Amongst the genes that are potentially activated by Fnr, large changes in transcript abundance were observed in genes required for the biosynthesis and activity of the cbb3-type heme-copper oxidase (Table 1). These include the fixNOP operon encoding the structural components of this oxidase and the maturation genes fixG, fixH, fixI and fixS, which in other bacteria is often organized as an operon16,17. In contrast, in H. seropedicae the fixG, fixH, fixI and fixS genes are dispersed in two distinct operons (Figure 4a), in which fixG and Hsero_3199 (fixH) appear to form an operon with the conserved putative transmembrane protein Hsero_3198 and fixI and fixS apparently form an operon with the heme biosynthesis gene hemN18 and Hsero_3206, which encodes a conserved hypothetical protein (Figure 4a). In agreement with the down-regulation observed in the absence of fnr, both the fixNOP and hemN-Hsero_3206-fixIS putative operons have well conserved Fnr-boxes located at positions −143 and −128 upstream of their respective translational start sites (Table 1). The Fnr-Box in the hemN promoter perfectly matches the consensus TTGAT-N4-ATCAA, while the fixN promoter Fnr-box, TTGAT-N4-GTCAA, has only one mismatch (underlined) (Table 1).
Fnr regulation of components of the electron transport chain (ETC) in H. seropedicae as determined by transcript profiling.
(a) Schematic representation of the probable organization of ETC branches in H. seropedicae based on the genome annotation. (b) Influence of Fnr on differential expression of genes represented in (a). FC indicates fold change.
Effect of fnr1, fnr2 and fnr3 mutations on expression of the fixNOP and hemN-Hsero_3206-fixIS operons in H. seropedicae.
(a) Schematic representation of genomic region encoding the fixNOP and hemN-Hsero_3206-fixIS operons. The genes and predicted functions in the locus are: Hsero_3198, transmembrane protein; Hsero_3199, FixH domain containing protein; fixG, iron-sulfur 4Fe-4S ferredoxin transmembrane protein; fixP, cbb3-type cytochrome c oxidase-subunit III; fixO, cbb3-type cytochrome oxidase-subunit II; fixN,cbb3- type cytochrome c oxidase, subunit I; fixS, nitrogen fixation protein P-type ATPase protein; fixI, cation transport P-type ATPase protein; Hsero_3206, conserved hypothetical protein; hemN, oxygen-independent coproporphyrinogen III oxidase. Black rectangles represent putative FNR-boxes. Genes are not drawn to scale. (b) β-Galactosidase activities of fixN::lacZ and hemN::lacZ fusions incubated for 3 hours under the oxygen concentrations of 2.0% (black bars), 4.0% (dark grey bars), 6.0% (light grey bars) and 20.8% (white bars). CTRL indicates the SmR1 strain carrying the vector plasmid pPW452 (which contains the lacZ gene without a promoter).
Apart from the cbb3-type heme-copper oxidase, genes encoding the other terminal respiratory oxidases in the H. seropedicae genome were not apparently differentially expressed in response to the presence of Fnr. Although the genome contains two copies of the coxBA operon encoding aa3-type oxidases, one of these (Locus Tags: Hsero_2311-Hsero_2312) did not appear to be expressed under our experimental conditions. The second coxBA operon (Hsero_4160-Hsero_4161) and its associated coxC (Hsero_4157) and coxG (Hsero_4159) genes do not appear to be Fnr-regulated. This was also the case for the cydAB genes encoding the bd-type oxidase and a bo3-type oxidase encoded by the cyoABCD operon. However, analysis of transcript abundance suggests that expression of several other components of the respiratory chain are subject to regulation by Fnr. Transcripts mapping to the petABC operon, which encodes ubiquinol-cytochrome c reductase (also known as the cytochrome bc1 complex or complex III) were down-regulated 7–11 fold in the Fnr mutant compared with the wild-type control (Table 1 and Figure 3b). In addition, significant differential expression was observed for genes encoding cytochrome c551/c552 (Hsero_1104) and cytochrome c553 (Hsero_0153) a c4 type cytochrome, that potentially acts as an electron donor to the cbb3-type heme-copper oxidase19 (Table 1 and Figure 3b). In addition to the apparent influence of fnr on the expression of c-type cytochromes (Table 1 and Figure 3), the Fnr protein(s) also appear to activate genes required for cytochrome c biogenesis. The H. seropedicae genome encodes system II-like machinery for cytochrome c maturation (Supplementary Fig. S6), including homologs of the ResB (CcsB) and ResC (CcsA) proteins, which are proposed to function in the handling of heme and its ligation to apocytochrome and the DsbD (CcdA) and ResA (CcsX) proteins that function in the reduction of the disulphide bond in the CXXCH heme binding site20,21. Transcript profiling revealed that genes encoding all four components were significantly down-regulated in the triple fnr deletion strain (Table 1). The dsbD gene is apparently located in an operon downstream of cutA, which is also implicated in cytochrome c biogenesis22,23. Transcripts mapping to the ndh gene, which encodes the non-coupling NADH dehydrogenase II enzyme, increased in the triple fnr mutant (Table 1), indicating that expression of this enzyme is repressed by Fnr as observed in other bacteria24,25. Perhaps to compensate for this, the expression of Hsero_4284, encoding the energy-conserving NADH dehydrogenase I is apparently activated 3-fold by Fnr (Table 1). Overall, in comparing the global expression pattern of genes involved in electron transport, it would appear that Fnr activates expression of genes involved in the heme-copper oxidase branch of the aerobic respiratory chain (Table 1 and Figure 3). This involves not only regulation of the expression of the cbb3-type oxidase, but also the cytochrome bc1 complex and maturation and expression of c-type cytochromes. Hence, Fnr is likely to influence the flow of electrons through the cytochrome c branch of the pathway in order to optimize energy generation. Additionally, the fnr genes may also control the composition of the quinone pool. Transcripts mapping to ubiF gene which encodes a 2-polyprenyl-3-methyl-6-methoxy-1,4-benzoquinol hydroxylase protein involved in the penultimate step of the ubiquinone biosynthesis pathway and a gene encoding an alternative 2-polyprenyl-6-methoxyphenol hydroxylase (Hsero_4190) also required for ubiquinone biosynthesis were significantly down regulated in the triple fnr deletion strain (Table 1 and Figure 3). In common with many other Proteobacteria, genes encoding a respiratory nitrate reductase, organized as a narGHJI-moaA operon, are also apparently up-regulated by Fnr in H. seropedicae, as transcripts mapping to this operon decreased 16–100 fold in the triple fnr deletion mutant (Table 1 and Supplementary Dataset 1). In addition, the nitrate/nitrate transporter encoded by narK1U operon and the nitrate-sensing two component regulatory system narXL are also strongly down-regulated in the fnr triple deletion (Table 1 and Supplementary Dataset 1). Although this may suggest the potential to utilise nitrate as a terminal electron acceptor, the function of this respiratory nitrate reductase in H. seropedicae is somewhat enigmatic, as various investigators have failed to demonstrate anoxic growth of this organism in the presence of nitrate1,4.
Amongst the global changes in transcript abundance, we observed differential expression of two of the three H. seropedicae fnr genes themselves. Whereas, the transcript abundance upstream of fnr3 did not significantly change in the triple deletion mutant, fnr1 and fnr2 were down-regulated 50-fold and 25-fold respectively (Table 1 and Supplementary Dataset 1). This suggests several possibilities that are not mutually exclusive: (a) Fnr3 is required to activate expression of fnr1 and fnr2, (b) Fnr1 and Fnr2 auto activate their respective promoters or (c) a combination of Fnr proteins is required to activate these promoters.
Analysis of Fnr regulation of genes encoding the cbb3-type respiratory oxidase
To confirm the involvement of Fnr in co-regulation of the fixNOP and hemN-Hsero_3206-fixIS operons we constructed fixN::lacZ and hemN::lacZ transcriptional fusions and analysed the expression of β-galactosidase in the various fnr mutant strains compared with the parental strain. Expression of the fixN::lacZ and hemN::lacZ fusions is apparently regulated by oxygen levels in the wild-type background since we observed a reduction in promoter activities upon exposure to increasing oxygen concentrations (Figure 4b). Consistent with the transcriptomics data (Table 1), both operons are apparently subject to regulation by Fnr. Notably, expression from the fixN and hemN promoters was significantly reduced in strains that lack either fnr1 or fnr3, but activity was equivalent to the parental strain in the fnr2 deletion strain MB2 (Figure 4b). This implies that both Fnr1 and Fnr3 are required to activate expression of the fixNOP and hemN-Hsero_3206-fixIS operons and that Fnr2 is not involved in the regulation of expression of the cbb3-type respiratory oxidase. Accordingly, under oxygen-limiting conditions, no growth penalty is observed for the MB2 strain (Supplementary Fig. S7).
Fnr influences the cytochrome content of H. seropedicae
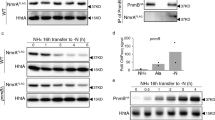
As the transcriptome analysis implicates Fnr as a regulator of genes involved in cytochrome c biogenesis, we compared the spectral features of wild-type and fnr deletion strains. We noticed that strains lacking the fnr1 gene were deficient in a pink pigment when cultured in liquid media (Figure 5a). To further explore this observation we analysed reduced minus oxidized spectra of protein extracts obtained from SmR1 and the fnr deletion strains (Figure 5b and Supplementary Fig. S8). Spectra of the wild type strain were consistent with the presence of c-type (α-band located around 550 nm) and b-type (α-band shoulder around 560 nm) cytochromes in the protein extract. Similar spectral features were found in strains lacking either fnr2 or fnr3. However, all strains lacking fnr1 appeared to be deficient in cytochrome content, which may account for the observed differences in culture pigmentation. In order to obtain further biochemical support for the spectral features observed, we stained protein extracts from SmR1 and fnr mutant strains for covalently bound heme (Figure 5c and Supplementary Fig. S8). In the wild-type strain SmR1, we detected five bands, which presumably represent c type cytochromes. The protein of approximately 34 KDa (band 1) could represent FixP by comparison with the heme staining profile of the fixN mutant strain, RAM21 (Supplementary Fig. S9) and by analogy with studies on the FixNOQP proteins from other bacteria26,27. Notably, the level of this protein was significantly diminished in strains lacking fnr1, consistent with decreased expression of the fixNOP operon observed in the transcriptome (Table 1 and Figure 3) and lacZ-fusion analysis (Figure 4). The bands 2, 4 and 5 may represent PetC, cytochrome c553 (Hsero_0153) and cytochrome c551/c552 (Hsero_1104), respectively, based on the apparent molecular masses and expression pattern of these proteins, which were identified as being activated by Fnr in the transcriptome analysis (Table 1 and Figure 3). All five c-type cytochromes, including band 4, were absent in strains lacking both fnr1 and fnr3 (MB13 and MB231). This is in agreement with the loss of the cytochrome α-band in the UV-visible difference spectra in strains lacking both the fnr1 and fnr3 genes (Figure 5b and Supplementary Fig. S8). Taken together with the data from transcription profiling, these results suggest that both Fnr1 and Fnr3 are necessary to maintain the level of c type cytochromes in H. seropedicae under microaerobic conditions.
Fnr proteins influence the cytochrome content in H. seropedicae.
(a) Bacterial suspensions of H. seropedicae SmR1 and fnr mutant strains. Cells, from 100 mL cultures, were collected by centrifugation and resuspended in 10 mL of buffer (100 mM NaCl and 50 mM Tris.HCl pH 7.5). (b) Reduced minus oxidized visible absorption spectra of protein extracts from H. seropedicae SmR1 and fnr mutant strains. For simplification, only the data for SmR1 (blue), MB1 (red), MB2 (green), MB3 (black) and MB231 (orange) are shown. (c) Heme stained gel of H. seropedicae protein extracts. Samples (50 μg protein per lane) from H. seropedicae SmR1 and fnr mutant strains were separated by 12% Tris-Tricine SDS-PAGE and stained for covalently bound heme with o-dianisidine. On the left the heme stained bands are labeled as 1, 2, 3, 4 and 5 from the top to the bottom of the gel. The apparent molecular masses of proteins (kDa) are indicated on the right. MW: Spectra™ Multicolor Broad Range Protein Ladder (Fermentas). The strains MB13, MB21 and MB23 gave similar spectra and heme stain profiles to MB231, MB1 and MB3, respectively (Supplementary Fig. S8).
Deletion of the three fnr genes impairs nitrogenase activity and growth on dinitrogen
In the analysis reported so far, strains were grown in minimal media containing a high concentration of fixed nitrogen, which represses nitrogen fixation in H. seropedicae. Since the cbb3-type heme-copper oxidase is known to have an important role as a terminal oxidase that supports nitrogen fixation under microaerobic conditions in the Rhizobacteriaceae28,29,30 and is subject to regulation by Fnr proteins, we were interested to determine if nitrogen fixation is influenced by the presence of the Fnr paralogs in H. seropedicae.
When fnr mutant strains were grown under N-deficient conditions (with 0.5 mM sodium glutamate) in semi-solid medium and tested for the ability to reduce acetylene as a measure of nitrogenase activity, no significant differences were observed in comparison with the wild-type strain (Figure 6a). The RAM21 strain ( fixN mutant) was also not deficient in acetylene reduction when grown under these conditions (Supplementary Fig. S10). However, since semi-solid medium enables bacteria to move towards optimal oxygen concentrations appropriate for growth, we sought a more rigorous method to determine the influence of limiting oxygen on nitrogenase activity in the mutant strains. When the fixN mutant strain RAM21 was grown in nitrogen-deficient liquid medium, under conditions of oxygen limitation (initial oxygen concentration of 5% in the gas phase) we observed that acetylene reduction was not severely compromised in comparison with the wild-type (Figure 6b). This suggests that the cbb3-type oxidase is not required to support nitrogenase activity in H. seropedicae under the oxygen-limiting conditions imposed in this experiment. In contrast, the nitrogenase activity of the MB231 mutant strain was severely impaired compared to the wild-type strain under these conditions (Figure 6b). Given the pleiotropic effects on expression of the electron transport components in the triple fnr deletion strain it is likely that the electron flux is insufficient to support nitrogenase activity in the fnr deletion strain under oxygen-limiting conditions. In agreement with this result, we observed that diazotrophic growth of the triple fnr mutant strain was also compromised in liquid N-free medium (Supplementary Fig. S11).
Influence of Fnr and FixN on nitrogenase activity.
The acetylene reduction assay was performed as described in Methods using strains grown in (a) semi-solid medium supplemented with 0.5 mM of sodium glutamate or (b) liquid medium supplemented with 0.5 mM of ammonium chloride under 5% initial oxygen in the gas phase. In (b) samples were taken from the culture to measure the growth curve (primary y axis) of SmR1 (black squares), RAM 21 (dark grey triangles) and MB231 (light grey circles). Black, dark grey and light grey bars (secondary y axis) indicate the nitrogenase activity of SmR1, RAM21 and MB231, respectively. Data represent the average of two independent experiments performed in duplicate. Error bars indicate standard deviations.
Discussion
In order to survive in rapidly changing environments and explore diverse habitats, many bacteria adjust the composition of their respiratory chains to cope with fluctuating oxygen concentrations. In many cases this involves regulation of the expression of terminal oxidases in order to optimize energy generation. The global transcriptional regulator Fnr and its various orthologs, provide a widespread mechanism for sensing oxygen and communicating this to the transcriptional apparatus in order to balance the levels of different terminal oxidases, according to prevailing environmental conditions. Accordingly, we observe that Fnr is required to activate the expression of genes required for the synthesis and activity of the high-affinity cbb3-type heme copper oxidase in H. seropedicae, as is the case in other Proteobacteria16,31. To balance respiratory requirements under oxygen-limiting conditions, Fnr and its orthologs commonly participate in negative regulation of the expression of other terminal oxidases, for example, the bd-type and bo3-type oxidases in E. coli and A. vinelandii32,33 and the bo3–type and CIO oxidases in Pseudomonas putida34. However, our transcriptomics data indicate that this is not the case in H. seropedicae. We only observe differential expression of the genes encoding the cbb3-type oxidase. Expression of the other terminal oxidases is not apparently affected by absence of the three H. seropedicae Fnr proteins. However, in contrast to other well-studied systems, the Fnr proteins in H. seropedicae appear to have a major influence on the composition of the complete electron transport chain that feeds electrons from NADH, through the ubiquinone pool to the cytochrome bc1 complex and onto the c-type cytochromes that are substrates for the heme-copper oxidases. This is clearly demonstrated by the depletion of c-type cytochromes and the down regulation of genes encoding the various components of this branch of the electron transport chain in the triple fnr mutant. Hence it would appear that the Fnr proteins play a major role in regulating the configuration of the H. seropedicae electron transport chain in order to exploit respiratory flexibility and optimize energy coupling in response to oxygen availability.
The cbb3-type heme copper oxidase is likely to be required for growth at very low (<0.5%) oxygen concentrations35 and can support symbiotic nitrogen fixation at nanomolar levels of dissolved oxygen (reviewed in 36). Nevertheless, the fixN insertion mutant of H. seropedicae RAM21 was competent to support nitrogenase activity under oxygen-limiting conditions implying that the cbb3-type oxidase is not required to support nitrogen fixation in this organism. Perhaps this result is not surprising, given that, to our knowledge, this oxidase is not required for nitrogen fixation in other free-living diazotrophs. By analogy with other nitrogen-fixing bacteria, it is possible that the bd-type oxidase supports nitrogenase activity in H. seropedicae. This oxidase is critical for microaerobic diazotrophy in Klebsiella pneumoniae37, it provides respiratory protection for nitrogenase in Azotobacter vinelandii38,39 and it is utilized as a terminal oxidase to support symbiotic nitrogen fixation in Azorhizobium caulinodans35. In contrast, the triple fnr deletion strain was compromised with respect to both nitrogenase activity and diazotrophic growth, which presumably reflects the major role played by Fnr in reconfiguring the electron transport chain under oxygen limiting conditions in H. seropedicae.
The results presented here do not provide a rationale for the existence of multiple Fnr proteins in H. seropedicae. Potentially, each ortholog may exhibit differential sensitivity to oxygen, recognize different DNA targets or have different propensities to dimerise under aerobic conditions. The transcript profiling reveals that expression of fnr3 is constitutive, whereas fnr1 and fnr2 expression is apparently positively controlled by one or more of the Fnr orthologs. The fnr1 gene, which is located close to the genes required for the maturation and activity of the cbb3-type oxidase, appears to play a critical role in regulating transcription of the fixNOP and hemN-Hsero_3206-fixIS operons and in controlling the expression of c-type cytochromes. Although, fnr3 also appears to be required to express the cbb3-oxidase, we cannot rule out the possibility that it is required to activate transcription of fnr1. In contrast, fnr2 does not appear to be required, either for expression of this oxidase or c-type cytochromes, under the experimental conditions employed here. Further detailed characterization of the three Fnr paralogs will be necessary in order distinguish their precise roles in gene regulation in H. seropedicae.
Methods
Bacterial strains and plasmids
H. seropedicae and E. coli strains and plasmids used are listed in Supplementary Table S1.
Growth conditions
E. coli strains were grown at 37°C in LB medium40. H. seropedicae strains were grown at 30°C in NFbHP-Malate medium41 supplemented with NH4Cl or 0.5 mM sodium glutamate. Appropriate antibiotics were used when required. For experiments requiring different oxygen concentrations, the air in the gas phase of Suba Seal® stoppered culture flasks was exchanged by injecting argon into the flasks for 30 minutes. To obtain different oxygen levels a given volume of air was injected back into the flask. The oxygen levels in the gas phase were verified by gas chromatography using a molecular sieve column and a TCD detector.
Identification and Analysis of H. seropedicae Fnr Orthologs
The sequences of Fnr1, Fnr2 and Fnr3 from H. seropedicae SmR1 were aligned with E. coli K12 substr. MG1655 Fnr (Ref seq: NP_415850.1) using Muscle software42. The presence of conserved domains in the Fnr homologs was investigated by submitting sequence search to the Pfam database (http://pfam.sanger.ac.uk/)43.
Phylogenetic Analysis
Amino acid sequence retrieval was performed by using a BLASTP search44 against the nonredundant NCBI database. The proteins used as queries were Fnr1, Fnr2 and Fnr3 from H. seropedicae, (Ref seq: YP_003776587.1, YP_003775788.1 and YP_003775945.1, respectively) FixK1 and FixK2 from B. japonicum (Ref seq: NP_772701.1 and NP_769397.1, respectively), Fnr from E.coli K12 substr. MG1655 (Ref seq: NP_415850.1) and CydR from A. vinelandii (Ref seq: YP_002799173.1). A limited number of sequences were selected, eliminating redundant information, while maintaining representative taxonomic diversity. All sequences selected (Supplementary Table S2) were checked to have the CRP-FNR superfamily protein signatures, CNMP_Binding _3 PS50042 and HTH_CRP_2 PS51063, proposed by PROSITE45. For phylogenetic tree reconstruction, an amino acid alignment was made using Muscle42. Maximum likelihood (ML) trees were derived using the JTT matrix-based model46 after bootstrapping 1,000 replicates of each original data set47 using the MEGA 5.05 software48.
Construction of H. seropedicae SmR1 fnr deletion and fixN insertional mutant strains
An allelic exchange strategy was used to generate derivatives of H. seropedicae SmR1 containing fnr orthologs deletions (Supplementary Fig. S2) and a tetracycline resistance cassette insertion into fixN (Supplementary Fig. S12). The primers used in this work are show in Supplementary Table S3. For construction of the allele exchange plasmids for fnr mutation the upstream and downstream regions of fnr1, fnr2 and fnr3 were amplified by PCR. These fragments were then ligated to generate fnr deletions, which were cloned into HindIII and BamHI sites of the suicide plasmid pSUP202, to generate pMBB1D, pMBB2D and pMBB3D (Supplementary Table S1). The nptI-sacB-sacR cartridge (from pMH1701) was then inserted into BamHI site. We generated three suicide plasmids: pMBB1DS for the 279 bp deletion of fnr1, pMBB2DS for the 276 bp deletion of fnr2 and pMBB3DS for the 267 bp deletion of fnr3 (Supplementary Table S1).
Conjugation was performed between E. coli S17.1 containing the plasmid of interest and H. seropedicae recipient strains. Conjugation was performed on NFbHP-Malate/LA (3:1) agar by mixing recipient and donor strains in two proportions (50/1 and 10/1). Transconjugants were selected on NFbHP-Malate agar supplemented with 20 mM NH4Cl and antibiotics. One mutant strain resulting from single cross-over was grown overnight in liquid NFbHP-Malate plus 20 mM NH4Cl without antibiotics at 30°C. After incubation 250 μL of the culture were plated on NFbHP-Malate agar, supplemented with 20 mM NH4Cl, 5% sucrose, 5 μg/mL nalidixic acid and 80 μg/mL streptomycin. Sucrose is toxic to bacteria that express the sacB gene, therefore only strains that lost the sacRB-KmR cassette by a second homologous recombination event could grow under these conditions. The mutant strains were analysed by PCR using primers (Supplementary Table S3) external to fnr1, fnr2 and fnr3. To construct double and triple H. seropedicae mutant strains the process described above was repeated using different allele exchange plasmids.
For fixN mutagenesis, the plasmid pHS17058H11 (Genopar Consortium)4 containing the fixN gene was subject to a transposition reaction using the EZ-Tn5 <TET-1> Insertion Kit (Epicentre). After confirmation of the TET-1 insertion into the fixN gene (pHS17058H11Tc), a cloramphenicol cassette from pTnMod-OCm plasposon was inserted into the plasmid, outside the fixN gene, to yield the plasmid pHS17058H11TcCm, which was then electro-transformed into H. seropedicae SmR1. A transconjugant resistant to tetracycline and sensitive to cloramphenicol was selected and named RAM21. The double recombinant was confirmed by DNA hybridization (Supplementary Fig. S12).
RNA isolation and RNAseq library construction
For total RNA extraction, we grew H. seropedicae SmR1 (wild-type) and MB231 strains (triple fnr mutant) under aerobic conditions to an optical density of 0.4 (cultures were shaken at 120 rpm in air) and then the cultures were switched to microaerobic conditions (initial oxygen concentration 2%) for 1.5 hours. After collection of the cells by centrifugation, the total RNA was isolated using the RNA RiboPure™-Bacteria kit (Ambion). Two rounds of enrichment were performed using MICROBExpress™ kit (Ambion) for removing ribosomal RNA from total RNA samples. Approximately 200 to 500 ng of mRNA enriched RNA were used for cDNA synthesis and library construction using the SOLiD™ Whole Transcriptome Analysis Kit (Life Technologies). The libraries were amplified by emulsion PCR using SOLiD ePCR kit and sequenced in a SOLiD 4 System (Life Technologies).
Read Mapping, Differential Expression Analysis and Fnr Binding Site Prediction
The reads were mapped against the H. seropedicae SmR1 genome (NC_014323) as reference using the CLC Genomics Workbench package. Read counts table was exported into the RobiNA software49 and both normalization and statistical evaluation of differential gene expression was performed by DESeq50 with a p-value cut-off of 0.05 using the Benjamini-Hochberg method for multiple testing correction51. Genes with fold change lower than three were excluded from the analysis. Genes with fold change lower than 3 or p-value slightly higher than 0.05 were included in the analysis when genome context indicated they are part of operon with genes differentially expressed according to the above parameters.
Potential Fnr-binding sites were located using the sequence motif search facilities of PEPPER52 (http://pepper.molgenrug.nl/index.php) and Virtual Footprinting53 (http://prodoric.tu-bs.de/vfp/vfp_promoter.php) using default search parameters.
Construction of transcriptional fusions
To study the expression of fixNOP and hemN-Hsero_3206-fixIS operons, the putative promoter regions were amplified by PCR, cloned into pTZ57R/T and then subcloned into PstI and BglII sites of pPW452 to generate the plasmids pPWPFN (fixN::lacZ fusion) and pPWPHN (hemN::lacZ fusion) (Supplementary Table S1). All constructs were verified by DNA sequencing.
β-Galactosidase activity
To analyse the activities of fixN::lacZ and hemN::lacZ fusions, strains were grown to an O.D 600 = 0.3, collected and then resuspended in NFbHP Malate liquid medium supplemented with 20 mM NH4Cl to an O.D600 = 0.05. The cells were incubated for 3 hours either under air or at initial concentrations of 2, 4 or 6% oxygen. After incubation samples were taken for β-galactosidase activity determination as described54. The results are expressed in Miller units (MU).
Acetylene reduction assay
The acetylene-reduction assay was used to determine nitrogenase activity on free-living cultures55,56. Ethylene formation was determined either by using a Varian Star 3400 CX gas chromatograph equipped with a Porapak N column.
Freshly grown cultures were used for inoculating NFbHP-Malate semi solid medium (0.17% agar) containing 0.5 mM sodium glutamate followed by eighteen hours incubation at 30°C. Acetylene (10%) was injected and after incubation for one hour at 30°C, 0.5 mL samples were collected for determination of produced ethylene by gas chromatography. The same procedure was used for assaying nitrogenase activity in liquid media, except that the cultures were collected by centrifugation (3220 g, for 4 min at room temperature), re-suspended in NFbHP-Malate supplemented with 0.5 mM of NH4Cl to an O.D of 0.2. The cells were incubated at 5.0% initial oxygen concentration in the gas phase as described. Nitrogenase activity is reported as nmol of C2H4 produced per minute per mg protein. Whole cell protein concentration was determined by the Bradford method57 after overnight lysis with 0.2 mM NaOH.
Reduced minus Oxidized Spectra and Heme Stain
For preparation of protein extracts H. seropedicae strains were cultured in 250 mL erlenmeyer flasks containing 50 mL of NFbHP-Malate supplemented with 20 mM NH4Cl. After overnight incubation, the cells were collected by centrifugation and re-suspended in 2 mL of sonication buffer (100 mM NaCl and 50 mM Tris.HCl pH 7.5). Cells were broken by sonication and cell debris was separated from the protein extract by centrifugation (6000 g, 30 min). The supernatant was collected and used for further analysis.
UV–visible difference spectra of 0.3 mg.mL−1 protein extracts were recorded in 1 cm path-length quartz cuvettes at room temperature on a Shimadzu UV-2501 PC spectrophotometer. Reduced-minus-oxidized difference spectra of the protein fractions were recorded by measuring the dithionite-reduced spectrum of the sample against the air-oxidized one.
For the covalently bound heme stain, 50 μg of the protein extract, prepared as described above, was loaded without boiling onto a 12% Tris-Tricine SDS-PAGE gel58 and stained using o-dianisidine59.
References
Baldani, J. I., Baldani, V. L. D., Seldin, L. & Dobereiner, J. Characterization of Herbaspirillum seropedicae gen. nov., sp. nov., a root-associated nitrogen fixing bacterium. Int. J. Sys. Bact. 36, 86–93 (1986).
Olivares, F. L., Baldani, V. L. D., Reis, V. M., Baldani, J. I. & Döbereiner, J. Occurrence of the endophytic diazotrophs Herbaspirillum spp. in roots, stems and leaves predominantly of Gramineae. Biol. Fertil. Soils. 21, 197–200 (1996).
James, E. K. & Olivares, F. L. Infection and colonization of sugar cane and other graminaceous plants by endophytic diazotrophs. Crit. Rev. Plan. Sci. 17, 77–119 (1998).
Pedrosa, F. O. et al. Genome of Herbaspirillum seropedicae strain SmR1, a specialized diazotrophic endophyte of tropical grasses. PLoS Genet. 7, http://dx.doi.org/10.1371/journal.pgen.1002064 (2011).
Körner, H., Sofia, H. J. & Zumft, W. G. Phylogeny of the bacterial superfamily of Crp-Fnr transcripitional regulators, exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol. Rev. 27, 559–592 (2003).
Kiley, P. J. & Beinert, H. Oxygen sensing by the global regulator FNR: the role of the iron-sulfur cluster. FEMS Microbiol. Rev. 22, 341–352 (1999).
Lazazzera, B. A., Beinert, H., Khoroshilova, N., Kennedy, M. C. & Kiley, P. J. DNA-binding and dimerization of the Fe-S containing FNR protein from Escherichia coli are regulated by oxygen. J. Biol. Chem. 271, 2762–2768 (1996).
Scott, C., Partridge, J. D., Stephenson, J. R. & Green, J. DNA target sequence and FNR-dependent gene expression. FEBS Lett. 541, 97–101 (2003).
Grabbe, R., Klopprogge, K. & Schmitz, R. A. Fnr is required for NifL-dependent oxygen control of nif gene expression in Klebsiella pneumoniae. J. Bacteriol. 183, 1385–1393 (2001).
Gutiérrez, D., Hernando, Y., Palacios, J. M., Imperial, J. E. & Ruiz-Argueso, T. FnrN controls symbiotic nitrogen fixation and hydrogenase activities in Rhizobium leguminosarum biovar viciae UPM791. J. Bacteriol. 179, 5264–5270 (1997).
Bobik, C., Meilhoc, E. & Batut, J. FixJ, a major regulator of the oxygen limitation response and late symbiotic functions of Sinorhizobium meliloti. J. Bacteriol. 188, 4890–4902 (2006).
Mesa, S. et al. Comprehensive assessment of the regulons controlled by the FixLJ-FixK2-FixK1 cascade in Bradyrhizobium japonicum. J. Bacteriol. 190, 6568–6579 (2008).
Nellen-Anthamatten, D. et al. Bradyrhizobium japonicum FixK2, a crucial distributor in the FixLJ-dependent regulatory cascade for control of genes inducible by low oxygen levels. J. Bacteriol. 180, 5251–5255 (1998).
Mesa, S., Hennecke, H. & Fischer, H.-M. A multitude of CRP/FNR-like transcription proteins in Bradyrhizobium japonicum. Biochem. Soc. Trans. 34, 156–159 (2006).
Tatusov, R. L. et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4, http://dx.doi.org/10.1186/1471-2105-4-41 (2003).
Cosseau, C. & Batut, J. Genomics of the ccoNOQP-encoded cbb3 oxidase complex in bacteria. Arch. Microbiol. 181, 89–96 (2004).
Koch, H. G., Winterstein, C., Saribas, A. S., Alben, J. O. & Daldal, F. Roles of the ccoGHIS gene products in the biogenesis of the cbb3-type cytochrome c oxidase. J. Mol. Biol. 297, 49–65 (2000).
Lieb, C., Siddiqui, R. A., Hippler, B., Jahn, D. & Friedrich, B. The Alcaligenes eutrophus hemN gene encoding the oxygen independent coproporphyrinogen III oxidase, is required for heme biosynthesis during anaerobic growth. Arch. Microbiol. 169, 52–60 (1998).
Chang, H. Y. et al. The diheme cytochrome c4 from Vibrio cholerae is a natural electron donor to the respiratory cbb3 oxygen reductase. Biochemistry 49, 7494–7503 (2010).
Goddard, A. D. et al. Comparing the substrate specificities of cytochrome c biogenesis Systems I and II. FEBS Journal 277, 726–737 (2010).
Simon, J. & Hederstedt, L. Composition and function of cytochrome c biogenesis System II. FEBS Journal 278, 4179–4188 (2011).
Crooke, H. & Cole, J. The biogenesis of c-type cytochromes in Escherichia coli requires a membrane-bound protein, DipZ, with a protein disulphide isomerase-like domain. Mol. Microbiol. 15, 1139–1150 (1995).
Yang, C. H., Azad, H. R. & Cooksey, D. A. A chromosomal locus required for copper resistance, competitive fitness and cytochrome c biogenesis in Pseudomonas fluorescens. Proc. Natl. Acad. Sci. USA 93, 7315–7320 (1996).
Meng, W., Green, J. & Guest, J. R. FNR-dependent repression of ndh gene expression requires two upstream FNR-binding sites. Microbiol. 143, 1521–1532 (1997).
Spiro, S., Roberts, R. E. & Guest, R. FNR-dependent repression of the ndh gene of Escherichia coli and metal ion requirement for FNR regulated gene expression. Mol. Microbiol. 3, 601–608 (1989).
Koch, H. G., Hwang, O. & Daldal, F. Isolation and characterization of Rhodobacter capsulatus mutants affected in cytochrome cbb3 oxidase activity. J. Bacteriol. 180, 969–978 (1998).
Zufferey, R., Preisig, O., Hennecke, H. & Thony-Meyer, L. Assembly and function of the cytochrome cbb3 oxidase subunits in Bradyrhizobium japonicum. J. Biol. Chem. 271, 9114–9119 (1996).
Batut, J. et al. fixK, a gene homologous with fnr and crp from Escherichia coli, regulates nitrogen fixation genes both positively and negatively in Rhizobium meliloti. EMBO J. 8, 1279–1286 (1989).
Mandon, K., Kaminski, P. A. & Elmerich, C. Functional analysis of the fixNOQP region of Azorhizobium caulinodans. J. Bacteriol. 176, 2560–2568 (1994).
Preisig, O., Anthamatten, D. & Hennecke, H. Genes for a microaerobically induced oxidase complex in Bradyrhizobium japonicum are essential for a nitrogen-fixing endosymbiosis. Proc. Natl. Acad. Sci. USA 90, 3309–3313 (1993).
Batut, J. & Boistard, P. Oxygen control in Rhizobium. Antonie van Leeuwenhoek 66, 129–150 (1994).
Cotter, P. A., Chepuri, V., Gennis, R. B. & Gunsalus, R. P. Cytochrome o (cyoABCDE) and d (cydAB) oxidase gene expression in escherichia coli is regulated by oxygen, ph and the fnr gene product. J. Bacteriol. 172, 6333–6338 (1990).
Wu, G. et al. Regulation of cytochrome bd expression in the obligate aerobe Azotobacter vinelandii by CydR (Fnr), sensitivity to oxygen, reactive oxygen species and nitric oxide. J. Biol. Chem. 275, 4679–4686 (2000).
Ugidos, A., Morales, G., Rial, E., Williams, H. D. & Rojo, F. The coordinate regulation of multiple terminal oxidases by the Pseudomonas putida ANR global regulator. Environ. Microbiol. 10, 1690–1702 (2008).
Kaminski, P. A., Kitts, C. L., Zimmerman, Z. & Ludwig, R. A. Azorhizobium caulinodans uses both cytochrome bd (quinol) and cytochrome cbb3 (cytochrome c) terminal oxidases for symbiotic N2 fixation. J. Bacteriol. 178, 5989–5994 (1996).
Marchal, K. & Vanderleyden, J. The “oxygen paradox” of dinitrogen-fixing bacteria. Biol. Fertil. Soils 30, 363–373 (2000).
Hill, S., Viollet, S., Smith, A. T. & Anthony, C. Roles for enteric d-type cytochrome oxidase in N2 fixation and microaerobiosis. J. Bacteriol. 172, 2071–2078 (1990).
Kelly, M. J. S., Poole, R. K., Yates, M. G. & Kennedy, C. Cloning and mutagenesis of genes encoding the cytochrome bd terminal oxidase complex in Azotobacter vinelandii mutants deficient in the cytochrome d complex are unable to fix nitrogen in air. J. Bacteriol. 172, 6010–6019 (1990).
Poole, R. K. & Hill, S. Respiratory protection of nitrogenase activity in Azotobacter vinelandii-roles of the terminal oxidases. Biosci. Rep. 17, 303–317 (1997).
Sambrook, J., Fritsch, E. F. & Maniatis, T. Molecular Cloning - a laboratory manual. Cold Spring Harbor, New York, Cold Spring Harbor Laboratory Press (1989).
Klassen, G., Pedrosa, F. O., Souza, E. M., Funayama, S. & Rigo, L. U. Effect of nitrogen compounds on nitrogenase activity in Herbaspirillum seropedicae SmR1. Can. J. Microbiol. 43, 887–891 (1997).
Edgar, R. C. MUSCLE, multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004).
Finn, R. D. et al. The Pfam families database. Nucleic Acids Res. 38, D211 (2010).
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST, a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
Sigrist, C. J. A. et al. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 38, D161 (2010).
Jones, D. T., Taylor, W. R. & Thornton, J. M. The rapid generation of mutation data matrices from protein sequences. Comput. App. Biosci. 8, 275–282 (1992).
Dopazo, J. Estimating errors and confidence intervals for branch lengths in phylogenetic trees by a bootstrap approach. J. Mol. Evol. 38, 300–304 (1994).
Tamura, K. et al. MEGA5, Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739 (2011).
Lohse, M. et al.RobiNA, a user-friendly, integrated software solution for RNA-Seq-based transcriptomics. Nucleic Acids Res. 40, W622 (2012).
Anders, S. & Huber, W. Differential expression analysis for sequence count data. Genome Biology 11, R106 http://dx.doi.org/10.1186/gb-2010-11-10-r106 (2010).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate, a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) 57, 289–300 (1995).
de Jong, A., Pietersma, H., Cordes, M., Kuipers, O. P. & Kok, J. PePPER: a webserver for prediction of prokaryote promoter elements and regulons. BMC Genomics 14, 299 http://dx.doi.org/10.1186/1471-2164-13-299 (2012).
Munch, R. et al. Virtual Footprint and PRODORIC: an integrative framework for regulon prediction in prokaryotes. Bioinformatics 21, 4187–4189 (2005).
Miller, J. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory, NY (1972).
Dilworth, M. J. Acetylene reduction by nitrogen-fixing preparations from Clostridium pasteurianum. Biochim. Biophys. Acta. 127, 285–294 (1966).
Schöllhorn, R. & Burris, R. H. Acetylene as a competitive inhibitor of N2 fixation. Proc. Natl. Acad. Sci. USA 58, 213–216 (1967).
Bradford, M. M. A rapid and sensitive method for the quantification of microgram quantities of protein utilization, the principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976).
Schägger, H. & von Jagow, G. V. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368–379 (1987).
Francis, R. T. & Becker, R. R. Specific indication of hemoproteins in polyacrylamide gels using a double-staining process. Anal. Biochem. 36, 509–514 (1984).
Acknowledgements
This work was supported by the Brazilian Program of National Institutes of Science and Technology-INCT/Brazilian Research Council-CNPq/MCT, Fundacão Araucária and CAPES. We would like to thank Leonardo Magalhães Cruz for help with the use of MEGA 5.05 software. We are also thankful to Roseli A. Prado, Julieta Pie, Marilza D. Lamour and Valter A. de Baura for technical assistance.
Author information
Authors and Affiliations
Contributions
M.B.B. conceived the work, designed and carried out experiments, analyzed the data and wrote the paper; M.Z.T.S. and H.F. carried out the construction and sequencing of RNA-seq library; R.W., M.B.R.S. and F.P. conceived the work and supervised the study; E.M.S., R.D. and R.A.M. conceived the work, supervised the study, designed experiments, analyzed the data and wrote the paper. All authors approved the final manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Information
Supplementary Information
Supplementary Dataset1
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Batista, M., Sfeir, M., Faoro, H. et al. The Herbaspirillum seropedicae SmR1 Fnr orthologs controls the cytochrome composition of the electron transport chain. Sci Rep 3, 2544 (2013). https://doi.org/10.1038/srep02544
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep02544
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.