Abstract
Breast cancer transcriptome acquires a myriad of regulation changes and splicing is critical for the cell to “tailor-make” specific functional transcripts. We systematically revealed splicing signatures of the three most common types of breast tumors using RNA sequencing: TNBC, non-TNBC and HER2-positive breast cancer. We discovered subtype specific differentially spliced genes and splice isoforms not previously recognized in human transcriptome. Further, we showed that exon skip and intron retention are predominant splice events in breast cancer. In addition, we found that differential expression of primary transcripts and promoter switching are significantly deregulated in breast cancer compared to normal breast. We validated the presence of novel hybrid isoforms of critical molecules like CDK4, LARP1, ADD3 and PHLPP2. Our study provides the first comprehensive portrait of transcriptional and splicing signatures specific to breast cancer sub-types, as well as previously unknown transcripts that prompt the need for complete annotation of tissue and disease specific transcriptome.
Similar content being viewed by others
Introduction
The process of breast cancer progression is accompanied by genomic alterations including inherited genetic variations, acquired genomic aberrations, changes in the splicing and transcriptome and resulting protein functions1,2,3. The recent transcriptome profiling studies highlighted the diversity and flexibility of genomic processes that allow a cancer cell to “tailor-make” specific functional units from the available exons of the gene4,5,6,7. The process of generating novel cancer-specific isoforms is driven by alterations at several layers such as alternative pre-RNAs, promoter usage and splicing and polyadenylation that alters coding regions and consequently, the function of the resulting proteins8,9,10. Therefore, understanding these regulatory elements is essential for a complete appreciation of the genomic contribution to the pathobiology of breast cancer. The relevance of differential splicing in human cancer is an evolving area of cancer biology. The complete annotation of all the transcripts associated with each cancer relevant gene in the human genome is still far from complete11,12,13. Consequently, distinguishing the isoforms that are generated due to natural transcriptomic dynamics from the ones that occur because of diseases such as cancer remains a great challenge. In addition, determining tissue-specific splice variants will be equally important for a better understanding of cancer specific splicing of genes14,15,16,17.
In breast cancer, Tenascin C (TNC) was one of the first genes identified to comprise an alternatively spliced region that induces focal adhesion and cell migration in stromal fibroblasts, periductal fibroblasts and residual myoepithelial cells18,19. Further microarray and qRT-PCR based studies reported genes including CD44, ESR1, ESR2, CALD1, COL6A3, LRRFIP2, PIK4CB and TPM1, that produce breast cancer specific splice-variants20. Interestingly, an overall up-regulation of splicing factors and remarkable changes in the exon models are widely observed in breast cancer21. Recent studies have identified specific variants of TP53, SYK, BRCA1 and MUC1 in breast cancer22,23,24,25,26,27, as well as splice variants of many genes28,29,30,31 that play important functional roles in tumor progression. Although focused studies on differential splicing of specific genes and microarray studies allowed us to identify many exons that undergo alterations in cancer20,21,22, the emerging RNA sequencing offers unprecedented approach to discover cancer specific isoforms, global transcriptomic alterations and post-transcriptional changes on large scale.
Here, we reveal the transcriptomic landscape of TNBC, non-TNBC and HER2-positive breast cancer in comparison to normal breast samples using massively parallel paired-end RNA sequencing. We determine the differentially spliced genes and resulting isoforms, differential promoter usage and expression of pre-RNA and coding regions between the normal and cancer breast tissues. More interestingly, breast cancer associated novel splice events and core sets of novel genes modified at the pre-RNA and splicing levels are also revealed. Together, this study provides the first comprehensive portrait of pre- and post- transcriptional changes and splicing signatures that are specific to TNBC, non-TNBC and HER2-positive breast cancers.
Results
Strategy to identify the pre- and post-transcriptional elements that underlie the transcriptomic diversity
The overview of all the analysis performed in this study is outlined in Figure 1A. We set out to define the significance of splicing in breast cancer subtypes using RNA-sequencing of TNBC, non-TNBC, HER2-positive breast cancers in comparison to human breast organoids (epithelium) samples derived from normal healthy women32 (mentioned throughout the manuscript as normal breast samples or NBS). The global statistics on the reads is presented in Supplemental Table 1. The 17 well-characterized individual human breast cancer tissues include six TNBC, six non-TNBC and five HER2-positive breast cancer samples5. The RNA sequencing of the samples was performed using the Illumina platform as outlined in our recent study5. We first mapped the reads to the Ensembl GRCh37.62 B (hg19) reference genome using RNA sequence aligner Tophat that aligns the reads across splice junctions independently of gene annotations33.
TNBC, non-TNBC and HER2-positive breast cancer RNA sequencing.
(A) Overview of the steps involved in the splicing and transcriptional regulatory elements that are specific to TNBC, non-TNBC and HER2-positive breast cancers in comparison to NBS using de novo assembled transcripts from RNA sequencing. (B) Total read distribution between NBS and cancer – higher proportion of intergenic reads is found in the cancer as compared to NBS. (C) Relative distribution of novel, identical to reference and mapping into the reference transcripts in the four studied groups. (D) Overlap of the novel isoforms between the three breast cancer subtypes.
The reference independent transcript reconstruction was performed using Cufflinks34,35,36. The isoforms identical to the Ensembl GRCh37.62 B reference genome (known isoforms) and the ones that comprise at least one novel splice junction (novel isoforms), are detected using cuffcompare program37. Once isoforms are isolated from all the assembled transfrags, we employed cuffdiff program to identify the differentially spliced genes between individual breast cancer subtypes against normal breast samples. Further, comparative analysis at the level of primary transcripts, promoter usage, spliced isoforms and coding regions provided a comprehensive overview of the transcriptional and post-transcriptional elements in breast cancer. To detect the TNBC, non-TNBC and HER2-positive breast cancer-specific splice events such as exon skip, exon inclusion, transcript start and termination and intron retention, we employed a direct exon model comparison analysis as well as multivariate analysis38.
De novo assembly of transcripts reveals high ratio of novel isoforms
The number of the de novo assembled transcripts and the corresponding genes in each sample and group are presented in Table 1. Examination of the read distribution and the reconstructed transcripts revealed several important observations. First, while about 73% of the NBS reads map into exons, this percent is only 58% in the breast cancer samples (Figure 1B). Second, in both cancer and NBS, higher proportion of novel, as compared to reference isoforms was estimated (Figure 1C). Third, among the novel isoforms that map within the reference, the breast cancer groups encompass a high percentage of novel junctions than the normal breast samples (Figure 1C). Fourth, the majority of the genes with novel isoforms appear to be identical between TNBC, non-TNBC and HER2-positive groups (Figure 1D, the reference-like isoforms and the overlap with NBS are shown on Supplemental Figure 1). Finally, the unsupervised clustering revealed that the NBS clustered together but distant to the breast cancer groups as expected (Supplemental Figure 2A and 2B). To eliminate the partially assembled transcripts and to focus on defining the splice signature of breast cancer, we restricted our further analysis to the isoforms that are similar to reference and novel isoforms with more than two exons.
Identification of the differentially spliced genes in breast cancer
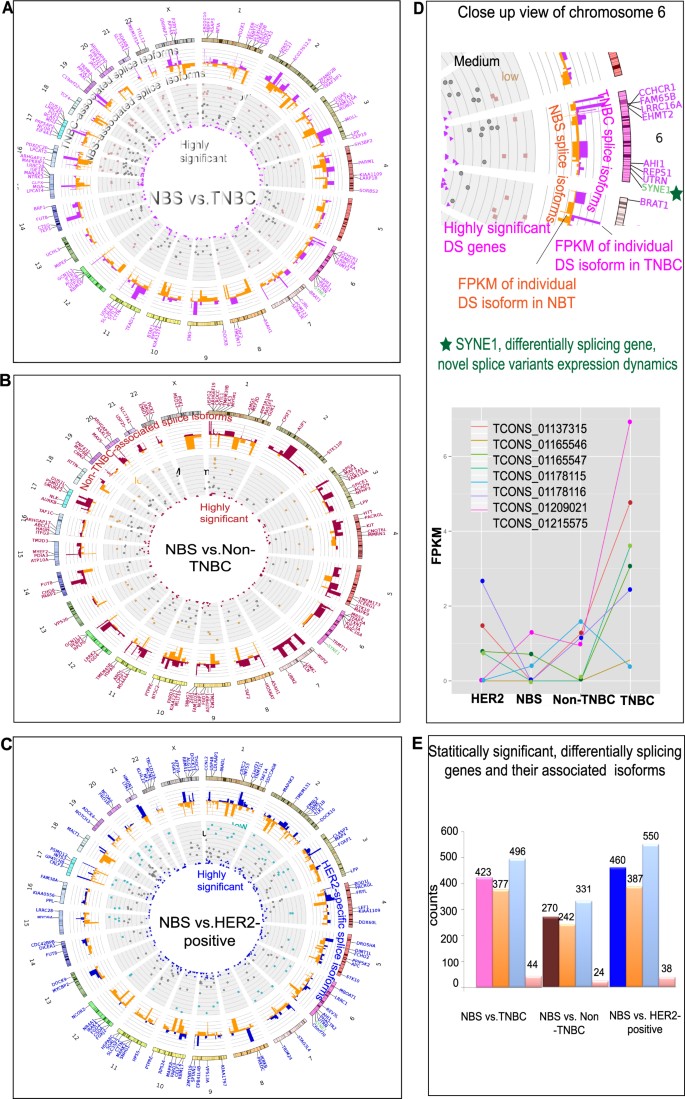
To identify the differences in splice ratios between the NBS and the different breast cancer subtypes, we employed Cuffdiff37, which calculates the changes in the relative splice abundances by quantifying the square root of the Jensen-Shannon divergence on all the primary transcripts that produce two or more isoforms (Supplemental Figure 3). It is essential to note that the distributions of genes, the primary transcripts and isoform FPKM are comparable between the samples that are taken for the differential splicing test (Supplemental Figures 2–5). When the NBS were compared against TNBC-subtype, 423 primary transcripts belonging to 377 genes, generating 496 novel isoforms, were found to be differentially spliced with the FDR and corrected p-value less than 0.05 (Figure 2A and 2E, Supplemental File 1). Similarly, comparisons of NBS against non-TNBC and HER2-positive breast cancers allowed us to identify 270 and 460 primary transcripts belonging to 242 and 387 differentially splicing genes, producing 331 and 550 novel isoforms, respectively (Figure 2B, 2C and 2E, Supplemental Figure 6, Supplemental Files 2 and 3). An example of differentially spliced gene is shown on Figure 2D, illustrating the different SYNE2 isoforms identified in NBS, TNBC, non-TNBC and HER2 positive samples. We discovered 39 genes (including, TAF2, PRKDC, PGK1, CHD8, TFAP2A and STK10, Supplemental File 4) that show statistically significant differential splicing in all the three breast cancer subtypes. When a similar differential splicing test was performed within the cancer groups, only few genes were differentially spliced among samples, indicating the similarity among the breast cancer subtypes and distinctiveness between normal breast and cancer (Supplemental Figure 7, Supplemental Files 5–7).
Differentially spliced genes and their associated isoforms between NBS and TNBC, non-TNBC and HER2-positive breast cancers.
(A-C) Circos plots representing the statistically significant, differentially spliced genes identified through pairwise comparisons of TNBC vs. NBS (A), non-TNBC vs. NBS (B) and HER2-positive vs. NBS (C). The genes shown as dots are coloured based on their Jensen-Shannon divergence test q value. The stacked histograms represent the abundance (FPKM) of specific differentially spliced isoforms that results from the primary transcripts. (D) Close view of chromosome 6 segment of TNBC vs. NBS comparison of differentially splicing genes, exemplified through SYNE1 novel splice variant expression dynamics shown as a line graph. Tcon numbers indicate the reassembled, distinct novel exon models of SYNE1. (E) Statistically significant differentially splicing genes and their associated isoforms – comparison with NBT.
Unraveling the splice signatures of TNBC, non-TNBC and HER2-positive breast cancers
We next investigated the differentially spliced (p-value<0.05, FDR<0.05) novel isoforms in the context of the same transcription start site (TSS) in the breast cancer subtypes by Jensen-Shannon divergence statistical test. The top 20 exclusively expressed in each cancer subtype isoforms are shown on Figure 3. These isoforms are sorted based on their preferential expression in one of the cancer subtypes versus very low or absent in the other two; all these transcripts were not detected in NBS. The distribution of the differentially spliced transcripts for the three cancer types is shown on Supplemental Figure 8; from them, 322, 246 and 368 isoforms are almost exclusively expressed in TNBC, non-TNBC and HER2-positive breast cancer samples, respectively and are not present in NBS (Supplemental Files 8–10). The majority of these cancer subtype specific isoforms comprise novel junctions and thus represent novel isoforms that have not been reported before (Supplemental Figure 8 shows the top twenty highly abundant isoforms that are not expressed in NBS and Supplemental File 11 (GTF) shows the exon models). The isoforms expressed exclusively in a cancer subtype and not present at all (FPKM = 0) in the other subtypes are presented in Supplemental Files 12–14.
TNBC, non-TNBC and HER2-positive breast cancers specific isoforms.
Heat map showing the top twenty novel junctions differentially expressed isoforms among the three cancer subtypes. Red color indicates high expression levels and green color indicates low expression levels. The top twenty are selected based on their abundance (FPKM values shown on the heat map) in TNBC (A), non-TNBC (B) and HER2-positive (C). The gene names are shown on the right and the closest emsembl transcript identifier is presented on the left of each heat map.
Other cancer specific isoforms included genes critical for cellular functions such as MTOR and MSI2 in the TNBC group, ZMYND19 and SEPT8 in non-TNBC and PRKDC and DIM1 in HER2-positive group (see Supplemental Files 8–10). We further evaluated whether the identified cancer specific isoforms comprise a functional open reading frame (ORF) by aligning the RNA of the novel isoforms against the human ORFeome 8.1 (http://horfdb.dfci.harvard.edu/). Notably, 18% (TNBC), 23% (non-TNBC) and 17% (HER2-positive) of the novel isoforms appear to comprise a fully functional open reading frame (Supplemental Table 2) revealing the possibility of expressed novel cancer specific proteins.
To experimentally validate the differential expression of the novel subtype specific isoforms, we selected novel isoforms exclusively expressed in each cancer group: EIF4EBP1 and MRPS15 for TNBC; NDUFA, PBX1 and MUTS1 for non-TNBC and AZIN, VAMP5 and ATP5G1 for HER2-positive group. Primers that bind to unique regions of these isoforms were designed (Supplemental File 15) and the qRT-PCR analysis and sequencing of the products was carried out. The expression levels detected by qRT-PCR were similar to the ones revealed through transcriptome sequencing and were higher in the cancer samples compared to low or absent in the NBS (Figure 4).
Significance of the primary transcript abundances in generation of cancer sub-type specific isoforms
Aberrant splicing and cancer specific splice variants represent emerging cancer biomarkers39,40. However, the fine-tuning of splicing cascade occurs at the level of primary transcript and promoter selection. Although the RNA sequencing of normal and cancer breast tissue captures the snap shot of a post-transcriptional state, the number and abundance of primary transcripts associated with a given gene can be derived from the sum of the abundances of the transcripts that share the same TSS. The changes in the relative abundance of the TSSs between NBS and TNBC, non-TNBC and HER2-positive breast cancers provides a list of cancer subtype-specific cellular choices at transcription regulation level (Figure 5A).
Identification of differential primary transcripts, promoter usage and promoter switch in breast cancers.
(A) The volcano plots show the statistically significant primary transcripts (in blue, corrected p < 0.05 and FDR < 0.05), identified in the comparisons of TNBC vs. NBS, non-TNBC vs. NBS and HER2-positive vs. NBS pair wise comparisons using cuffdiff program. (B) The relative abundance of all the primary transcripts associated with TFAP2A, a gene that is involved in differential promoter usage in TNBC, non-TNBC and HER2-positive breast cancers. All the primary transcripts (TSSs) of TFAP2A and their abundances are shown. The primary transcripts (TSS) that produce isoforms identical to a known ensembl transcript are shown in different colour other than black. The novel isoforms that share at least one splice junction with ENST00000489805 isoform of TFAP2A are shown in black.
All reconstructed genes comprised two or more isoforms and more than 50% (TNBC: 11394, non-TNBC: 7107, HER2-positive: 8300 genes) qualified for the two-tailed t-test in Cuffdiff. To eliminate the primary transcripts that arise due to impartial built, the transcripts that comprised at least 5–8 exons and were similar to the reference were included in these analyses. The differential expression analysis in the TNBC group revealed 1219 up- and 159 down-regulated TSS groups as compared to NBS (Supplemental File 16). In the non-TNBC group, 650 and 355 TSS groups were up and down-regulated, respectively and 1333 and 211 TSS groups were up- and down-regulated in the HER2-positive samples (Supplemental File 17 and 18). There were 161 genes deregulated in all three types of breast cancer at the primary transcript level (Supplemental File 19). In comparison to differential splicing, the number of genes regulated through differential primary transcript expression appears to be higher. Therefore, the primary transcript expression appears to be a prevalent mechanism contributing to the isoform diversity in cancer. Several important gene groups, encoding transcription factors, histone modifiers, protein kinases and receptors, are found to be deregulated in all the three comparisons.
Differential promoter usage and promoter switch
We next investigated the differential promoter usage by grouping the primary transcripts of a gene based on the promoter used. This was followed by testing changes in the isoform abundance by measuring the square root of the Jensen-Shannon divergence that occur within and between these groups in normal and cancer breast samples. There were 138, 83 and 178 distinct promoter switching genes in TNBC, non-TNBC and HER2-positive groups, respectively, as compared to NBS (Supplemental Table 3, Supplemental Files 20–22). Interestingly, only five genes (HSPA8, MTAP, CTDP1, TFAP2A and DTX4) that employ distinct promoters were shared among the three cancer groups (Supplemental Figure 9).
We next investigated the potential promoter switch regulation for the genes comprised from more than one transcript initiating from distinct genomic loci. Initially, we separated the novel and the known isoforms associated with genes that utilize distinct promoters, then identified the isoforms with altered coding sequence due to promoter switch. There were 75, 44 and 152 coding region-altered transcripts resulting from promoter switch events in TNBC, non-TNBC and HER2-positive breast cancer subtypes, respectively (Supplemental Table 3, Supplemental File 23–25).
For selected genes, the promoter switch and the posttranscriptional splice regulation was investigated in details at the level of the individual gene. In many cases, we observed multiple transcripts with functional ORFs resulting from the same primary transcript. An example is transcription factor AP-2-alpha (TFAP2A). We identified 12 distinct primary transcripts (TSS I – TSS XII) that produced 20 different TFAP2A isoforms, including five known (Figure 5B). Among them, only twelve isoforms encoded fully functional ORFs (potential protein product). Twelve novel isoforms were found in TNBC group and four TSSs (TSS I, TSS II, TSS VII and TSS XI) appear to encode the predominant species in TNBC group. Although similar isoforms were seen in the NBS, they differed in their expression levels and coding sequences (Supplemental Figure 10). The combined expression of different transcripts originating from the same TSS differed significantly between TNBC and NBS. Our data show that TFAP2A expression differences between TNBC and NBS result from differences in the pre-mRNA amounts, differential promoter usage, as well as differential regulation at post transcriptional level (i.e. splicing). Apart from TFAP2A, several other candidates that employ promoter switching to produce cancer specific isoforms were identified (Supplemental File 26).
Pathways influenced by deregulated genes in breast cancer
To determine the impact of all the above described transcriptomic modifications on biological functions and pathways, we performed Ingenuity Pathway Analysis (IPA) (Supplemental Figures 11–13). In TNBC, the pathways most severely deregulated at the level of differential promoter usage and promoter switch included cell-to-cell signaling and interaction, cellular movement, cellular development, system development and immunological response (Supplemental File 27). In contrast, when the posttranscriptional splice deregulation was examined (differential expression of isoforms resulting from the same TSS), genes from cell death, cell cycle pathway and cellular function and maintenance were predominant. Similar analysis for the non-TNBC and HER2-positive groups revealed cell-to-cell signaling and interaction as one of the main pathways deregulated at transcriptional level in all three breast cancer subtypes; whereas the posttranscriptional splice deregulation affected mostly cell death, cell morphology and post-translational modifications pathways (Supplemental File 28 and 29).
Finally, the investigation of whether there is a core set of genes that are consistently and significantly deregulated in their primary transcript abundances, promoter usage/switching and post-transcriptional splicing, outlined fourteen genes in TNBC including DYRK1A, MSI2, MLL5, ABCG1 and PHF16 (Supplemental Figure 14A, Supplemental File 30). Similarly, we found FGD4, NCAPD2, KIAA0664, TIAA1217 and SNHG7 to be significantly deregulated at the level of primary transcript, promoter usage and splicing in the non-TNBC subtype (Supplemental Figure 14B, Supplemental File 31). In the case of HER2-positive, 26 such core genes are found and they include DICER1, CASP10, SORL1, PPP1R12A, INPP4B and ATP11B (Supplemental Figure 14C, Supplemental File 32).
Exon skipping and intron retention are predominant breast cancer specific splicing events
Almost 90% of multi-exon human genes undergo alternative splicing during development, cell differentiation and disease4,41. The alternative exon selection manifests between subtle and “all or nothing” mechanisms of specific exons expression. We compared the splice profiles of the 17 breast cancer samples individually against normal breast samples, as well as merged breast cancer subtypes against merged normal breast samples. Events of exon skipping, mutually exclusive exon, alternative start, stop and intron retention, as compared to NBS, were annotated using a multivariate analysis of transcript splicing program, MATS38 (Figure 6A, Supplemental Table 4–6). MATS provides a statistical framework that determines the junction counts supporting the inclusion or the exclusion of specific splice events in cancer against NBS. In the TNBC group, we identified 2898 and 2038 exon skipping and intron retention events, respectively. In TNBC, 1549 mutually exclusive exons, 446 transcripts start and 443 transcript stop site changes were observed. Intron retention and exon skipping appear to be the most predominant splice events in all three breast cancer subtypes. The complete annotation of splice events that are specific to TNBC, non-TNBC and HER-positive group is presented in Supplemental File 33 to 35.
Annotation of novel splice events.
(A) Peaks graph showing the inclusion or exclusion of exons that occur in the individual breast cancer subtypes in comparison to NBS. The exact inclusion or exclusion event count is shown on the relevant peak. (B) An example of “switch like” exon occurrence in XBP1, encoding potent transcription factor. In normal breast samples sample, the junctions and reads support the possibility of two types of isoforms that include or exclude exon 2 in XBP1. In contrast, in all six non-TNBC breast cancer type, the reads encode the entire five exons, supporting only the potent DNA binding domain intact splice variant expression.
The inclusion of exon and intron in cancer appears to be the predominant splice event in all three types of breast cancer (Figure 6A, Supplemental Files 36–38). Among breast cancer specific events that occur in all the samples of the subgroups, we often observed events such as switch-like exon, defined by the absence of reads supporting one of the compared conditions. For example, we detected reads supporting two exon models of XBP1, a key transcription factor with a critical role in anti-estrogen responsiveness in breast cancer cells: inclusion of all intact exons from the genome reference, thus encoding a whole length functional protein and alternative isoform that is lacking exon 2. Exon 2 is not in frame and its elimination leads to a premature stop codon generation early in the newly formed isoform, likely subjected to a Nonsense mediated mRNA decay (NMD). This exon 2 excluding isoform appears to be expressed in NBS, but in none of the six non-TNBC samples (Figure 6B). Another interesting observation on XBP1 is higher prevalence of the spliced isoform XBPs that is related to endoplasmic reticulum (ER) stress and unfolded protein response (UPR) in the breast cancer samples compared to the NBS. Of note, although this prevalence was observed in all three breast cancer groups, the greatest abundance of XBP1s was seen in the non-TNBC group.
Similar to XBP1 exclusive exon presence in cancer vs NBS is seen for other genes, such as breast cancer anti-estrogen resistance 1 (BCAR1), exon 6 and high mobility group nucleosome-binding domain-containing protein 3 (HMGN3), exon 6. In the later two examples however, in contrast to XBP1, the exclusion of the exon does not lead to NMD. In both cases, a suggested mechanism of action is through overexpression in the cancer cells of functional protein, which is suppressed in the NBS through a mechanism of exon exclusion. Finally, to supply confidence in the MATS outcomes, we cross-compared them with the direct exon models generated by cufflinks and cuffcompare – the majority of the novel splice events detected by MATS were reflected in the cufflinks output.
Another interesting finding revealed by combined application of MATS and direct exon modeling is the formation of novel isoforms with new exon assembly. We observed such novel isoforms in several genes, including Cyclin-dependent kinase 4 (CDK4), La-related protein 1 (LARP1), PH domain leucine-rich repeat-containing protein phosphatase 2 (PHLPP2) and Gamma-adducin (ADD3) in all the cancer samples of a subtype (Supplemental Figures 15 and 16). To experimentally validate the assembly and expression of the novel isoforms, we designed unique primers and performed RT-PCR and subsequent sequencing analysis. We were able to identify the expected hybrid exon assembly in the corresponding breast cancer samples, thus validating the RNA-seq findings and increasing the confidence of the analysis (Figure 7A–7C).
Validation of novel cancer specific isoforms at cDNA and protein level.
(A) CDK4 novel isoform identified in non-TNBC validated by RT-PCR. The left side panel shows the relative abundance of all the primary transcripts. The red arrow points to a pie chart that shows the relative abundance of all the isoforms that originate from the TSS129057. The lower bar chart shows the relative abundance of isoforms that are generated from the TSSs shown in the middle panel. To indicate the origin of the isoforms, the bars are color coded similar to their primary transcript color. The right side panel shows a novel isoform that is formed through a junction merging two CDK4 isoforms, ENST00000257904 and ENST00000552862 and skipping the first non-coding exon of ENST00000257904. The novel isoform does not change coding sequence. The RT-PCR gel electrophoresis is shown on the right. (B) LARP1 novel isoform identified in non-TNBC samples by RNA sequencing and validated by RT-PCR (gel on the right, the box indicates the region that was amplified by RT-PCR). (C) PHLPP2 novel isoform identified in TNBC samples by RNA sequencing and validated by RT-PCR (gel on the right, the box indicates the region that was amplified by RT-PCR). (D) Validation of novel isoforms identified in cancer samples in breast cancer cell lines. LARP1 RT-PCR product was detected in MCF7 only and PHLPP2 was detected in MCF7 and HS578T; in contrast the CDK novel isoform was detected in all eight screened cell lines. (E) LARP1 novel protein isoform validation by Western blot analysis in breast cancer cell lines. An additional band, corresponding to the predicted novel isoform of 1096AA (as compared to 1019 in the wild type) is identified by LARP1 specific antibody. Congruent with RT-PCR results, the novel longer LARP1 isoform was detected in MCF-7 and not in the remaining tested breast cancer cell lines. (F) PHLPP2 novel protein isoform validation by Western blot analysis in breast cancer cell lines. The novel shorter (1256AA) as compared to the wild type (1323AA) isoform was identified in MCF7 and HS578T cell lines, in line with RT-PCR results.
For all three genes listed above, the novel hybrid isoforms contained exon combinations of two or more known isoforms. For instance, the newly identified isoform of CDK4 includes the first exon of ENST00000547853, which is added as a first exon to the ENST00000257904, comprising seven exons through a novel junction and skipping an exon located in the 5 prime untranslated region from ENST0000257904. Of note, the skipped exon is known to be involved in the translation of CDK4 by p53 and TGFbeta42,43. It is also notable that the RNA-seq analysis revealed this exon model in three of the six non-TNBC samples, predominantly expressed in all three of them. The Sanger validation confirmed the presence of the novel CDK4 isoform in all three RNA-seq positive samples and in one additional non-TNBC sample, thus confirming the prevalence expression in the non-TNBC group (four out of six, see Figure 7A).
Similarly, a novel hybrid isoform was identified for LARP1 (Figure 7B, Supplemental Figure 16). LARP1 is an RNA-binding protein that regulates negatively RAS-MAPK pathway and is shown to be involved in cell division, migration and apoptosis44,45. The human LARP1 gene has 15 different isoforms, from which only one - ENST00000336314 – is known to be expressed at protein level. The N-terminal region of the novel LARP1 isoform discovered in our study retains the exon model of ENST00000336314 except the first exon. That first exon appears to be similar to the first exon of another isoform, ENST00000518297 and encodes 145 (as compared to 68 encoded by the first exon of ENST00000336314) amino acids of the N-terminal region, proximal to the RNA binding domain (AA397-487). RT-PCR and Sanger sequencing confirmed the presence of the novel hybrid isoforms in four out of the 6 non-TNBC samples in which it was originally detected by RNA-seq. Similarly, novel hybrid isoforms, including altered UTRs, were identified and confirmed by Sanger sequencing for the tumor suppressor PHLPP246 (Figure 7C, Supplemental Figures 15 and 16) and the membrane skeletal associated protein ADD3 (Supplemental Figures 15 and 16)47.
In addition to the studied breast cancer samples, we screened for the presence of these “novel hybrid isoforms” in several breast cancer cell lines including ZR75, MCF-7, SKBR3, SUM159, BT549 and HS578T (Figure 7D) in an attempt to identify a reproducible model system for further biological characterization of the novel isoforms. Of note, the novel hybrid isoform of CDK4 was present in all the cell lines whereas ADD3 and LARP1 were detected only in MCF7 cell line. The hybrid PHLPP2 isoform was detected in MCF7 and HS578T cell lines.
Finally, we compared the exon assembly of all the assembled transcripts without including a statistical cutoff or overlapping with MATS output and outlined the exons that are under cancer-specific splice control (Supplemental Tables 7 and 8, Supplemental Files 39–41). This allowed us to inspect and report all the cancer-related splice changes in TNBC, non-TNBC and HER2-positive breast cancers.
We next set to confirm the expression of the novel hybrid isoforms of LARP1 and PHLPP2 on protein level (Supplemental Figure 15). For LARP1, the hybrid protein isoform is estimated to be 1096AA long (as compared to the 1019AA of the closest isoforms ENST00000336314). As expected, a band corresponding to ENST00000336314 was detected by Western blot in all three tested breasted cell lines (MCF7, ZR-75 and HS578T, Figure 7E). In MCF-7, one additional band, corresponding to the longer hybrid LARP1 isoform was present. As noted above, MCF7 was the only cell line expressing the LARP1 hybrid cDNA; thus, Western blot completely agreed with the RT-PCR and RNA sequencing data.
According to our RNA-seq findings, PHLPP2 hybrid isoform is expected to encode a protein of 1256AA (as compared to the 1323AA of the closest isoform ENST00000568954). Concurring with the RT-PCR results, bands corresponding to both ENST00000568954 and the shorter novel hybrid isoform were detected in MCF-7 and HS578T cell lines by Western Blot; none of these bands was present in the ZR-75 (Figure 7F). In MCF-7, one additional longer band was detected by our Western blot. We could not identify reads corresponding to such an elongated PHLPP2 isoform among the TNBC, non-TNBC and HER-2-positive samples; thus, the detected longer protein might potentially represent MCF-7 specific isoform. Of note, the only PHLPP2 isoform known to be expressed at protein level so far was ENST00000568954.
Discussion
Stringently regulated mechanisms such as transcription, splicing, poly-adenylation, RNA editing, post-translational modification and proteolysis enable the generation of multiple functional variants of an individual gene. In cancer, many of these mechanisms are seized to favor the malignant state. Massive parallel RNA sequencing analysis allows us to explore the cancer-related changes that occur at the stage of transcription, pre-RNA, splicing and editing and to outline isoforms that are specific for given cancer subtype. Cancer specific splice variants of genes that control the cell proliferation and DNA damage (e.g. FGFR2, BRCA1, FHIT), adhesion, invasion (CD44, MST1R), angiogenesis (VEGF) and apoptosis (BCL10, CASP2) have been reported in the last decade39. Therapeutic approaches that target these specific variants are proving to be effective in the clinic. For instance, specific antibody against a domain coded by exon 6 of a specific splice isoform, CD44v6 is used in radiotherapy, highlighting the urgent need to explore other promising target niches for cancer treatment31.
Here, for the first time, we provide an overview on all transcriptomic and splicing changes in TNBC, non-TNBC and HER2-positive breast cancers in comparison to NBS using RNA sequencing analysis. RNA sequencing is a powerful tool that allows deciphering of multiple layers of transcriptome regulation, including promoter selection, transcription, splicing and RNA editing. The notion of breast cancer specific exons and splice variants of one gene is recent and most of the studies so far are either focused on individual genes or utilize preselected splice-sensitive exon expression arrays28,48,49,50,51. When we compared the differentially spliced genes identified by our unbiased global RNA sequencing approach, followed by reference independent assembly, we validated many previously reported breast cancer specific alternatively spliced genes such as FGFR2, NOTCH3, SYNE2, TLK1 and UTRN (Supplemental Files 42–48)39,45,46,47,48.
At the moment of our analysis, no parallel exon array data targeting the cancer subtypes chosen by our study was available in the Gene expression Omnibus database; however we found a dataset (GSE33692, Affymetrix Human Exon 1.0 ST Array) that includes three normal breast samples, nine ductal carcinoma in situ (DCIS) and 10 invasive breast cancer patient tissue52. The overlap between the RNA sequencing based alternatively spliced genes and the genes identified from the comparative analysis of normal vs. DCIS or IBS are 33.4% and 36%, respectively (Supplemental Figure 17), revealing breast cancer associated differentially spliced genes regardless of the specific cancer subtype (Supplemental Files 49 and 50).
Notably, we found significant overlap with a comparative exon array study on splicing changes between TNBC and HER2-positive subtypes in comparison to normal tissue. In the exon array study, 3283 and 1976 exons were found to be over-expressed in TNBC and HER2 positive breast cancer compared to normal breast samples, respectively48. Among those, 560 genes in the TNBC group and 333 in the HER2-positive group were found to undergo mutually exclusive and exon inclusion events by our study (Supplemental Files 42). These findings clearly validate the overlapping gene sets, but also underlie the strength of the RNA-seq approach allowing unbiased identification of not only exon-altering changes, but also novel splicing events such as hybrid isoforms, intron retention and partial exon inclusion, that could not be identified through array technologies. Thus, this study for the first time provides a portrait of all the novel isoforms specific to TNBC, non-TNBC and HER2-positive breast cancers. The vast majority of the identified novel isoforms that comprise an ORF are predicted to be degraded through NMD due to the generation of an early stop codon located upstream of the last exon. However, approximately 5% of these ORF comprising novel transcripts contain all the attributes of a functional ORF, including properly located polyadenylation signal and thus may encode novel expressed protein isoforms – we have validated such novel, alternative size proteins for LARP1 and PHLPP2.
The comparison of our alternative splicing gene set against previously reported breast cancer cell lines28 (Supplemental Files 44) and mouse primary tumors with different metastatic capabilities49,50 indicated overlapping alternative spliced genes (Supplemental Files 45-46). This is not surprising since, in contrast to the global RNA-sequencing approach, these exon array studies include preselected probe sets and comprise only limited number of genes.
Apart from outlining numerous specific targets for individual focused studies, our results suggested several previously unacknowledged expression-regulation mechanisms. Notable example is the “exon-switch like” mechanism (e.g. XBP1), which leads to elevated expression of fully functional wild type protein in the cancer samples compared to some proportion of alternative/degraded protein in the normal tissues. The expression pattern of some of these proteins (XBP1, BCAR1) is correlated with the invasiveness or the level of malignancy of different breast tumors. Thus, the observed cancer specific “exon-switch like” may represent alternative mechanism for expressional up-regulation that may account for their cancer-associated elevated levels. Another notable example is promoter switching. Promoter switching is a transcription regulatory mechanism that is still not completely defined, but its significance is increasingly acknowledged. For instance, the expression of breast and small cell lung cancer specific RNA aromatase (CYP19) splice variant is recently reported to be regulated through promoter switching and it is proposed to be a therapeutic intervention point53,54. Our datasets highlighted multiple genes that are regulated through differential promoter usage or promoter switch in a cancer subtype specific manner. Further investigation is required to forward these discoveries into clinical use.
The average number of exons per transcript in our de novo assembly was between eight and ten and did not differ from the current estimations on the human transcriptome. However, most of the identified breast cancer specific isoforms were combinations of previously unknown exon assembly, suggesting that our study reveals largely unknown transcriptomic landscape. It is essential to keep in mind that the transcriptional and splicing dynamics of various tissues, including the breast, are still being annotated. Therefore, translating our results into clinical use would require validation of the cancer specific isoforms on a large scale and detailed annotation of tissue-specific transcriptomic variability.
Methods
Human patient samples
Dr. Suzanne Fuqua (Baylor College of Medicine) provided the human breast cancer tissue RNA samples. Dr. Kornelia Polyak (Dana-Farber Cancer Institute and Harvard Medical School) provided the human breast organoids (epithelium) samples (NBS). All of the human samples were used in accordance with the IRB procedures of Baylor College of Medicine and Dana-Farber Cancer Institute and Harvard Medical School, respectively. The breast tumor types, TNBC, Non-TNBC and HER2-positive, were classified on the basis of RNA sequencing FPKM abundance5 and immunohistochemical and RT-qRT-PCR classification (data not shown).
Illumina Genome sequencing RNA sequencing library preparation
Large and small ribosomal RNA (rRNA) was removed from total RNA using RiboMinus Eukaryote Kit (Invitrogen, Carlsbad, CA). Five micrograms of total RNA were hybridized to rRNA-specific biotin labeled probes at 70°C for 5 minutes. The rRNA-probe complexes were then removed by streptavidin-coated magnetic beads. The rRNA-free transcriptome RNA was concentrated by ethanol precipitation. The cDNA synthesis and DNA library construction for all the seventeen samples were performed as described5.
Read alignment and transcript assembly
The paired end raw reads were aligned using the TopHat version 1.2.0 that allows two mismatches in the alignment. The aligned reads were assembled into transcripts using cufflinks version 2.0.0. The alignment quality and distribution of the reads were estimated using SAM tools. From the aligned reads, the de novo transcript assembly was performed to capture the major splice rearrangements and novel variations that occur in the transcriptomes of TNBC, Non-TNBC and HER2- positive breast cancers in comparison to NBS using cufflinks version 1.3.036. In addition, Advanced Reference Annotation Based Transcript (RABT) Assembly was also performed to check whether including faux reads would enhance our chances of novel isoform discovery. The cuffcompare program was used to identify transcripts that are identical to the reference human genome (the Ensembl GRCh37.62 B (hg19) reference genome). Further analysis and novel isoform call was performed through the reconstructed transfrags that comprise novel splice junctions and share at least one splice junction with a reference transcript. The very low abundant transcripts were identified by binning the transcripts according to their FPKM and the transcripts with FPKM below 0.3 were eliminated from further analysis. All the analyses presented in this manuscript are performed using only two categories of transcripts: transcripts that are identical to reference and transcripts that comprise novel junctions. The global statistics, which includes the distributions of FPKM scores across samples and the dendogram that shows the relationship between the samples based on the reconstructed transcripts, were analyzed using cummeRbund package of cufflinks suite of programs. The average exon number was in the reassembled transcripts is comparable to the human genome reference average.
Discovery of differential splicing, primary transcript and promoter usage
The transcripts that are similar to the reference and the novel splice junctions were chosen to identify the genes that undergo statistically significant differential splicing between each breast cancer subtype as opposed to NBS using the most recent cuffdiff program that allows us to test several samples as a group. Although the manual inspection and binning of the transcripts based on their abundances emphasizes the heterogeneity in the expression abundances, it is difficult to predict the significance of it due to the lack of read depth and complete coverage across the transcripts. Therefore, the samples that belong to TNBC, Non-TNBC and HER2-positive cancers were given as a group against the three normal breast samples group for the cuffdiff analysis (p-values and FDR below 0.05) that reports statistically significant differential splicing, primary transcripts and promoter usage. In addition, the genes that undergo promoter switching are also examined for differentially expressing promoters by investigating the genes that comprise isoforms with distinct transcript start sites.
Annotation of Novel Splice events
In order to annotate all the novel splice events that occur in TNBC, Non-TNBC and HER2-positive cancers in comparison to NBS, we used recently releases program Multivariate Analysis of Transcript Splicing (MATS)38. Additionally, for consistency checking and independent validation we used an in-house built program (http://genomics.jhu.edu/software/ASproFile/) to compare the exon models between isoforms assembled with the program cufflinks for the normal and cancer samples (as mentioned earlier, only the isoforms that are similar to reference and isoforms that comprise novel splice junctions were considered) and determine the splicing differences indicative of exon inclusion, exclusion, alternative 5′, 3′ and intron retention events.
GO and IPA analysis
To associate cellular functions with the set of differential splicing, pre-RNA expression, promoter switching and genes, we used Database for Annotation, Visualization and Integrated Discovery (DAVID http://david.abcc.ncifcrf.gov/) and ingenuity pathway analysis (IPA, Ingenuity® Systems, www.ingenuity.com).
RT-PCR validation
Initially, the isoforms associated with the statistically significant differential spliced genes were identified. Isoforms for individual validation were selected among the ones expressed only in TNBC, non-TNBC and HER2-positive in comparison with normal breast samples. It is important to note that these isoforms are detected in other breast cancer types and absent only in NBS. Primers were designed to amplify the whole transcripts using unique for the isoform regions. A second set of validation candidates were chosen from the list of novel isoforms that are discovered through the novel splice event annotations. When we inspected the exon model of the isoforms that undergo exon skipping or inclusion in breast cancer compared to NBS, we detected several new isoforms comprising novel junctions that combine partial exon models of two distinct isoforms of the same gene. We selected these “novel hybrid isoforms” and designed unique primers that would amplify only this particular newly discovered isoforms using qRT-PCR. The amplified products were then gel purified and sequenced using Sanger’s sequencing. Similar experimental validation was performed using the same primers in various breast cancer cell lines.
For qRT-PCR, total RNA was isolated using RNeasy Midi kit (Qiagen, Cat No # 75144) and cDNA was synthesized with SuperScript II reverse transcriptase (Invitrogen) using 1 μg of total RNA and oligo dT primer. qRT-PCR was performed with the gene-specific primers listed in Supplemental File 15, using a CRX96 Real Time System (BioRad), Hercules, CA. The levels of RNA expression of all the genes were normalized against the expression levels of cyclophillin B RNA.
Immunoblot analysis
For the Western Blot Analysis, MCF-7, ZR-75 and HS578T cells were washed three times with PBS and incubated in a lysis buffer (50 mmol/L Tris-HCl [pH 7.5], 120 mmol/L NaCl, 1% Triton X-100, 1X protease inhibitor mixture (Roche), 1 mmol/L sodium vanadate on ice for 30 min. Cell lysates containing equal amounts of protein were resolved on 14% SDS-PAGE and transferred to nitrocellulose membranes. Antibodies for LARP1 and PHLPP2 were purchased from Santa Cruz Biotechnology (Cat## SC-102006, SC-137663). Membranes were probed with respective antibodies and detected by means of enhanced chemiluminescence.
Comparing the RNA sequencing and microarray based differentially splicing genes associated with breast cancer
To compare the results of splicing analysis our RNA-sequencing data with published microarray data, we searched for datasets (datasets that used Affymetrix Human Exon 1.0 ST Array, GPL5175 platform in GEO) that contained breast cancer patient samples hybridized onto an exon array. We downloaded the GSE33692 dataset that contained patient samples from three normal breast samples, nine ductal carcinoma in situ (DCIS) and 10 invasive breast cancer (IBC)52. Using GeneSpring GX, the differentially spliced genes between normal and cancer samples (DCIS & IBC) were identified using cut off of Benjamini Hochberg FDR of 0.05 and splice index above 0.5. There are 8223 genes between normal and IDC and 7570 genes between normal and DCIS to be differentially spliced. Subsequently, the statistically significant spliced genes from our RNA-sequencing studies were overlapped with the results from the microarray analysis. Since the samples in the microarray couldn’t be classified into specific subtypes, all the differentially splicing genes discovered in our RNA sequencing study (i.e. NBS vs. TNBC, Non-TNBC and HER2-positive) were compared with the microarray compassion. There are 468 genes overlapping between normal vs. IBC from microarray studies and RNA-sequencing studies. Similarly, we found 434 genes overlapping form normal vs. DCIS comparison with RNA-sequencing data. Additionally, we also compared subtype specific data from RNA-seq studies with microarray results. Comparison of normal vs. IDC gave 213, 145 and 260 overlapping genes for TNBC, non-TNBC, HER-2 comparisons respectively. Similarly, comparison of normal vs. DCIS gave 195, 135 and 238 genes for TNBC, non-TNBC and HER-2 comparisons respectively.
References
Stratton, M. R. & Wooster, R. Hereditary predisposition to breast cancer. Curr Opin Genet Dev 6, 93–97 (1996).
Turnbull, C. & Rahman, N. Genetic predisposition to breast cancer: past, present and future. Annu Rev Genomics Hum Genet 9, 321–345 (2008).
Robertson, L. et al. BRCA1 testing should be offered to individuals with triple-negative breast cancer diagnosed below 50 years. Br J Cancer 106, 1234–1238.
Wang, E. T. et al. Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470–476 (2008).
Eswaran, J. et al. Transcriptomic landscape of breast cancers through mRNA sequencing. Sci Rep 2, 264 (2012).
Curtis, C. et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486, 346–352.
Mercer, T. R. et al. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat Biotechnol 30, 99–104 (2012).
Carninci, P. et al. The transcriptional landscape of the mammalian genome. Science 309, 1559–1563 (2005).
Carninci, P. Tagging mammalian transcription complexity. Trends Genet 22, 501–510 (2006).
Strausberg, R. L. & Levy, S. Promoting transcriptome diversity. Genome Res 17, 965–968 (2007).
Pennisi, E. Why do humans have so few genes? Science 309, 80 (2005).
Johnson, J. M., Edwards, S., Shoemaker, D. & Schadt, E. E. Dark matter in the genome: evidence of widespread transcription detected by microarray tiling experiments. Trends Genet 21, 93–102 (2005).
Birney, E. et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816 (2007).
Grosso, A. R. et al. Tissue-specific splicing factor gene expression signatures. Nucleic Acids Res 36, 4823–4832 (2008).
Matlin, A. J., Clark, F. & Smith, C. W. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol 6, 386–398 (2005).
Heinzen, E. L. et al. Tissue-specific genetic control of splicing: implications for the study of complex traits. PLoS Biol 6, e1 (2008).
Lopez, A. J. Alternative splicing of pre-mRNA: developmental consequences and mechanisms of regulation. Annu Rev Genet 32, 279–305 (1998).
Tsunoda, T. et al. Involvement of large tenascin-C splice variants in breast cancer progression. Am J Pathol 162, 1857–1867 (2003).
Guttery, D. S., Shaw, J. A., Lloyd, K., Pringle, J. H. & Walker, R. A. Expression of tenascin-C and its isoforms in the breast. Cancer Metastasis Rev 29, 595–606.
Thorsen, K. et al. Alternative splicing in colon, bladder and prostate cancer identified by exon array analysis. Mol Cell Proteomics 7, 1214–1224 (2008).
Shapiro, I. M. et al. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet 7, e1002218 (2011).
Okumura, N., Yoshida, H., Kitagishi, Y., Nishimura, Y. & Matsuda, S. Alternative splicings on p53, BRCA1 and PTEN genes involved in breast cancer. Biochem Biophys Res Commun 413, 395–399 (2011).
Wang, L. et al. Alternative splicing disrupts a nuclear localization signal in spleen tyrosine kinase that is required for invasion suppression in breast cancer. Cancer Res 63, 4724–4730 (2003).
Ghosh, A., Stewart, D. & Matlashewski, G. Regulation of human p53 activity and cell localization by alternative splicing. Mol Cell Biol 24, 7987–7997 (2004).
Lixia, M., Zhijian, C., Chao, S., Chaojiang, G. & Congyi, Z. Alternative splicing of breast cancer associated gene BRCA1 from breast cancer cell line. J Biochem Mol Biol 40, 15–21 (2007).
Orban, T. I. & Olah, E. Emerging roles of BRCA1 alternative splicing. Mol Pathol 56, 191–197 (2003).
Ng, W., Loh, A. X., Teixeira, A. S., Pereira, S. P. & Swallow, D. M. Genetic regulation of MUC1 alternative splicing in human tissues. Br J Cancer 99, 978–985 (2008).
Lapuk, A. et al. Exon-level microarray analyses identify alternative splicing programs in breast cancer. Mol Cancer Res 8, 961–974 (2010).
Dutertre, M., Vagner, S. & Auboeuf, D. Alternative splicing and breast cancer. RNA Biol 7, 403–411 (2010).
Ferreira, E. N. et al. Alternative splicing enriched cDNA libraries identify breast cancer-associated transcripts. BMC Genomics 11 Suppl 5, S4.
Brinkman, B. M. Splice variants as cancer biomarkers. Clin Biochem 37, 584–594 (2004).
Wu, Z. J. et al. Gene expression profiling of human breast tissue samples using SAGE-Seq. Genome Res 20, 1730–1739.
Trapnell, C., Pachter, L. & Salzberg, S. L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009).
Garber, M., Grabherr, M. G., Guttman, M. & Trapnell, C. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat Methods 8, 469–477 (2011).
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7, 562–578 (2012).
Roberts, A., Pimentel, H., Trapnell, C. & Pachter, L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics 27, 2325–2329 (2011).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28, 511–515 (2010).
Shen, S. et al. MATS: a Bayesian framework for flexible detection of differential alternative splicing from RNA-Seq data. Nucleic Acids Res 40, e61 (2012).
Venables, J. P. Aberrant and alternative splicing in cancer. Cancer Res 64, 7647–7654 (2004).
Germann, S., Gratadou, L., Dutertre, M. & Auboeuf, D. Splicing programs and cancer. J Nucleic Acids 2012, 269570.
Castle, J. C. et al. Expression of 24,426 human alternative splicing events and predicted cis regulation in 48 tissues and cell lines. Nat Genet 40, 1416–1425 (2008).
Ewen, M. E. p53-dependent repression of cdk4 synthesis in transforming growth factor-beta-induced G1 cell cycle arrest. J Lab Clin Med 128, 355–360 (1996).
Miller, S. J., Suthiphongchai, T., Zambetti, G. P. & Ewen, M. E. p53 binds selectively to the 5' untranslated region of cdk4, an RNA element necessary and sufficient for transforming growth factor beta- and p53-mediated translational inhibition of cdk4. Mol Cell Biol 20, 8420–8431 (2000).
Nykamp, K., Lee, M. H. & Kimble, J. C. elegans La-related protein, LARP-1, localizes to germline P bodies and attenuates Ras-MAPK signaling during oogenesis. RNA 14, 1378–1389 (2008).
Burrows, C. et al. The RNA binding protein Larp1 regulates cell division, apoptosis and cell migration. Nucleic Acids Res 38, 5542–5553.
Brognard, J., Sierecki, E., Gao, T. & Newton, A. C. PHLPP and a second isoform, PHLPP2, differentially attenuate the amplitude of Akt signaling by regulating distinct Akt isoforms. Mol Cell 25, 917–931 (2007).
Naydenov, N. G. & Ivanov, A. I. Adducins regulate remodeling of apical junctions in human epithelial cells. Mol Biol Cell 21, 3506–3517.
Andre, F. et al. Exonic expression profiling of breast cancer and benign lesions: a retrospective analysis. Lancet Oncol 10, 381–390 (2009).
Bemmo, A. et al. Exon-level transcriptome profiling in murine breast cancer reveals splicing changes specific to tumors with different metastatic abilities. PLoS One 5, e11981.
Dutertre, M. et al. Exon-based clustering of murine breast tumor transcriptomes reveals alternative exons whose expression is associated with metastasis. Cancer Res 70, 896–905.
Mercatante, D. R., Bortner, C. D., Cidlowski, J. A. & Kole, R. Modification of alternative splicing of Bcl-x pre-mRNA in prostate and breast cancer cells. analysis of apoptosis and cell death. J Biol Chem 276, 16411–16417 (2001).
Knudsen, E. S. et al. Progression of ductal carcinoma in situ to invasive breast cancer is associated with gene expression programs of EMT and myoepithelia. Breast Cancer Res Treat 133, 1009–1024.
Zhou, J., Gurates, B., Yang, S., Sebastian, S. & Bulun, S. E. Malignant breast epithelial cells stimulate aromatase expression via promoter II in human adipose fibroblasts: an epithelial-stromal interaction in breast tumors mediated by CCAAT/enhancer binding protein beta. Cancer Res 61, 2328–2334 (2001).
Demura, M. et al. Changes in aromatase (CYP19) gene promoter usage in non-small cell lung cancer. Lung Cancer 73, 289–293 (2011).
Acknowledgements
This work is supported by the McCormick Genomic and Proteomics Center.
Author information
Authors and Affiliations
Contributions
R.K. conceived the project and directed all aspects of the breast cancer transcriptome project. J.E., A.H. and R.K. designed the experiments, analyzed the data and wrote the main manuscript text. D.C., P.M. and S.G. designed experiments, generated results and analyzed the data. S.D.R. and K.O. performed RT-qRT-PCR validation experiments and provided reagents. S.A.W.F. and K.P. provided reagents and biological insights. LF, SSN and AH provided assistance for the data analyses and the manuscript preparation.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplemental Figures and Tables
Supplementary Information
Supplementary Files
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Eswaran, J., Horvath, A., Godbole, S. et al. RNA sequencing of cancer reveals novel splicing alterations. Sci Rep 3, 1689 (2013). https://doi.org/10.1038/srep01689
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep01689
This article is cited by
-
Retained introns in long RNA-seq reads are not reliably detected in sample-matched short reads
Genome Biology (2022)
-
Development of alternative splicing signature in lung squamous cell carcinoma
Medical Oncology (2021)
-
Loss of CTNNB1 exon 3 in sclerosing angiomatoid nodular transformation of the spleen
Virchows Archiv (2021)
-
Identification of novel alternative splicing biomarkers for breast cancer with LC/MS/MS and RNA-Seq
BMC Bioinformatics (2020)
-
Splice variants of RAS—translational significance
Cancer and Metastasis Reviews (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.