Abstract
The proton motive force (PMF) is bio-energetically important for various cellular reactions to occur. We developed PMF-photogenerating mitochondria in cultured mammalian cells. An archaebacterial rhodopsin, delta-rhodopsin, which is a light-driven proton pump derived from Haloterrigena turkmenica, was expressed in the mitochondria of CHO-K1 cells. The constructed stable CHO-K1 cell lines showed suppression of cell death induced by rotenone, a pesticide that inhibits mitochondrial complex I activity involved in PMF generation through the electron transport chain. Delta-rhodopsin was also introduced into the mitochondria of human neuroblastoma SH-SY5Y cells. The constructed stable SH-SY5Y cell lines showed suppression of dopaminergic neuronal cell death induced by 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP), an inducer of Parkinson's disease models, which acts through inhibition of complex I activity. These results suggest that the light-activated proton pump functioned as a PMF generator in the mitochondria of mammalian cells and suppressed cell death induced by inhibition of respiratory PMF generation.
Similar content being viewed by others
Introduction
The proton motive force (PMF) is an intracellular energy supply for various bio-reactions in all living organisms. Nerve actions are carried out by the PMF, various cellular substrates are transported by the PMF, bacterial flagella are driven by the PMF and ATP is synthesized by the PMF across the cytoplasmic membranes of prokaryotic cells and the inner mitochondrial membranes of eukaryotic cells. The PMF is also generated from light energy by the thylakoid membranes of chloroplasts and the cytoplasmic membranes of hyperhalophilic bacteria. Delta-rhodopsin (dR) has been known as a light-driven proton transporter expressed in Haloterrigena turkmenica1,2,3. It can be functionally expressed in Escherichia coli under normal growth conditions3, whereas bacteriorhodopsin (bR), which has been well studied, shows low proton transport activity even when expressed in E. coli under optimal growth conditions4,5. Expression of dR produced a light-driven PMF across the membranes of E. coli vesicles and, recently, we photogenerated ATP using vesicles co-expressing dR and ATP synthetase6. It is well accepted among biologists that mitochondria originated from bacteria belonging to the proteo-bacterium genus, which includes E. coli. In this study, we aimed to express dR in mammalian mitochondria to produce a light-driven PMF photogenerating ability like that in chloroplasts. Using constructed stable cell lines expressing dR in the mitochondrial membrane, protons were transported from the mitochondrial matrix to the inner mitochondrial membrane space through the action of dR activated by light irradiation. As a result, the PMF across the mitochondrial membrane was increased by light irradiation and prevented cell death induced by inhibitors of respiratory PMF generation in the mitochondria of mammalian cells.
Results
Mitochondrial-specific expression of delta-rhodopsin in CHO-K1 cells
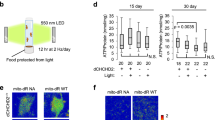
The gene encoding dR was cloned into a targeting vector for expression in mitochondria and the construct was transfected into CHO-K1 cells to make stable CHO-K1 cell lines expressing dR in mitochondria. We confirmed the over-expression of dR protein in these cells using western blot analysis. The expression of dR protein in transfected CHO-K1 cells presented as a band of the calculated molecular weight (26 kDa); in contrast, this band was not present in lysates from mock cells (Fig. 1a). To characterize the distribution of dR in mitochondria, we examined the subcellular localization of dR using biochemical methods. Western blotting of subcellular fractions revealed that dR was present in the mitochondrial fraction, similar to the mitochondrial outer membrane protein Tom20 (Fig. 1b). These results clearly showed the specific expression of dR in the mitochondria in CHO-K1 cells. We also examined subcellular localization of dR in the transfected cells using immunofluorescence microscopy. As shown in Figure 1c, exogenous dR was co-stained with mitochondrial marker, MitoTracker Red CMXRos.
Mitochondrial-specific expression of dR in CHO-K1 cells.
(a) The expression of dR was confirmed using Western blotting. The 26-kDa band was observed in dR transfectants. (b) Western blot analysis after subcellular fractionations of dR expressed CHO-K1 cells. The dR protein was fractionated to mitochondria fraction, same as outer mitochondrial membrane protein, Tom20. (c) Delta-rhodopsin-pCMV/myc/mito vector was transfected into CHO-K1 cells. Z-stack confocal analysis of these cells for MitoTracker Red CMXRos and myc tag immunofluorescence were performed and merged images were indicated. Z-stacks were taken in steps of micrometer. The top to middle sections of the Z-stacks are shown. Scale bar, 20 μm. Exogenous delta-rhodopsin was co-stained with MitoTracker Red CMXRos in CHO-K1 cells.
Function of mitochondrial dR in CHO-K1 cells
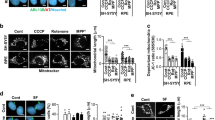
The function of mitochondrial dR in stable CHO-K1 cell lines was evaluated. To clarify the function of dR in the mitochondria of CHO cells, CHO cells expressing dR in mitochondria were treated with rotenone, a pesticide that inhibits mitochondrial complex I activity. As shown in Figure 2, rotenone treatment of CHO cells resulted in increased LDH release, which was correlated with the increase in cell death. When CHO cells possessing the mitochondrial dR were light-irradiated, LDH release was significantly decreased compared with that in cells not subjected to light irradiation (p < 0.05) (Fig. 2a). As a negative control, we performed the same experiments using normal CHO-K1 cells. The LDH release of the light-irradiated CHO-K1 cells showed no significant difference compared to no light irradiation (Fig. 2b). These results indicate that light-activated dR forms a PMF across the inner mitochondrial membrane to partially compensate for the inhibition of complex I activities.
The function of mitochondrial dR in CHO-K1 cells.
(a) Rotenone-induced LDH release from stable CHO-K1 cell lines expressing dR in mitochondria. LDH release from light-irradiated CHO-K1 cells expressing dR in mitochondria was significantly decreased compared with that in cells with no light irradiation (p < 0.05). (b) Rotenone-induced LDH release from CHO-K1 cells. LDH release from the light-irradiated CHO-K1 cells represented no significant difference compared to that of no light irradiated CHO-K1 cells.
Application of photoenergetic mitochondria to neuronal cells
We applied PMF-photogenerating mitochondria to human neuroblastoma SH-SY5Y cells. The stable SH-SY5Y cell line expressing dR in mitochondria was screened and specific dR expression in mitochondria was confirmed by biochemical fractionation (Supplementary Fig. a and Fig. b). The cell line was treated with 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP), an inducer of cell death that has been used to induce models of Parkinson's disease (PD) and which acts through inhibition of complex I of the electron transport chain. PD is characterized by progressive dopaminergic neuronal cell death in the substantial nigra7. SH-SY5Y cells are dopaminergic neuroblastoma cells and are frequently used as cellular models for PD, because MPTP is taken up by dopaminergic SH-SY5Y cells through the dopamine transporter after conversion to MPP+ and inhibits mitochondrial complex I activity. As shown in Figure 3a, cell death induced by MPTP was significantly suppressed upon light irradiation of SH-SY5Y cells expressing dR in mitochondria compared with non-irradiated cells (p < 0.05). As a negative control, we performed the same experiments using normal SH-SY5Y cells. The MPTP-induced cell death was no significant difference upon light irradiation in these cells compared to non-irradiated cells (Fig. 3b). This result suggests that the activation of PMF-photogenerating mitochondria introduced into neuronal SH-SY5Y cells suppressed dopaminergic neuronal cell death in an MPTP model of PD.
MPTP-induced LDH release from stable SH-SY5Y cell lines expressing dR in mitochondria.
(a) MPTP-induced cell death was significantly suppressed upon light irradiation in these cells compared with non-irradiated cells (p < 0.05). (b) MPTP-induced cell death showed no significant difference between light irradiation and non-irradiation.
Discussion
We constructed a new PMF-photogenerating system by targeting an archaebacterial light-driven proton pump in mammalian mitochondria. In this study, we showed that this system works in neuronal cells. It is presumed that this system has the potential to be a tool for understanding the molecular mechanisms underlying PD, one of the most common progressive neurodegenerative disorders. Recently, mitochondrial dysfunction has been revealed as pathological feature associated with PD7. Inhibitors of mitochondrial complex activity such as rotenone and MPTP used in this study produce features of a Parkinsonian syndrome when administered to animals. Complex I deficiency has been observed in PD patients8,9. However, it is unclear whether mitochondrial dysfunction is associated with the onset of PD and if it promotes neuronal dysfunction and degeneration. dR activity in mitochondria could enable control of the mitochondrial PMF via changes in light intensity. On–off switching control of PMF could thus be available by scheduling of light irradiation. Furthermore, the immediate effect of light would have advantages for real-time imaging. If PMF-photogenerating mitochondria are effective in delaying the onset of a Parkinson's disease model in vivo, that would indicate that reduced mitochondrial PMF is associated with the early stage of PD.
Although targeting of light-driven proton pumps derived from microorganisms to mitochondria has not been carried out previously, the optogenetic approach, defined as the combination of genetic and optical methods to control the function of well-defined events in specific cells of living tissue10 has been broadly applied in research11,12,13,14. For example, in the neuroscience field, light–gated proteins from microorganisms, such as channel rhodopsin, bacteriorhodopsin and halorhodopsin have been expressed in the cytoplasmic membranes of neuronal cells and optically controlled. Optogenetic studies also had been applied to control the motion of individual mammals by activation of excitatory neurons using optical fibers15,16. Using these optical techniques, the photoenergetic mitochondria developed in this study could offer the possibility of a new treatment for PD.
Bioprocesses that consume ATP regenerated by microbes as an energy source for industrial chemical productions have already been carried out. In our recent study using E. coli inside-out membranes expressing dR and FoF1-ATP synthase, ATP was photosynthesized and supplied to hexokinase as a model of ATP-consuming bioproduction6. The photoenergetic mitochondria developed in this study must have potential to be applied to light-driven bio-production using eukaryotic host strains (e.g. mammalian, plant, insect and fungi).
From a wider perspective, light-driven PMF formation by mitochondria could have an impact on our understanding of the functional integration of mitochondria and chloroplast. This new technology is expected to be used in a broad range of scientific fields.
Methods
Antibodies
The following antibodies were used: mouse anti-Myc Tag, clone 4A6 (Millipore, Billerica, MA); Tom20 (F-10) antibody (Santa Cruz Biotechnology, Santa Cruz, CA); α-tubulin (DM1A) antibody (Sigma-Aldrich, St. Louis, MO). Anti-rabbit IgG, HRP-linked whole antibody, donkey (GE Healthcare, Waukesha, WI); anti-mouse IgG, HRP-linked whole antibody, sheep (GE Healthcare, Waukesha, WI).
Construction of expression plasmids for transfection and screening dR stable transfectants
The dR gene was amplified from genomic DNA of H. turkmenica (JCM 9743, Riken BRC, Japan) using forward and reverse primers containing SalI and XhoI sequences, respectively. The SalI/XhoI fragment containing dR was isolated and ligated into the mitochondrial targeting vector pCMV/myc/mito (Life Technologies, Carlsbad, CA) digested with the same endonucleases. The resultant recombinant plasmid, pCMV/myc/mito–dR, was used for further studies. The entire nucleotide sequence was confirmed by DNA sequencing. Vector containing the dR gene was transfected into CHO-K1 cells using Polyfect Transfection reagent (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Clones that survived in 0.6 mg/ml Geneticin (Life Technologies) were isolated and maintained in D-MEM medium (Wako, Osaka, Japan) supplemented with 10% fetal bovine serum (FBS) containing 0.6 mg/ml Geneticin at 37°C in 5% CO2. Vector containing the dR gene was also transfected into SH-SY5Y cells using Lipofectoamine 2000 reagent (Life Technologies) and clones were isolated and maintained in the same condition as CHO-K1 cells.
Immunofluorescence microscopy
Cells were cultured on a glass-bottom dish and incubated with MitoTracker Red CMXRos (Life Technologies) for 30 min. The cells were fixed in 4% paraformaldehyde made using phosphate-buffered saline (PBS) for 15 min at room temperature. Cells were stained with mouse anti-Myc Tag antibody and with the secondary antibody: mouse Alexa 488 (Life Technologies). Samples were mounted with VECTASHIELD mounting media with DAPI (Vector Laboratories, Burlingame, CA) for imaging. Images were acquired by confocal microscope (FV1000; Olympus).
Subcellular fractionation
A classical method for subcellular fractionation was used for cultured cells. In brief, cells were homogenized in ice-cold buffer (50 mM Tris·HCl, pH 7.4/150 mM NaCl) plus protease inhibitor mix (Roche, Bazel, Switzerland) by using a motor-driven Teflon homogenizer. Homogenates containing equal amounts of protein were centrifuged at 1,000 × g for 10 min at 4°C to obtain a crude nuclear pellet and postnuclear supernatant. The postnuclear supernatant was further centrifuged at 13,000 × g for 30 min at 4°C to obtain the cytoplasmic fraction (supernatant) and crude mitochondrial fraction (pellet). The supernatant is considered to be cytoplasmic fraction with enriched in α-tubulin and the final mitochondrial fraction is considered to be a mitochondrial fraction with enriched in the mitochondria outer membrane protein Tom20.
Western blotting
A standard protocol17,18 was used with minor modifications. Proteins were separated using SuperSep Tris-Glycine gels (Wako) and transferred onto a polyvinylidene difluoride membranes (Millipore). The same amounts of protein based on the protein concentration of each sample were loaded into lanes. For Western blotting, nonspecific binding was blocked with 5% skim milk (Wako) in PBS containing 0.1% Tween 20 (BioRad, Hercules, CA). The blots were then incubated with primary antibodies overnight at 4°C. For detection of both monoclonal and polyclonal antibodies, appropriate peroxidase-conjugated secondary antibodies were used in conjunction with Novex ECL (Life Technologies) to obtain images using an LAS-3000 imager (Fujifilm, Tokyo, Japan).
Rotenone treatment of CHO cells and LDH assay
The dR stable transfectant CHO-K1 cells were treated with 1 μM of Rotenone (Sigma-Aldrich, St. Louis, MO). After 24h, the cells were irradiated with white light beam (0.438 mW/cm2) in growth chamber (NK System, Biomulti incubator LH 30 8CT) at 37°C for 2 h and then kept it for 30min in normal culture condition. LDH activity was determined with LDH assay kits, CytoTox 96, obtained from Promega Co. (Madison, WI, USA) according to the manufacturers' instructions.
MPTP treatment of SH-SY5Y cells and LDH assay
dR stable transfectant SH-SY5Y cells were treated with 750 μM MPTP (Sigma-Aldrich, St. Louis, MO). After 24 h, the cells were light irradiated under the same condition as described in rotenone treatment. LDH activity was determined with the LDH assay kits.
References
Ihara, K. et al. Evolution of the archaeal rhodopsins: evolution rate changes by gene duplication and functional differentiation. J Mol Biol 285, 163–174 (1999).
Mukohata, Y., Ihara, K., Tamura, T. & Sugiyama, Y. Halobacterial rhodopsins. J Biochem 125, 649–657 (1999).
Kamo, N. et al. A light-driven proton pump from Haloterrigena turkmenica: functional expression in Escherichia coli membrane and coupling with a H+ co-transporter. Biochem Biophys Res Commun 341, 285–290 (2006).
Braiman, M. S., Stern, L. J., Chao, B. H. & Khorana, H. G. Structure-function studies on bacteriorhodopsin. IV. Purification and renaturation of bacterio-opsin polypeptide expressed in Escherichia coli. J biol chem 262, 9271–9276 (1987).
Nassal, M., Mogi, T., Karnik, S. S. & Khorana, H. G. Structure-function studies on bacteriorhodopsin. III. Total synthesis of a gene for bacterio-opsin and its expression in Escherichia coli. J biol chem 262, 9264–9270 (1987).
Hara, K. Y., Suzuki, R., Suzuki, T., Yoshida, M. & Kino, K. ATP photosynthetic vesicles for light-driven bioprocesses. Biotechnol Lett 33, 1133–1138 (2011).
Exner, N., Lutz, A. K., Haass, C. & Winklhofer, K. F. Mitochondrial dysfunction in Parkinson's disease: molecular mechanisms and pathophysiological consequences. EMBO J 31, 3038–3062 (2012).
Schapira, A. H. et al. Mitochondrial complex I deficiency in Parkinson's disease. Lancet 1, 1269 (1989).
Parker, W. D. Jr., Parks, J. K. & Swerdlow, R. H. Complex I deficiency in Parkinson's disease frontal cortex. Brain Res 1189, 215–218 (2008).
Deisseroth, K. Optogenetics. Nat Methods 8, 26–29 (2011).
Zhang, F. et al. Multimodal fast optical interrogation of neural circuitry. Nature 446, 633–639 (2007).
Han, X. & Boyden, E. S. Multiple-color optical activation, silencing and desynchronization of neural activity, with single-spike temporal resolution. PLoS One 2, e299 (2007).
Gradinaru, V. et al. Molecular and cellular approaches for diversifying and extending optogenetics. Cell 141, 154–165 (2010).
Chow, B. Y. et al. High-performance genetically targetable optical neural silencing by light-driven proton pumps. Nature 463, 98–102 (2010).
Gradinaru, V. et al. Targeting and readout strategies for fast optical neural control in vitro and in vivo. J Neurosci 27, 14231–14238 (2007).
Hira, R. et al. Transcranial optogenetic stimulation for functional mapping of the motor cortex. Journal Neurosci Methods 179, 258–263 (2009).
Sawamura, N. et al. Site-specific phosphorylation of tau accompanied by activation of mitogen-activated protein kinase (MAPK) in brains of Niemann-Pick type C mice. J Biol Chem 276, 10314–10319 (2001).
Sawamura, N., Sawamura-Yamamoto, T., Ozeki, Y., Ross, C. A. & Sawa, A. A form of DISC1 enriched in nucleus: altered subcellular distribution in orbitofrontal cortex in psychosis and substance/alcohol abuse. Proc Natl Acad of Sci USA 102 (2005).
Acknowledgements
This study was financially supported by the High-Tech research center (TWIns), the Consolidated Research Institute of Advanced Science and Medical Care (ASMeW), the Global COE ‘Practical Chemical Wisdom' projects and the Leading Graduate Program in Science and Engineering, Waseda University. This study was also financially supported by Grant-in-Aid for Scientific Research on Innovative Areas (“Matryoshka-type evolution,” 23117001) from the Ministry of Education, Sports, Culture, Science and Technology (MEXT). Hara KY was supported by a Grant-in-Aid for Young Scientists (B) (22760608).
Author information
Authors and Affiliations
Contributions
K.Y.H., T.W. and N.S. designed the study; T.W. and N.S. performed experiments; K.K. and T.A. contributed new reagents/analytic tools and discussed the results; K.Y.H. and N.S. wrote the manuscript; N.S. supervised the study; All authors read and approved the final manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Hara, K., Wada, T., Kino, K. et al. Construction of photoenergetic mitochondria in cultured mammalian cells. Sci Rep 3, 1635 (2013). https://doi.org/10.1038/srep01635
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep01635
This article is cited by
-
Engineering yeast with a light-driven proton pump system in the vacuolar membrane
Microbial Cell Factories (2024)
-
Light-driven activation of mitochondrial proton-motive force improves motor behaviors in a Drosophila model of Parkinson’s disease
Communications Biology (2019)
-
The Neuroprotective Effect of Thalidomide against Ischemia through the Cereblon-mediated Repression of AMPK Activity
Scientific Reports (2018)
-
Mitochondrial cereblon functions as a Lon-type protease
Scientific Reports (2016)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.