Abstract
Light chain (AL) amyloidosis is characterized by the misfolding of immunoglobulin light chains, accumulating as amyloid fibrils in vital organs. Multiple reports have indicated that amyloidogenic light chains internalize into a variety of cell types, but these studies used urine-derived proteins without indicating any protein sequence information. As a result, the role of somatic mutations in amyloidogenic protein internalization has not been yet studied. We characterized the internalization of AL-09, an AL amyloidosis protein into mouse cardiomyocytes. We also characterized the internalization of the germline protein κI O18/O8, devoid of somatic mutations and three AL-09 restorative mutations (I34N, Q42K and H87Y) previously characterized for their role in protein structure, stability and amyloid formation kinetics. All proteins shared a common internalization pathway into lysosomal compartments. The proteins caused different degrees of lysosomal expansion. Oregon green (OG) labeled AL-09 showed the most rapid internalization, while OG-Q42K presented the slowest rate of internalization.
Similar content being viewed by others

Introduction
Light chain (AL) amyloidosis is a protein misfolding disease characterized by the secretion of monoclonal immunoglobulin light chains that misfold and deposit as amyloid fibrils1. Cardiac involvement is found in approximately 50% of patients with systemic AL amyloidosis, leading to congestive heart failure and death2. Cardiomyocytes are the main cells affected in the heart3. Median survival for AL amyloidosis patients is 12–40 months, but in cases of advanced disease, the median survival is only 6 months4.
Immunoglobulin light chains are composed of a variable domain (VL) and a constant domain (CL)5. For many years, AL amyloidosis researchers were guided to only use the VL in their studies after a report that stated that amyloid deposits from AL amyloidosis patients were formed primarily by the VL and small regions of the CL6. In 2009, evidence from Vrana et al. demonstrated with a large cohort of AL amyloidosis patient samples that full length proteins are present in the amyloid deposits of these patients7, challenging the previous assumption. The function of the CL in the pathophysiology of AL amyloidosis is not well established. Only recently, the role of CL in thermal stability and aggregation has been characterized for a λ6a full length AL protein and its corresponding VL and CL proteins8. This has prompted our laboratory to explore the behavior of full length AL proteins and compare it to studies previously conducted with their corresponding VL domains.
Though it is assumed that deposition of amyloid fibrils in plaques causes stress to vital organs, smaller soluble oligomeric species populated by amyloidogenic proteins are considered to be the main cause of AL amyloidosis pathophysiology9. It has been demonstrated that soluble amyloidogenic light chains can be internalized into cardiac fibroblasts10 via pinocytosis11 and that the presence of amyloidogenic light chains can impair cardiomyocyte cell function12. It has also been recently reported that light chains from AL patients trigger programmed cell death via the p38α MAPK pathway13. All these studies were conducted with urine-derived AL proteins from which the protein sequences and the oligomerization state of the light chain species being internalized were not reported.
Our laboratory is interested in the role that somatic mutations play in the pathophysiology of AL amyloidosis. We have previously reported the structure, stability and amyloid formation properties of VL AL-09, a protein from a cardiac AL amyloidosis patient. This protein has 7 somatic mutations with respect to its germline sequence κI O18/O8, three of these somatic mutations are non-conservative changes (N34I, K42Q and Y87H) all located within or adjacent the dimer interface. The protein VL κI O18/O8 is more stable than AL-09 and adopts a canonical dimer structure. In contrast, VL AL-09 adopts an altered dimer interface. The mutation VL AL-09 H87Y restores all thermodynamic stability and delays amyloid formation back to germline levels. In addition, VL AL-09 H87Y also restores the dimer interface structure. VL AL-09 I34N partly restores thermodynamic stability and has an intermediate effect on amyloid formation kinetics. Interestingly, VL AL-09 Q42K is as unstable as AL-09 but presents delayed amyloid formation kinetics compared to the kinetics observed for VL κI O18/O8 and VL AL-09 H87Y14. In cell culture, we have shown that the presence of monomeric and dimeric VL AL-09 has cytotoxic effects on mouse cardiomyocytes compared to VL kI O18/O815.
We hypothesize that non-conservative somatic mutations in AL proteins will play a role in protein internalization equivalent to the role these mutations play in protein stability and amyloidogenic potential. In this paper, we studied the role of somatic mutations in full length protein internalization into cardiomyocytes. We determined that all full length proteins adopted the expected β-sheet structure followed by circular dichroism. Using HL-1 mouse cardiomyocytes, we studied protein internalization using confocal microscopy. The five proteins all internalize into lysosomal compartments, although exhibited differences in the rates of internalization with very large degrees of variability within the same cell culture condition. Rates of internalization correlated with rates of amyloid formation for the corresponding VL protein.
Results
Full length light chains are folded as β-sheets
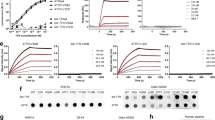
All full length proteins we studied were produced recombinantly, expressed, purified and characterized to confirm that they were properly folded before we used them for cell culture. All proteins are able to adopt the expected β-sheet structure (Figure 1). The proteins were unable to refold reversibly, so no thermodynamic parameters were calculated. To demonstrate that OG labeling did not affect the structure of full length light chains, Far UV-CD spectra were recorded for unlabeled and OG-labeled full length light chains. OG labeling caused only minimal changes in the Far UV-CD spectrum compared to the Far UV-CD spectrum of the corresponding unlabeled light chains.
Far UV CD spectra of OG labeled AL proteins show native β-sheet structure.
Data is shown as mean residue ellipticity (MRE) as a function of wavelength. Proteins display the minimum at around 218 nm, characteristic of β-sheets. Experiments were done with 20 μM protein concentration in 10 mM Tris-HCl pH 7.4 at 4°C.
Cellular internalization of light chains optimized with live cell imaging and cellular fractionation
To determine the appropriate time frame for protein internalization, live cell imaging experiments were conducted with OG-AL-09, showing that internalization happens within 24 hours. We also observed that after cellular internalization began, cells appeared to undergo blebbing and started to change shape (data not shown). The protein clustered around what appeared to be the nucleus. However, the cells began to die after 24 hours. This cell death could have been caused by the constant exposure to the beam of light and/or the difficulty of maintaining sterile conditions throughout the course of the experiment using our live cell imaging setup.
Cellular fractionation experiments with HL-1 cardiomyocytes incubated with the five proteins used in this study were performed to further pinpoint the subcellular location where the proteins internalize into cardiomyocytes. These experiments showed all light chain proteins in the nuclear/cellular debris fraction and both the soluble and insoluble lysosomal fractions. In order to determine that the fractionation yielded pure organelle fractions, western blots using anti-LAMP2 (lysosomal marker) and anti-lamin (nuclear marker) antibodies were performed. We observed reactivity to LAMP2 in all fractions and to lamin in both the insoluble lysosomal fraction and the nuclear/debris fraction. Both organelle markers showed the strongest reactivity to the fractions corresponding to the expected organelle fraction, indicating that while we accomplished some level of cellular fractionation, part of the fractions remained mixed (data not shown).
Light chains internalize into cardiomyocytes and traffic into lysosomes
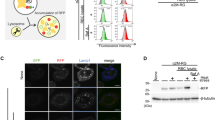
To determine the subcellular compartmentalization of the internalized light chain proteins, we utilized confocal microscopy and the organelle dyes specific for nucleus (DAPI) and lysosomal compartments (lysotracker). After 24 hours, OG-AL-09 was found to be internalized into the majority of the cardiomyocytes imaged. OG-I34N showed also most cells with internalized protein. OG-H87Y and OG-Q42K also showed internalization, but not to the same degree as OG-AL-09 (Figure 2).
Internalization in cardiomyocytes is observed at different degrees with 1 μM OG-labeled light chain proteins after 24 hours of incubation.
Cardiomyocytes incubated in the presence of different light chains were imaged using confocal microscopy. Oregon green labeled protein (green), lysotracker (red), DAPI (blue) and merged images show protein internalization in some cells at 24 hours. Scale bar is 50 μm.
At 24 hours, the cultures with the different proteins presented some level of variability. Cells incubated with OG-AL-09 and OG-I34N had the least variability in the percentage of cells with internalized protein (supplemental table 1). OG-κI O18/O8 displayed the greatest variability in both percentage of cells with internalized protein as well as lysosomal expansion (defined as a phenotypic transformation towards a more macrophage-type phenotype, with an increase in the overall volume of lysosomal compartments observed within one cell16) across multiple areas of a single cell culture time point (Figure 3). Cells treated with OG-κI O18/O8 after 48 hours of incubation formed clusters, each of which displayed increasing levels of lysosomal expansion compared to Q42K at 24 h (Figure 2).
OG-κI O18/O8 48 hour internalization is not uniform within a single sample and amongst cell populations within the same cell culture.
Different degrees of internalization and lysosomal expansion of OG-κI O18/O8 at the 48 hour time point. Cells showing little internalization are on the left and the degree of internalization increases going across to the right. Scale bar is 50 μm.
Optical sectioning of the cells and examination of the Z-series performed with all proteins at all time points confirmed colocalization of internalized proteins with lysosomes (Figure 4).
Cells incubated with OG-κI O18/O8 followed by confocal microscopy z-series showing that the protein is internalized into lysosomal compartments.
Panels corresponding to slices 9–18 of a 20 slice z-series of OG-κI O18/O8 internalization at 48 hours, verifying that the protein is in fact internalized. OG-κI O18/O8 (green), lysotracker (red), DAPI (blue) and merged images show internalization of OG-κI O18/O8 and colocalization with lysotracker. Scale bar is 50 μm.
After 48 hours (Figure 5), we observed an overall increase in lysosomal expansion for all proteins. However, there were still distinct sub-populations of cells with separate internalization kinetics. The lysosomal compartments congregated around the nucleus.
Increased protein internalization in cardiomyocytes with 1 μM OG-labeled light chain proteins after 48 hours of incubation.
Cardiomyocytes incubated in the presence of different light chains were imaged using confocal microscopy. Oregon green labeled protein (green), lysotracker (red), DAPI (blue) and merged images. Proteins colocalize with lysotracker. Scale bar is 50 μm.
Some light chain mutants present slow cellular internalization
At 72 hours (Figure 6), we observed mixed lysosomal expansion with OG-AL-09 and OG-I34N, with only a subset of cells with significant lysosomal expansion. At the same time, more cells incubated with OG-I34N showed significant increase in protein internalization compared to 48 hours. At 72 hours, OG-Q42K- and OG-H87Y-treated cells presented an internalization profile similar to the internalization profile observed for OG-AL-09 after 24 hours of incubation. At 72 hours, the number of cells with internalized protein has reached a plateau for most proteins and presented the smallest amount of variability (Figure 7).
Increased internalization in cardiomyocytes is observed with 1 μM OG-labeled light chain proteins after 72 hours of incubation.
Cardiomyocytes incubated in the presence of different light chains were imaged using confocal microscopy. Oregon green labeled protein (green), lysotracker (red), DAPI (blue) and merged images. The internalization displayed by each protein is stronger than at previous time points. Scale bar is 50 μm.
Percentage of cells with internalized protein as a function of time.
All proteins show an increase in cellular internalization as a function of time with large variability during the 24 h incubation time. Data was gathered from all the images taken at the different time points and have been displayed as a fraction of cells with internalized protein. Error bars correspond to the standard deviation. Supplementary table 1 shows the actual numbers of cells utilized for each time point and protein.
Light chain internalization of mutant light chains correlates with their rate of amyloid formation
The largest difference in protein internalization occurs at 24 hours, although there is large standard deviation among the different images analyzed and the data did not reach statistical significance (Figure 7). We have found a correlation between the percent of cells that internalize a given OG-labeled protein after 24 hours of incubation and the rate of amyloid fibril formation calculated for the corresponding VL protein (Figure 8). The correlation is not perfect given the fact that the percentage of cells with internalized protein do not show a linear correlation with regards to the mutations found in the proteins, but the values follow the same trend. The lysosomal area found in all the cells treated with the different proteins and characterized in supplemental table 1 was quantified (Figure 9). OG-κI O18/O8 after 24 h of incubation presents the largest variability in lysosomal area. At 48 h, OG-AL-09 presents the largest lysosomal area with a large variability between cells. The only significant differences are found between the lysosomal area of protein OG-H87Y and OG-Q42K at 24 h (p < 0.001). The interesting thing about OG-Q42K at 24 h is that this protein presents the smallest percentage of cells that internalize protein (supplementary table S1 online), but from those cells with internalized protein, all have a large lysosomal area comparable to those proteins with large lysosomal area or larger than other proteins.
Correlation between rate of fibril formation of VL protein and number of cells with internalized OG-labeled light chain protein after 24 hours of incubation.
The rate of fibril formation of VL proteins shows a strong correlation with the number of cells that have internalized light chains after 24 hours of incubation. The y axis corresponding to the fraction of cells with internalized protein is not extended to zero to highlight the correlation between these two parameters.
Quantification of lysosomal area over total number of cells as a function of incubation time.
OG-AL-09 presents the largest lysosomal area with a large variability between cells. The only significant differences are found between the lysosomal area of protein OG-H87Y and OG-Q42K at 24 h (p < 0.001). The lysosomal area was calculated using imageJ software working with the same images used to quantify cells with internalized protein in supplementary table 1. Error bars are the standard deviation of lysosomal area measured on at least three different images for each protein.
Cardiomyocytes live cell population remains fairly constant during light chain internalization experiments
We calculated the percentage of live cells in the presence of the different light chain proteins over time (Supplementary Fig. S1 online) to make sure that cell viability was comparable for all proteins within the time frame of the internalization experiments. We observed that over time, live cell population remained fairly constant with the exception of some sudden loss of cell viability with AL-09 at 24 hours and I34N at 12 hours. These cells recovered (probably due to cell growth) over time. Overall, the proteins at these concentrations with the growth conditions used for cellular internalization were not toxic enough to greatly disturb the cell populations within the 72 hours incubation time of this experiment.
Discussion
The main events in the pathophysiology of AL amyloidosis involve protein misfolding, amyloid formation and deposition in target organs, such as the heart, kidneys and liver. The current understanding of the mechanism that leads to organ damage in AL amyloidosis is limited. In the present study, we demonstrate that full length light chains are able to internalize into cardiomyocytes and co-localize with lysosomal compartments. OG-κI O18/O8, the germline protein without somatic mutations, showed the most diverse lysosomal expansion profile: from cells with no internalization and diffused lysosomal compartments to cells with large amounts of protein internalized and significant lysosomal expansion (Figure 3). The proteins OG-AL-09 and OG-I34N caused the greatest lysosomal expansion in the cell. Our current study follows up our earlier study where we describe the cytotoxicity of amyloidogenic light chains, utilizing HL-1 cardiomyocytes as our cell culture model15. The most cytotoxic protein found in our previous study (followed by cellular viability derived from MTT assay and caspase activity) was VL AL-09.
Lack of reversibility in our thermal denaturation experiments precluded us from calculating any thermodynamic parameters for the full length proteins used in this study. Previously our laboratory studied the thermodynamic stability and fibril formation properties of VL AL-09, κI O18/O8, AL-09 H87Y, I34N and Q42K17. We found that VL AL-09 and VL Q42K were the least thermodynamically stable proteins compared to VL κI O18/O8. VL I34N had intermediate restored stability, while VL H87Y was as stable as κI O18/O8. The kinetics of amyloid fibril formation (measured as the time needed to reach completion of the fibril formation reaction) followed a similar trend with the exception of VL Q42K that has low thermodynamic stability but presents delayed amyloid formation kinetics even compared to VL κI O18/O8. In this study, the percent of cells with internalized protein at 24 h correlates with the corresponding kinetics of VL amyloid formation (Figure 8), although it does not follow a linear trend. This would suggest that the somatic mutation K42Q (present in OG-AL-09, OG-H87Y and OG-I34N) not only promotes fibril formation but also increases the rate of cellular internalization. Our results suggest that thermodynamic stability and amyloid formation may not always correlate, but amyloid formation kinetics correlates with other properties associated with the misfolding phenotype, such as cellular internalization and lysosomal expansion. At 24 h the only protein that has formed amyloid fibrils in vitro is VL AL-0917. The other proteins included in this study presented delayed kinetics of amyloid formation compared to AL-09: VL I34N (72 h); VL κI O18/O8 (216 h); VL H87Y (264 h); and VL Q42K (336 h). While aggregation has not occurred within 24 h with any of these other proteins, we propose that at 24 h these proteins may already be populating species within the amyloid formation pathway that may play a role in internalization and possibly toxicity.
In general, we observed a heterogeneous response to protein exposure within cellular populations. This was most evident with cells incubated with OG-κI O18/O8 where subpopulations of cells show a wide range of protein internalization. We observed low to medium range level of heterogeneity with OG-Q42K and homogeneous levels observed with OG-AL-09. One way to explain the heterogeneous populations observed within one cell culture is based on the assumption that some cells reacted strongly to the presence of the protein while some did not. It is possible that the individual cells protein homeostasis status at the time when protein was added to the media defined their competency for internalization and subsequent phenotypic transformation18.
Over the course of the internalization experiment (72 hours), all of the restorative mutants would follow AL-09 trend of lysosomal expansion at different rates. Our results are in agreement with previous reports where an amyloidogenic light chain protein was internalized into renal mesangial cells creating divergent phenotypes, including lysosomal expansion16.
Some earlier observations indicated that human light chains are capable of forming amyloid fibrils upon incubation with lysosomal extracts and enzymes19,20. The only evidence we have of protein aggregation comes from our cellular fractionation data where a large amount of protein is found in the insoluble lysosomal extracts. At this point, we have no evidence that the light chains are aggregating as amyloid fibrils in the lysosomal compartments of HL-1 cardiomyocytes. The small amount of material recovered from these fractionation experiments precluded us from performing a more extensive characterization of possible aggregated material.
Nuclear localization was apparent in our live cell imaging (data not shown) but not very evident with our confocal microscopy experiments. It is possible that what we saw in live cell imaging was an expanded lysosomal compartment with perinuclear localization. The cellular fractionation data did not provide additional information due to the incomplete organelle fractionation we experienced. Alternatively, the additional stress derived from the constant exposure to light in the live cell imaging could have caused the nuclear localization of light chains as one of the ultimate signs of cellular damage. This suggests that nuclear localization is an alternative destination for misfolding-prone proteins, as has been shown for polyglutamine repeat proteins21,22,23. Further studies with organelle markers and additional stressors could help us elucidate if there are two parallel internalization pathways or if lysosomal localization is the only place where amyloidogenic light chains internalize.
Future internalization experiments with amyloid fibrils and multiple oligomerized states of these different proteins would shed more light on protein internalization and light chain induced cell death. Cellular internalization studies using VL proteins would help to understand the role of the constant domain in the internalization process. More extensive studies analyzing the early events in cellular internalization would also increase our understanding of the mechanism of endocytosis used by these proteins. For example, previous studies have shown that light chains internalize via a receptor-mediated mechanism into renal mesangial cells24 while fibroblasts internalize light chains via pinocytosis11. Our studies have been conducted using HL-1 cardiomyocytes, which are an immortalized mouse cell line derived from the AT-1 mouse atrial cardiomyocyte tumor lineage25,26. The HL-1 cell line was established from an AT-1 subcutaneous tumor excised from an adult female Jackson Laboratory-inbred C57BL/6J mouse. These cells can be serially passaged, yet they maintain the ability to contract and retain differentiated cardiac morphological, biochemical and electrophysiological properties26. Ultrastructural characteristics typical of embryonic atrial cardiac muscle cells were found consistently in the cultured HL-1 cells. Future studies using primary cardiomyocytes derived from adult mice (and eventually human cardiomyocytes) would be essential for comparison, so that we could determine whether light chain internalization characteristics are dependent on the type of protein, the type of cells, and/or the laboratory procedure from which the cells are derived.
Methods
Protein preparation
The VL sequences for κI O18/O8 and AL-09 have been deposited in GenBank under nucleotide accession numbers EF640313 and AF490909, respectively. There is only one κ CL sequence (protein accession number P01834). We mutated C214S at the end of the CL domain to avoid the formation of non-native disulfide bonds. The expression vectors for full length AL-09 H87Y, I34N and Q42K were generated from AL-09 vector via site directed mutagenesis. The expression vector pET12a containing the genes corresponding to the full length proteins were transformed into Escherichia coli Rosetta Gami cells and purified from inclusion bodies as described previously27. Protein purity was verified with SDS-PAGE. Circular dichroism spectroscopy (CD) was used to determine the secondary structure and the thermal stability of all full length proteins on a Jasco Spectropolarimeter 810 (JASCO Inc, Easton, MD) as described previously27. Protein identity was confirmed using mass spectrometry and western blots as previously reported14. Proteins were flash frozen with 10% glycerol (Sigma) and stored at −80°C. Whenever we use the names AL-09, κI O18/O8, H87Y, I34N and Q42K, we are referring to full length proteins.
Cell culture
Mouse HL-1 cardiomyocytes were obtained from Dr. William Claycomb25. These cells are designed to retain the phenotypic characteristics of adult cardiomyocytes in an immortalized cell line26. HL-1 cardiomyocytes were cultured as previously described using Claycomb media (SAFC Global)15,26. Supplemented Claycomb media contains 10% fetal bovine serum (Sigma), 1% L-glutamine (Invitrogen), 1% Penicillin/Streptomycin (Invitrogen), 1% norepinephrine and 0.2% Anti-fungizone (BAL).
For imaging experiments, cells were counted and added to 35 mm dishes with a 10 mm glass coverslip opening at the bottom (MatTek) at a concentration of 1 × 104 cells/mL. After four hours, supplemented Claycomb media was added for a total volume of 2 mL.
Confocal microscopy
An LSM 510 confocal microscope was used to image the cells with a 63× DIC lens (1.2 n.a.) using a water immersion objective. Laser wavelengths of 364, 488 and 543 nm were used. For z-series, 20 0.5 μm thick slices were taken. The images were captured using Zeiss LSM Image Version 3.2SP2. The images were prepared using ImageJ version 1.43 u.
Live cell imaging
Live cell imaging was performed on a 5 Live microscope (Zeiss) over a 24 hour period with a 63× DIC lens (1.4 n.a.) using an oil immersion objective. Images were taken every 5 minutes.
Cellular fractionation
Each protein was thawed and dialyzed for 10 minutes in 10,000 molecular weight cut off (MWCO) membrane against water to remove glycerol. Cells were grown at a concentration of 1 × 108 cells/mL on p100 plates (Fisher). After 24 hours, 1 μM protein was added to plates. Cells were homogenized and fractionated based on the protocol from Graham28.
Oregon green labeling
The proteins (20 μM) were thawed and dialyzed with a 10,000 MWCO membrane in water for 10 minutes to remove glycerol. The membrane was transferred to a phosphate buffer saline (PBS) solution for four hours. 1 mg of Oregon green (OG) 488 (Invitrogen) was solubilized in 100 μL DMSO and added to the sample for a desired degree of labeling of 4 molecules/protein. OG was incubated with the protein overnight at 4°C in the dark. Free OG was removed from the labeled protein solution by consecutive rounds of protein concentration and dilution with 10 mM Tris-HCl pH 7.4 through a 10,000 MWCO centrifugal filter.
To calculate the degree of labeling, the absorbance of the OG-labeled protein was measured at 280 and 496 nm. The absorbance of the protein was calculated with the equation:

The concentration of protein was then calculated from the absorbance using Lambert-Beer's equation. The degree of labeling in calculated by the following equation:

Protein internalization experiments
Cells were plated at a concentration of 1 × 104 cells/mL and allowed to grow overnight before the protein was added to the media. OG-labeled protein was added to the cells at a concentration of 1 μM in a total of 2 mL of supplemented Claycomb media. 2 hours before the desired time point, 1 μL of 1 mM lysotracker (Invitrogen) was added to 1 mL of unsupplemented Claycomb media and added to the cells. At the desired time points, cells were fixed using 4% formaldehyde and stained using 300 μM 4′, 6-diamidino-2-phenylindole (DAPI) (Invitrogen). 1 mL of Claycomb media was added to prevent dry out. Cells with OG, DAPI and lysotracker alone were imaged as controls. No spectral bleed-through was observed with the OG, DAPI and lysotracker alone control images. Absence of spectral bleed through is also evident by the existence of unique green labeling in most of our images (possibly reflect non-specific binding of OG-labeled protein and OG dye to the plasma membrane). Based on these controls, lysotracker and OG merged images (yellow from green and red merge) denote true colocalization of protein in lysosomal compartments.
Lysosomal expansion was calculated using ImageJ by calculating the total area labeled with lysotracker per cell. The average was calculated for all cells in an image and then divided by total number of cells in a given image. The images used for these calculations were the same images used to calculate the number of cells that had internalized protein (supplementary table 1).
Cellular viability
Live cells were counted using the Invitrogen Cell Countess. Briefly, cells were plated at a concentration of 5 × 104 cells/mL in 24-well plates (Corning). 1 μM protein was added after 24 h of culture. At the designated time points, cells were removed from plates using 0.05% Trypsin-EDTA (Invitrogen). Cells were pelleted and resuspended in 100 μL supplemented Claycomb media. 10 μL of resuspended cells were mixed with 10 μL of Trypan blue dye (Invitrogen) and the number of live and dead (blue) cells was obtained using the Countess system, following company protocol.
References
Falk, R. H., Comenzo, R. L. & Skinner, M. The systemic amyloidoses. N Engl J Med 337, 898–909 (1997).
Falk, R. H. Diagnosis and management of the cardiac amyloidoses. Circulation 112, 2047–2060 (2005).
Kieninger, B. et al. Amyloid in endomyocardial biopsies. Virchows Arch 456, 523–532 (2010).
Kumar, S. K. et al. Recent improvements in survival in primary systemic amyloidosis and the importance of an early mortality risk score. Mayo Clinic proceedings 86, 12–18 (2011).
Schroeder, H. W., Jr & Cavacini, L. Structure and function of immunoglobulins. J Allergy Clin Immunol 125, S41–52 (2010).
Olsen, K. E., Sletten, K. & Westermark, P. Fragments of the constant region of immunoglobulin light chains are constituents of AL-amyloid proteins. Biochem Biophys Res Commun 251, 642–647 (1998).
Vrana, J. A. et al. Classification of amyloidosis by laser microdissection and mass spectrometry-based proteomic analysis in clinical biopsy specimens. Blood 114, 4957–4959 (2009).
Klimtchuk, E. S. et al. The critical role of the constant region in thermal stability and aggregation of amyloidogenic immunoglobulin light chain. Biochemistry 49, 9848–9857 (2010).
Liao, R. et al. Infusion of light chains from patients with cardiac amyloidosis causes diastolic dysfunction in isolated mouse hearts. Circulation 104, 1594–1597 (2001).
Trinkaus-Randall, V. et al. Cellular response of cardiac fibroblasts to amyloidogenic light chains. Am J Pathol 166, 197–208 (2005).
Monis, G. F. et al. Role of endocytic inhibitory drugs on internalization of amyloidogenic light chains by cardiac fibroblasts. Am J Pathol 169, 1939–1952 (2006).
Brenner, D. A. et al. Human amyloidogenic light chains directly impair cardiomyocyte function through an increase in cellular oxidant stress. Circ Res 94, 1008–1010 (2004).
Shi, J. et al. Amyloidogenic light chains induce cardiomyocyte contractile dysfunction and apoptosis via a non-canonical p38alpha MAPK pathway. Proceedings of the National Academy of Sciences of the United States of America 107, 4188–4193 (2010).
Baden, E. M. et al. Altered dimer interface decreases stability in an amyloidogenic protein. J Biol Chem 283, 15853–15860 (2008).
Sikkink, L. A. & Ramirez-Alvarado, M. Cytotoxicity of amyloidogenic immunoglobulin light chains in cell culture. Cell Death Dis 1, e98 (2010).
Keeling, J., Teng, J. & Herrera, G. A. AL-amyloidosis and light-chain deposition disease light chains induce divergent phenotypic transformations of human mesangial cells. Lab Invest 84, 1322–1338 (2004).
Baden, E. M., Randles, E. G., Aboagye, A. K., Thompson, J. R. & Ramirez-Alvarado, M. Structural insights into the role of mutations in amyloidogenesis. J Biol Chem 283, 30950–30956 (2008).
Balch, W. E., Morimoto, R. I., Dillin, A. & Kelly, J. W. Adapting proteostasis for disease intervention. Science 319, 916–919 (2008).
Epstein, W. V., Tan, M. & Wood, I. S. Formation of "amyloid" fibrils in vitro by action of human kidney lysosomal enzymes on Bence Jones proteins. J Lab Clin Med 84, 107–110 (1974).
Tan, M. & Epstein, W. Polymer formation during the degradation of human light chain and Bence-Jones proteins by an extrct of the lysosomal fraction of normal human kidney. Immunochemistry 9, 9–16 (1972).
Iwata, A. et al. Increased susceptibility of cytoplasmic over nuclear polyglutamine aggregates to autophagic degradation. Proceedings of the National Academy of Sciences of the United States of America 102, 13135–13140 (2005).
von Mikecz, A. PolyQ fibrillation in the cell nucleus: who's bad? Trends Cell Biol 19, 685–691 (2009).
Yang, W., Dunlap, J. R., Andrews, R. B. & Wetzel, R. Aggregated polyglutamine peptides delivered to nuclei are toxic to mammalian cells. Hum Mol Genet 11, 2905–2917 (2002).
Teng, J. et al. Different types of glomerulopathic light chains interact with mesangial cells using a common receptor but exhibit different intracellular trafficking patterns. Lab Invest 84, 440–451 (2004).
White, S. M., Constantin, P. E. & Claycomb, W. C. Cardiac physiology at the cellular level: use of cultured HL-1 cardiomyocytes for studies of cardiac muscle cell structure and function. Am J Physiol Heart Circ Physiol 286, H823–829 (2004).
Claycomb, W. C. et al. HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proceedings of the National Academy of Sciences of the United States of America 95, 2979–2984 (1998).
McLaughlin, R. W., De Stigter, J. K., Sikkink, L. A., Baden, E. M. & Ramirez-Alvarado, M. The effects of sodium sulfate, glycosaminoglycans and Congo red on the structure, stability and amyloid formation of an immunoglobulin light-chain protein. Protein Sci 15, 1710–1722 (2006).
Graham, J. M. Isolation of lysosomes from tissues and cells by differential and density gradient centrifugation. Current protocols in cell biology Chapter 3, (2001).
Acknowledgements
We thank Amy Cadwallader, Laura Sikkink and Tanya Poshusta for assistance with the cloning of the expression vectors, Kristi Simmons, Eugenia Trushina and Jonathan S. Wall for their critical reading of the manuscript, Eugene Krueger and Janey Hsu for excellent technical assistance and the Ramirez-Alvarado laboratory for their assistance with this project. This project was supported by the National Institutes of Health R01 grant GM 071514, National Institutes of Health ARRA administrative supplement GM 071514S1, the Mayo Foundation and the generosity of amyloidosis patients and their families.
Author information
Authors and Affiliations
Contributions
M.R.A. designed the experiments, analyzed data and wrote the paper R.T.L. performed some experiments, analyzed data and assisted with the initial stages of the writing of the paper. O.O.O. performed some experiments, analyzed data and assisted with the writing of the paper. E.G.R. performed some of the initial experiments, analyzed data and assisted with the writing of the paper. K.G.H. performed some experiments. A.C.D.C. performed some experiments.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-ShareALike 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Levinson, R., Olatoye, O., Randles, E. et al. Role of mutations in the cellular internalization of amyloidogenic light chains into cardiomyocytes. Sci Rep 3, 1278 (2013). https://doi.org/10.1038/srep01278
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep01278
This article is cited by
-
Light chain amyloidosis induced inflammatory changes in cardiomyocytes and adipose-derived mesenchymal stromal cells
Leukemia (2020)
-
Solid-state NMR chemical shift assignments for AL-09 VL immunoglobulin light chain fibrils
Biomolecular NMR Assignments (2017)
-
Pathophysiology and treatment of cardiac amyloidosis
Nature Reviews Cardiology (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.