Abstract
Most of DNA strand exchange reactions in vitro are based on toehold strategy which is generally nonequilibrium and intracellular strand exchange mediated by proteins shows little sequence specificity. Herein, a new strand exchange promoted by equilibrious DNA conformational switching is verified. Duplexes containing c-myc sequence which is potentially converted into G-quadruplex are designed in this strategy. The dynamic equilibrium between duplex and G4-DNA is response to the specific exchange of homologous single-stranded DNA (ssDNA). The SER is enzyme free and sequence specific. No ATP is needed and the displaced ssDNAs are identical to the homologous ssDNAs. The SER products and exchange kenetics are analyzed by PAGE and the RecA mediated SER is performed as the contrast. This SER is a new feature of G4-DNAs and a novel strategy to utilize the dynamic equilibrium of DNA conformations.
Similar content being viewed by others
Introduction
DNA strand exchange is a vital biological process in homologous gene recombination, DNA replication and the repair of DNA damages1. The chromosomes are connected to one another through strand exchange during meiosis process2. Double-strand breaks in the replication of lesion-containing DNA are principally repaired through strand exchange reaction (SER)3. SER technologies have been applied in genetic diagnoses, such as single nucleotide polymorphism and gene-based physiological differences which response to human diseases4. In engineering filed, DNA SERs can be employed as an excellent principle in the designing of digital logical circuits. The strategy naturally incorporates biomolecules for large-scale circuitry, such as general debugging tools, parallel circuit preparation and a new abstraction hierarchy5. In the initial step of a strand exchange reaction, a single-stranded DNA (ssDNA) is recognized by its homologous double-stranded DNA (dsDNA). Then, a complementary strand is exchanged and paired with the ssDNA to form a heteroduplex product3.
In vitro, most of the SERs are based on the Toehold Exchange strategy6. Toehold constructs mediate fast and reversible strand displacement and they could be used in single-base discrimination at a suitable temperature7. In toehold systems, the displaced DNA strands are different from the target strands and newly formed hybrids tend to be more stable. However, the specificity of this strategy is compromised for long strands, except near the melting temperature7. DNA SERs can be stimulated by cationic comb-type copolymers through the formation of stable three-stranded intermediates8. Although this SER intermediate is accumulated as fast as 50000-folds at 37°C by the polymers and this protocol could be applied in genetic diagnoses, it shows little DNA sequence selectivity4. In vivo SERs are catalyzed by the RecA protein in Escherichia coli and Rad51 protein in eukaryotes9,10. Dmc1 and Rhp51, which belong to the RecA family, also show an efficient stimulation of SERs11,12. Proteins that mediate strand exchange possess an ATP-dependent activity, meaning that energy input is essential for SER. Recombinase-promoted SERs occur in a sequence-independent manner because the proteins can bind to ssDNA without selectivity, leading to the non-specificity of genetic recombination13.
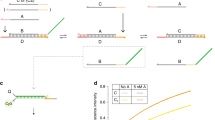
Another SER developed by Liu group employed the hidden toehold strategy showed an ATP responsive aspect. They constructed a DNA bulge-loop structure hidden among the duplex and this structure could be bound with ATP, leading to the structure alteration of the bulge-loop, which stimulated the strand exchange14. In this work, we presented an enzyme-free, physiological temperature strand exchange promoted by the conformational switching between duplexes and G-quadruplexes and this SER indicated an equilibrium state since the displaced ssDNAs were identical to the homologous ssDNAs. Which meant the reaction was maintained equilibrium throughout the process because the reactant DNAs was the same to the product DNAs. No ATP was needed in this type of SER and the stability of DNAs was not changed. The strand exchange specifically occurred in the presence of c-myc sequence and its mutated type mu-c-myc sequence (Figure 1A), which could form G-quadruplexes under the SER condition, while the random sequence with the same lengths were not exchanged (Figure 1B).
The SERs between dsDNA (label-free) and ssDNA (Cy5-labeled) in the presence of c-myc (A) and in the absence of c-myc (B).
The c-myc sequence is colored by green with its complementary strand, while the random sequence is colored by yellow. The blue indicates the same additional sequences for both A and B. The complementary strands (ssDNAs) are labeled by Cy5 at 5′ ends. The Cy5 labeled complementary strand of c-myc sequence is identical to the replaced single strand. The green cube indicates the G-quadruplex.
The c-myc sequence used in this paper was located in the promoter regions of c-MYC oncogenes. Human c-MYC was the second proto-oncogene to be identified and encoded a basic transcription factor (c-Myc protein). Three MYC homologues including Burkitt's lymphoma (c-MYC), neuroblastoma (N-MYC) and small cell lung cancer (L-MYC) were activated in over half of human cancers, where they regulated critical pathways that contribute to tumorigenesis15,16. The c-myc gene affected diverse cellular processes involved in cell growth, cell proliferation, apoptosis and cellular metabolism17. C-myc and mu-c-myc were G-rich regions of chromosomes that maintained dynamic equilibrium between duplexes and G-quadruplexes in presence of the complementary strands and these G-rich sequences were predominantly in duplex form since duplexes were more stable than G-quadruplexes18,19. This G-quadruplex-forming sequence could be folded into G-quadruplex both in vivo and in vitro, regulating up to 90% of total c-MYC transcription20. And this SER indicated a potential biological function of c-myc.
Results
C-myc promoted SERs
Two sets of DNA sequences, 24 bp, 48 bp, or 72 bp dsDNAs and 5′-Cy5-labeled 24 nt, 48 nt, 72 nt ssDNAs were employed in the c-myc promoted SERs. The ssDNAs were complementary to the strands containing c-myc and random sequences in the dsDNAs. One set had the c-myc sequence at the core region of the strands, while the other set had random sequence at the same position. The invading ssDNAs were Cy5 labeled, including the c-myc complementary sequences, c-myc sequences and random sequences. After the incubation of dsDNAs with homologous ssDNAs, we found that when the invading ssDNAs contained the complementary strands of c-myc, the dsDNAs with c-myc sequence were obviously exchanged, while the random dsDNAs were only weakly exchanged (Figure 2, left panel). Unexpectedly, when the invading strands contained c-myc sequences, the dsDNAs were not exchanged at all (Figure 2, right panel). These results were obtained in sodium phosphate buffer and analyzed by native PAGE. We altered the sodium phosphate buffer to TE (Tris-HCl, EDTA) and the similar SERs were observed (Figure S1). All the SER samples were incubated in TE buffer instead of sodium phosphate while other conditions were identical. We found that the c-myc containing dsDNAs were significantly exchanged, but the random sequences were weakly exchanged.
The SERs between dsDNAs and the homologous ssDNAs.
In left panel, the invading strands contain the complementary strand of c-myc. Lane 1: SERs of the random sequences; lane 2: SERs of the c-myc containing sequences; lane 3: dsDNAs without c-myc; lane 4: dsDNAs with c-myc; lane 5: ssDNAs without c-myc; lane 6: ssDNAs with c-myc-com. In right panel, the invading strands contain the c-myc sequence. Lane 1: SERs of the random sequences; lane 2: SERs of the c-myc containing sequences; lane 3: dsDNAs without c-myc; lane 4: dsDNAs with c-myc; lane 5: ssDNAs without c-myc; lane 6: ssDNAs with c-myc. All the SERs were performed at 20 μM dsDNA and 20 μM ssDNA, in the solution containing 200 mM KCl, 20 mM sodium phosphate buffer, pH 7.4. And the incubation times for 24 bp and 24 nt, 48 bp and 48 nt, 72 bp and 72 nt were 3 hours, 30 hours, 60 hours, respectively. The SERs systems were maintained at 37°C.
G-quadruplex formation in duplexes
We proposed that this new SER was triggered by the G-quadruplex formation in duplex DNA (Figure 1A). C-myc and the complementary sequence kept dynamic equilibrium between G-quadruplex and double strand, but the dsDNA was predominant. The c-myc containing dsDNA was simultaneously exchanged by the homologous ssDNA as soon as the formation of G-quadruplex.
The CD spectrum of c-myc and mu-c-myc sequences (24 nt single strands) was recorded in the presence of KCl (Figure S2), both of them presented a positive peak at around 260 nm and a negative peak at around 240 nm, indicated the formation of G-quadruplex21. In Figure S3 and Figure S4, the CD spectra of dsDNAs containing c-myc and mu-c-myc sequences and the random dsDNAs were recorded. It showed significant difference between the random sequences and the c-myc sequences. Then the CD curves of the latter were subtracted by the former, series CD spectra were obtained with a positive peak at around 260 nm and a negative peak at around 230 nm. They were similar to that of G-quadruplex formed by c-myc ssDNAs. These CD data might indicate the formation of transient G-quadruplex in dsDNAs.
To further confirm the formation of G-quadruplex in double stranded DNA, we employed the native PAGE containing 40% PEG200 to identify G-quadruplex (Figure 3). The dsDNAs containing random sequences and c-myc (2c-myc) sequences were Cy5 labeled and incubated in 40% PEG200 buffer. PEG200 created molecular crowding environment in the buffer and the molecular crowding was en essential factor for the formation of stable G-quadruplex in dsDNAs, since the transient G-quadruplex was not stable and quickly dissociated in normal buffer22. In the results, 24 bp, 48 bp and 72 bp, there were only dsDNA bands for random sequences, while there were obvious G-quadruplex bands for the c-myc sequences, regardless of the Cy5 labeled on the c-myc strands (Figure 3, lane 2) or the complementary strands (Figure 3, lane 3). These results indicated that the transient state G-quadruplex could be formed in normal buffer and stable G-quadruplex could be formed in 40% PEG buffer.
The native PAGE analysis of G-quadruplex formation in Cy5 labeled double stranded DNAs.
Lane 1: dsDNAs of random sequences; lane 2: dsDNAs containing Cy5 labeled c-myc complementary strands; lane 3: dsDNAs containing Cy5 labeled c-myc strands; lane 4: ssDNAs of random sequences; lane 5: ssDNAs of c-myc complementary strands; lane 6: ssDNAs of c-myc strands. 20% PAGE was employed with the addition of 40% PEG200 (v/v), all the samples were incubated in 200 mM KCl, 20 mM sodium phosphate buffer, pH 7.4. And the incubation times for 24 bp and 24 nt, 48 bp and 48 nt, 72 bp and 72 nt were 3 h, 30 h, 60 h, respectively. The incubation temperature was 37°C. The G-quadruplexes formed in duplexes were labeled by red panes.
Kenetics of the c-myc promoted SERs
To further investigate the kinetics of c-myc stimulated SERs, we measured the concentrations of SER products along with increased time under the identical incubated condition. The incubation times of the 24 bp and 24 nt, 48 bp and 48 nt and 72 bp and 72 nt were increased from 0 to 7 hours, 33 hours and 72 hours at 37°C, respectively. Then, native PAGE was employed to analyze the SER products (Figure S5). The bands were integrated to calculate the SER product concentrations based on the dsDNA markers. All of the kinetics experiments were repeated at least 3 times and the averages were reported (Figure 4). The product concentrations were gradually increased along with the time increase about the c-myc containing sequences, while the random sequences were hardly exchanged. The difference of the SER product concentrations and the SER rate between the c-myc sequences and the random sequences showed a manner of strand length dependent. The probability of the formation of secondary structures for random sequence ssDNAs which could hinder the strand exchange rate was excluded. The Tm results for all the random ssDNAs showed no obvious values and were flat (Figure S6). And the denaturing PAGE gel was employed to analyze the mobility of the Cy5 labeled complementary single stranded DNAs (Figure S7). The differences between the ssDNAs with the same lengths were observed under the denatured condition, similar with the native PAGE gel results.
The kenetics results of the SERs extracted from the PAGE gel electrophoresis.
All the SER samples were adjusted to 20 μM dsDNA and 20 μM ssDNA, buffered by 200 mM KCl, 20 mM sodium phosphate, pH 7.4 and the systems were maintained at 37°C. The incubation times of 24 bp and 24 nt (A), 48 bp and 48 nt (B), 72 bp and 72 nt (C) were gradiently increased from 0 to 7 h, 33 h, 72 h, with the increments of 1 h, 3 h, 3 h, respectively. And the SERs product concentrations were recorded versus time increasing, based on DNA markers.
Tm values of dsDNAs
To confirm the conformational switching between duplex and G-quadruplex leading to the specific strand exchange, we excluded the decreased stability of dsDNAs containing c-myc. We tested the Tm values of each duplex at 200 mM KCl and 20 mM Na2HPO4/NaH2PO4 (pH 7.4). The Tm for: random 24 bp, Tm = 74.5°C; c-myc 24 bp, Tm = 75.5°C; random 48 bp, Tm = 78.5°C, c-myc 48 bp, Tm = 83.3°C; random 72 bp, Tm = 80.7°C, c-myc 72 bp, Tm = 86.2°C (Figure S8). These results indicated that the double strands with c-myc were even more stable then strands with the random sequence; this additional stability was produced by the G-rich c-myc sequence in the presence of the C-rich complementary strands. This indicated that the specific SERs were not caused by unstable dsDNAs.
Considering the Tm values of dsDNAs might be too high to evaluate correctly, we used low salt concentration buffer to perform the melting experiments. All the dsDNAs and SER samples including random sequences, c-myc sequences, mu-c-myc sequences, mis-c-myc sequences and 2c-myc sequence were buffered by 20 mM KCl, 20 mM NaCl and 20 mM TE, pH 7.4. The normalized UV-melting experiments were carried out (Figure S9) and the Tm values were obtained (Table 1). At low salt conditions, the Tm values of c-myc sequences were still higher than that of random sequences. The Tm values of SER samples (dsDNAs added exchanged ssDNAs) were the similar with the dsDNAs and the Tm values of mu-c-myc sequences were the similar with that of c-myc sequences. 2c-myc 72 bp Tm was much higher than c-myc 72 bp. While the Tm values decreased much when there was 1 mismatch in the dsDNAs (mis-c-myc sequences).
The various SERs
The main SER expriments were performed in the buffer of 200 mM KCl and 20 mM Na2HPO4/NaH2PO4 (pH 7.4). To investigated the effect of monovalent cations and crowding agents on the c-myc promoted SERs, series of SERs were carried out at only 5 mM TE, 5 mM Li+, 5 mM Na+, 5 mM K+, 5 mM K+ in 40% PEG200 (crowding agent), the incubation time and temperature were the same to the main SERs (Figure 5A). And the SER product concentrations were presented as histograms (Figure 5B, C, D). We found that the monovalent cations and the PEG200 significantly affected the SERs. In the c-myc sequences, only TE buffer, 5 mM Li+ and 5 mM Na+ had little effect on the SERs, while 5 mM K+ significantly promoted the SERs. When the SER buffer contained 5 mM K+ and 40% PEG200, both the c-myc sequences and random sequences were obviously exchanged.
The SERs performed in various conditions.
(A) The native PAGE analysis of the sequences with or without c-myc; lane 1: SERs of random sequences, lane 2: SERs of c-myc sequences, both under the condition of 5 mM TE; lane 3: SERs of random sequences, lane 4: SERs of c-myc sequences, both under the condition of 5 mM TE, 5 mM Li+; lane 5: SERs of random sequences, lane 6: SERs of c-myc sequences, both under the condition of 5 mM TE, 5 mM Na+; lane 7: SERs of random sequences, lane 8: SERs of c-myc sequences, both under the condition of 5 mM TE, 5 mM K+; lane 9: SERs of random sequences, lane 10: SERs of c-myc sequences, both under the condition of 5 mM TE, 5 mM K+ and 40% PEG200; lane 11: dsDANs markers of random sequences, lane 12: dsDNAs markers of c-myc sequences, both under the condition of 5 mM TE. All the samples pH were 7.4, the SERs times were 3 h, 30 h, 60 h for 24 bp and 24 nt, 48 bp and 48 nt, 72 bp and 72 nt, respectively. And the system temperature was maintained at 37°C. (B), (C), (D) The histograms of the SERs product concentrations of 24 bp and 24 nt, 48 bp and 48 nt, 72 bp and 72 nt, respectively.
The main SERs were performed with single c-myc, double c-myc (2c-myc 72 bp, 72 nt) was used as a control (Figure S10 A). This SER was carried out at 200 mM KCl, 20 mM sodium phosphate, pH 7.4, incubated at 37°C for 60 h. The histogram (Figure S10 B) indicated that both single and double c-myc significantly promoted the SERs and the efficiency of double c-myc was a little higher than that of single c-myc.
Another control experiment was performed by single base mutation in c-myc (mu-c-myc, Table S1). Although one of the G bases in c-myc was mutated into A, the mu-c-myc single-stranded DNA formed a G-quadruplex, similar to c-myc under the condition of 200 mM KCl and 20 mM Na2HPO4/NaH2PO4 (pH 7.4). After incubation with homologous ssDNAs, the mu-c-myc-containing dsDNAs presented a similar stimulation of strand exchange with the c-myc sequence (Figure S11). The c-myc-promoted SERs were compared with another type of strand exchange, which was caused by single base mismatch (Figure S12). We used the strands containing the single-base-mutation (mu-c-myc) sequence to pair with the normal complementary strands and then determined whether the mis-dsDNAs were exchanged with Cy5-labeled strands containing the complementary strand of mu-c-myc. Interestingly, the c-myc-promoted SERs were as efficient as the SERs involving the single-base-mismatch.
The possibility of the dissociation of double strands caused by the c-myc sequence was also excluded, by using circular dichroism (CD) to monitor the dsDNA signals (Figure S3, Figure S4). The CD spectra demonstrated that both the duplex with or without c-myc sequence existed as whole and stable double strands. Similar results were observed via PAGE. As shown in lane 3 and lane 4 of Figure 2, both were stable double strands, without any dissociation. The dsDNAs including c-myc sequences and random sequences were both in duplex DNA formation (Figure S13). The random dsDNAs and c-myc dsDNAs were analyzed by native PAGE and stained by EB with DNA markers. Both bands were the same with the same bp, indicating that the random dsDNAs and c-myc dsDNAs were in duplex formation.
RecA mediated SERs
RecA mediated SERs of the c-myc containing sequences and the random sequences were performed as the contrast of the conformational switching promoted SERs, since RecA was reported to stimulate the SERs23. The SERs were carried out in 1X RecA reaction buffer, 70 mM Tris-HCl, 10 mM MgCl2, 5 mM DTT, pH 7.5. And 3 mM ATP was added as the energy supply, because the release of the heteroduplex from the protein-DNA complexes needed ATP24. The RecA stimulated SERs presented no obvious DNA sequence selectivity, as described previously (Figure 6A). Both the c-myc containing sequences and the random sequences were significantly exchanged in the presence of RecA protein. From the gel results and the histogram (Figure 6B), we found that the SERs were promoted by both protein and DNA conformational switching. The product bands showed little difference between the dsDNAs with and without c-myc sequence. And the protein mediated SER product concentration decreased with the increase of DNA strand length. The RecA mediated SERs in the absence of ATP were carried out as the contrast, all the SER conditions were the same except the addition of ATP (Figure S14). As a result, no SER product was observed in the contrast.
(A) The SERs between dsDNAs and Cy5-labeled ssDNAs mediated by Rec A. Lane 1: random 24 bp, nt; lane 2: c-myc 24 bp, nt; lane 3: random 48 bp, nt; lane 4: c-myc 48 bp, nt; lane 5: random 72 bp, nt; lane 6: c-myc 72 bp, nt. (B) The histogram of protein mediated SERs product concentrations extracted from PAGE results.
Discussion
As shown in Figure 2 and Figure S11, the SERs between the dsDNAs and Cy5-labeled ssDNAs of c-myc (mu-c-myc) and random sequence containing DNAs showed significant differences under identical incubation conditions. The c-myc sequence effectively promoted the SERs compared with the random sequence. These SERs occurred spontaneously without the catalysis of recombination proteins, although the 72 bp duplex was much stable during the incubation process. Both the dsDNA markers with and without c-myc sequence maintained duplexes under the incubated condition and the duplexes were more stable than G-quadruplexes. Considering the dynamic equilibrium between the duplex and G-quadruplex, this dynamic process was most likely a response to the specific SERs. This is the first observation of DNA strand exchange promoted by conformational switching between duplex and G-quadruplex.
C-myc was G-rich sequence that potentially formed into G-quadruplex in presence of the complementary strand, even in long double stranded DNA (Figure 1). The G-quadruplex formed in duplexes was in transient state, not stable and quickly converted into duplexes. The complementary strand of c-myc was in loop when the transient G-quadruplex was formed. Then the homologous ssDNA was exchanged simultaneously to afford the SER product. Although the transient G-quadruplex in duplex was difficult to be observed, it could be fixed into stable structure in the molecular crowding buffer and then be distinguished from duplexes in PAGE analysis.
The concentrations of SER products showed time-dependent patterns in the kenetics results. The concentrations of the SER products demonstrated exponential growth with increased time, consistent with a previous report25. These results showed obvious differences between the sequences containing c-myc and random base pairs with the same oligonucleotide lengths. The random 24 bp and 24 nt also showed a certain amount of exchange, which could have been achieved through the sequential displacement pathway stimulated by incubation temperature26. However, the 24 bp duplex containing c-myc showed much higher exchange efficiency. The SER products of 48 bp and 48 nt, 72 bp and 72 nt showed high specificity with or without the c-myc sequence. Considering the random sequences might be formed into secondary structures, a denaturing PAGE gel of the complementary strands (ssDNAs) was carried out. The mobility of the ssDNAs in denaturing gel was very similar to that of native gel, indicating the different mobility of the same strand length might be correlated with DNA sequences. The insertion of the c-myc sequence into dsDNAs significantly promoted the homologous ssDNA exchange, while the exchange of dsDNA with random sequence was near zero. But in toehold and poly-cations stimulation strategy, it was hard to control the specificity and these protocols were usually used in short oligonucleotides.
The Tm values of c-myc containing sequences were higher than random sequences resulted from the much higher content of GC base pairs of the former. But this more stable dsDNAs were even more facile to be exchanged than random sequences. The CD spectra demonstrated both the c-myc containing and random sequences were double-stranded DNAs without any dissociation. The displaced single-stranded DNAs were identical to the homologous DNAs, so the c-myc promoted SER products were the same to the reactant DNAs. The whole SER processes were at equilibrium state and no ATP was needed. This SER was carried out at physiological temperature (37°C) and achieved a high sequence specificity compared with the toehold strategy and the protein mediated SERs.
Various monovalent cations were used in the buffers to carry out the c-myc promoted SERs, the results indicated that the cations significantly affected the SER efficiency. C-myc sequence was not able to form stable G-quadruplex in only TE buffer, Li+ buffer or Na+ buffer, while K+ ion increased the stability of G-quadruplex. Then the SER product concentrations in K+ buffer were much higher than that of the other ions. These also verified the G-quadruplex formation induced by K+ in duplexes. PEG200 was used as the crowding agent, it raised the SER product concentrations of both the c-myc sequences and random sequences, since the crowding agents significantly decreased the stability of all duplexes (Figure S15)22. The Tm values of the dsDNAs obtained in the buffer containing 40% PEG200 showed dramatic decrease compared with the results in Figure S9. And this decreased stability of duplexes led to the significant strand exchange of random sequences.
Double c-myc was used as a control of the SER, but the SER product concentration was increased only a little. These indicated that the SER was affected by the stability of G-quadruplex rather then other factors. The SERs promoted by c-myc and mu-c-myc were very similar because they possessed the similar ability to form G-quadruplex and both the duplex Tm values were the same. But when the mu-c-myc duplexes contained a mismatch in the strands (mis-c-myc), the SER efficiency was much higher than the c-myc sequences. This SER was promoted by the instability of duplexes resulted from the unpaired bases. The possibility of SER process via triplex formation was also excluded. The Tm values of SER samples (dsDNAs + ssDNAs) were compared with the duplexes (dsDNAs) and the results were almost the same (Table 1, Figure S9). If the triplex DNAs were formed in the SER process, the Tm values of SER samples would be increased. Moreover, no triplex DNA was observed in all the PAGE expriments, indicated that the SER process was not via triplex formation27.
The RecA protein was able to bind any single stranded DNA, regardless of sequence identity and then pair with the homologous dsDNA to generate heteroduplexes. In this process, the release of RecA from the heteroduplex product required ATP hydrolysis28. A contrast was performed in the absence of ATP and no SER product was observed, demonstrating the energy requirement in protein mediated SERs. These protein-mediated SERs required energy input. In contrast, the SERs promoted only by equilibrious conformational switching showed high DNA sequence specificity and this exchange could occur spontaneously without ATP. These results indicated that the SERs promoted by c-myc were both highly specific and highly efficient and that the SERs were stimulated by a DNA conformational switch.
The present study demonstrated a new DNA strand exchange reaction promoted by c-myc sequence through DNA conformational switching. The SER was enzyme-free and equilibrium state. No recombinase or ATP was needed for this SER and the displaced strands were identical to the homologous strands. The c-myc-promoted strand exchange was highly specific and at physiological temperature compared with protein mediated SERs and toehold strategy. This is another new feature of G-quadruplex. Finally, this DNA conformational switching stimulated SER might be a nature process in vivo and biological significance.
Methods
Nucleic acids
33 oligonucleotides were designed, including 15 with c-myc sequence, 9 with mu-c-myc sequence and 9 with random sequences. The sequences of oligonucleotides were presented in Table S1. Lengths of the oligonucleotides were 24 nt, 48 nt, 72 nt; the 24 nt was the core region of 48 nt and 72 nt and the rest parts of them were the same random sequences. Every oligonucleotide with c-myc sequence had the relative random sequence as the contrast. The displaced single-stranded DNAs were Cy5 labeled or label free. The Cy5 labeled oligonucleotides were purchased from TaKaRa (Dalian) and the label free oligonucleotides were purchased from Invitrogen (Shanghai).
Preparation of SERs samples
Every oligonucleotide was quantified by measuring the UV absorbance at 260 nm. 1 M LiCl, 1 M NaCl, 1 M KCl, 1 M TE (Tris-HCl, EDTA, pH 7.4) and 1 M Na2HPO4/NaH2PO4 (pH 7.4) were used as the storage buffer solutions. In the label-free double-stranded DNAs, the complementary strands of the c-myc containing strands and random strands were 20% excess (1.2 N), when the invading strands (Cy5 labeled ssDNAs) contained the complementary strands of c-myc; while the c-myc containing strands were 20% excess (1.2 N) when the invading strands (Cy5 labeled ssDNAs) contained the c-myc sequence. The Cy5-labeled dsDNAs markers were 20% excess (1.2 N) strands with or without c-myc paired with their Cy5-labeled complementary strands. DNAs were annealed at 95°C for 5 min and then slowly cooled to room temperature over 5 hours, buffered by 20 mM Na2HPO4/NaH2PO4 or 20 mM TE (pH 7.4).
CD spectra and Tm values
The CD spectra and melting temperature (Tm) measurements of dsDNAs and ssDNAs were taken by Chirascan CD Spectrometer (Applied Photophysics). DNA samples were buffered by 200 mM KCl, 20 mM Na2HPO4/NaH2PO4 (pH 7.4); 20 mM KCl, 20 mM NaCl, 20 mM TE, pH 7.4. The CD spectra and Tm values of 24 bp and 24 nt, 48 bp and 48 nt, 72 bp and 72 nt were recorded at 5 μM, 2 μM, 2 μM, respectively. Each dsDNA and ssDNA was melted with the temperature increasing and the UV datas were recorded at 260 nm.
SER processes
Each dsDNA (label free) was adjusted to 20 μM and the related ssDNA (Cy5 labeled) was added at the same concentration. In the buffer of 200 mM KCl, 20 mM Na2HPO4/NaH2PO4 (pH 7.4), the incubation was carried out at 37°C with variety time. The incubation times of 24 bp and 24 nt, 48 bp and 48 nt, 72 bp and 72 nt were gradiently increased from 0 to 7 h, 33 h, 72 h, respectively, with the increments of 1 h, 3 h, 3 h. In the buffer of 200 mM KCl, 20 mM TE (pH 7.4), the incubation times of 24 bp and 24 nt, 48 bp and 48 nt, 72 bp and 72 nt were 3 h, 30 h, 60 h, respectively. After that, the SERs were stopped by cooling to 4°C and then the samples were added bromophenol blue as loading buffer. 20% PAGE gel was employed to electrophoresed the samples at 70 V voltage for 8 h. The SERs products were scanned and integrated by Pharos FX Molecular Imager (Bio-Rad). All those gels were scanned with fluorescent detector except the gel stained by EB in Figure S13. And all the SERs product concentrations were calculated based on the dsDNA markers (20 μM).
Various SERs
The main SERs were the invading ssDNAs (Cy5 labeled ssDNAs) containing the complementary strands of c-myc, while the SERs in which the invading ssDNAs containing the c-myc (G4) sequence were also performed under the same conditions. The G-quadruplex formation in duplex DNA was analyzed by native PAGE containing 40% PEG200. The Cy5 labeled dsDNAs 24 bp and 24 nt, 48 bp and 48 nt, 72 bp and 72 nt were adjusted to 20 μM, buffered by 200 mM KCl, 20 mM soudium phosphate, 40% PEG200, pH 7.4, incubated for 3 h, 30 h, 60 h, respectively. Then PAGE containing 40% PEG200 was employed to analyze the DNA samples at 70 V voltage for 15 h.
The mutated c-myc (mu-c-myc) promoted SERs and mismatch c-myc (mis-c-myc) induced SERs were all carried out at the same conditions as the main SERs. The 2c-myc sequence (72 nt) contained two c-myc sequence that could potentially form two G4 in the duplex DNA, leading to the SER of the complementary strand. This SER was performed at 20 μM dsDNA and 20 μM, in the buffer of 200 mM KCl, 20 mM soudium phosphate, pH 7.4. The samples were incubated at 37°C for 60 hours.
The series of SERs in various monovalent cations (5 mM TE, 5 mM Li+, 5 mM Na+, 5 mM K+, 5 mM K+ in 40% PEG200) were performed under the condition of 5 mM TE, pH 7.4. The incubation times form 24 bp and 24 nt, 48 bp and 48 nt, 72 bp and 72 nt were 3 h, 30 h, 60 h, respectively. Every SER samples were performed at least 3 times and the average values were obtained.
Protein mediated SER
RecA protein was purchased from New England Biolabs (Lot: 0011202). The RecA mediated SERs were performed at 10 μM dsDNAs, 10 μM ssDNAs and 5 μM RecA, in the buffer containing 70 mM Tris-HCl, 10 mM MgCl2, 5 mM DTT, pH 7.5. 3 mM ATP was added as the energy supply. RecA was incubated with ssDNAs for 10 min and then the homologous dsDNAs were added for further 10 min incubation at 37°C. The SERs were stopped by addition of 5% SDS and 100 mM EDTA. 5× bromophenol blue was used as loading buffer, then samples were analysed by 12% PAGE gel at 70 V voltage for 8 h. The control experiment of RecA mediated SERs was carried out at the same condition in the absence of ATP. And the results were analyzed by Pharos FX Molecular Imager (Bio-Rad).
References
Eggleston, A. K., Mitchell, A. H. & West, S. C. In Vitro Reconstitution of the Late Steps of Genetic Recombination in E. coli. Cell 89, 607–617 (1997).
Neale, M. J. & Keeney, S. Clarifying the mechanics of DNA strand exchange in meiotic recombination. Nature 442, 153–158 (2006).
Ragunathan, K., Joo, C. & Ha, T. Real-Time Observation of Strand Exchange Reaction with High Spatiotemporal Resolution. Structure 19, 1064–1073 (2011).
Kim, W. J., Sato, Y., Akaike, T. & Maruyama, A. Cationic comb-type copolymers for DNA analysis. Nat. Mater. 2, 815–820 (2003).
Qian, L. & Winfree, E. Scaling Up Digital Circuit Computation with DNA Strand Displacement Cascades. Science 332, 1196–1201 (2011).
Zhang, D. Y. & Winfree, E. Control of DNA Strand Displacement Kinetics Using Toehold Exchange. J. Am. Chem. Soc. 131, 17303–17314 (2009).
Zhang, D. Y., Chen, S. X. & Yin, P. Optimizing the specificity of nucleic acid hybridization. Nat. Chem. 4, 208–214 (2012).
Kim, W. J., Akaike, T. & Maruyama, A. DNA Strand Exchange Stimulated by Spontaneous Complex Formation with Cationic Comb-Type Copolymer. J. Am. Chem. Soc. 124, 12676–12677 (2002).
Gupta, R. C., Golub, E. I., Wold, M. S. & Radding, C. M. Polarity of DNA strand exchange promoted by recombination proteins of the RecA family. Proc. Natl. Acad. Sci. USA 95, 9843–9848 (1998).
New, J. H., Sugiyama, T., Zaitseva, E. & Kowalczykowski, S. C. Rad52 protein stimulates DNA strand exchange by Rad51 and replicationproteinA. Nature 391, 407–410 (1998).
Sehom, M. G., Sigurdsson, S., Bussen, W., Unger, V. M. & Sung, P. Human meiotic recombinase Dmc1 promotes ATP-dependent homologous DNA strand exchange. Nature 429, 433–437 (2004).
Murayama, Y., Kurokawa, Y., Mayanagi, K. & Iwasaki, H. Formation and branch migration of Holliday junctions mediated by eukaryotic recombinases. Nature 451, 1018–1022 (2008).
Nara, T., Hamada, F., Namekawa, S. & Sakaguchi, K. Strand Exchange Reaction in Vitro and DNA-Dependent ATPase Activity of Recombinant LIM15/DMC1 and RAD51 Proteins from Coprinus cinereus. Biochem. Biophys. Res. Commun. 285, 92–97 (2001).
Xing, Y., Yang, Z. & Liu, D. A Responsive Hidden Toehold To Enable Controllable DNA Strand Displacement Reactions. Angew. Chem. Int. Ed. 50, 11934–11936 (2011).
Rounbehler, R. J. et al. Tristetraprolin Impairs Myc-induced Lymphoma and Abolishes the Malignant State. Cell 150, 563–574 (2012).
Murphy, M. J., Wilson, A. & Trumpp, A. More than just proliferation: Myc function in stem cells. TRENDS in Cell Biology 15, 128–137 (2005).
Boxer, L. M. & Dang, C. V. Translocations involving c-myc and c-myc function. Oncogene 20, 5595–5610 (2001).
Phan, A. T. & Mergny, J. L. Human telomeric DNA: G-quadruplex, i-motif and Watson–Crick double helix. Nucleic Acids Res. 30, 4618–4625 (2002).
Neaves, K. J., Huppert, J. L., Henderson, R. M. & Edwardson, J. M. Direct visualization of G-quadruplexes in DNA using atomic force microscopy. Nucleic Acids Res. 37, 6269–6275 (2009).
Gallo, A. et al. Structure of Nucleophosmin DNA-binding Domain and Analysis of Its Complex with a G-quadruplex Sequence from the c-MYC Promoter. J. Biol. Chem. 287, 26539–26548 (2012).
Mathad, R. I., Hatzakis, E., Dai, Z. & Yang, D. c-MYC promoter G-quadruplex formed at the 5′-end of NHE III 1 element: insights into biological relevance and parallel-stranded G-quadruplex stability. Nucleic Acids Res. 39, 9023–9033 (2011).
Zheng, K., Chen, Z., Hao, Y. & Tan, Z. Molecular crowding creates an essential environment for the formation of stable G-quadruplexes in long double-stranded DNA. Nucleic Acids Res. 38, 327–338 (2009).
Singh, P., Patil, K. N., Khanduja, J. S., Kumar, P. S., Williams, A., Rossi, F., Rizzi, M., Davis, E. O. & Muniyappa, K. Mycobacterium tuberculosis UvrD1 and UvrA Proteins Suppress DNA Strand Exchange Promoted by Cognate and Noncognate RecA Proteins. Biochemistry 49, 4872–4883 (2010).
Heijden, T., Modesti, M., Hage, S., Kanaar, R., Wyman, C. & Dekker, C. Homologous Recombination in Real Time: DNA Strand Exchange by RecA. Mol. Cell. 30, 530–538 (2008).
Choi, S. W., Kano, A. & Maruyama, A. Activation of DNA strand exchange by cationic comb-type copolymers: effect of cationic moieties of the copolymers. Nucleic Acids Res. 36, 342–351 (2007).
Reynaldo, L. P., Vologodskii, A. V., Neri, B. P. & Lyamichev, V. I. The Kinetics of Oligonucleotide Replacements. J. Mol. Biol. 297, 511–520 (2000).
Taniguchi, Y. & Sasaki, S. An efficient antigene activity and antiproliferative effect by targeting the Bcl-2 or surviving gene with triplex forming oligonucleotides containing a W-shaped nucleoside analogue (WNA-βT). Org. Biomol. Chem. 10, 8336–8341 (2012).
Volodin, A. A., Bocharova, T. N., Smirnova, E. A. & Camerini-Otero, R. D. Reversibility, Equilibration and Fidelity of Strand Exchange Reaction between Short Oligonucleotides Promoted by RecA Protein from Escherichia coli and Human Rad51 and Dmc1 Proteins. J. Biol. Chem. 284, 1495–1504 (2009).
Acknowledgements
The authors thank the National Basic Research Program of China (973 Program)(2012CB720600, 2012CB720603), the National Science of Foundation of China (No. 91213302, 30973605), the National Grand Program on Key Infectious Disease (2012ZX10003002-014). Supported by Program for Changjiang Scholars and Innovative Research Team in University (IRT1030).
Author information
Authors and Affiliations
Contributions
Z.G.W. and X.Z. designed experiments and wrote the manuscript; Z.G.W. carried out most experiments. All authors discussed the results.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Wu, Z., Xie, X., Li, P. et al. Equilibrious Strand Exchange Promoted by DNA Conformational Switching. Sci Rep 3, 1121 (2013). https://doi.org/10.1038/srep01121
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep01121
This article is cited by
-
DNA Self-assembly Catalyzed by Artificial Agents
Scientific Reports (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.