Abstract
Arthropods from four genetically modified (GM) maize hybrids (coleopteran resistant, coleopteran and lepidopteran resistant, lepidopteran resistant+herbicide tolerant and coleopteran resistant and herbicide tolerant) and non-GM varieties were sampled during a two-year field assessment. A total number of 363 555 arthropod individuals were collected. This represents the most comprehensive arthropod dataset from GM maize, and together with weed data, is reasonable to determine functional groups of arthropods and interactions between species. Trophic groups identified from both phytophagous and predatory arthropods were previously considered non-target organisms on which possible detrimental effects of Bacillus thuringiensis (Bt) toxins may have been directly (phytophagous species) or indirectly (predators) detected. The high number of individuals and species and their dynamics through the maize growing season can predict that interactions are highly correlational, and can thus be considered a useful tool to assess potential deleterious effects of Bt toxins on non-target organisms, serving to develop biosafety risk hypotheses for invertebrates exposed to GM maize plants.
Design Type(s) | randomized complete block design • genetic modification design |
Measurement Type(s) | Arthropoda • animal damage • weed coverage |
Technology Type(s) | animal trap • visual observation method |
Factor Type(s) | toxin • animal damage resistance • genetic transformation • biological replicate |
Sample Characteristic(s) | chernozem |
Machine-accessible metadata file describing the reported data (ISA-Tab format)
Similar content being viewed by others
Background & Summary
This works presents the data underlying the studies in published papers1–3. In the USA, genetically modified (GM) maize was grown on 38 M ha in 2016 (ref. 4). Throughout EU countries maize is one of the major crops and is cultivated annually on approximatively 13 million hectares, representing 13% of the total cultivated area in the EU and about 8% of maize production area worldwide5. The two most potent insect pests of non-GM maize crops in Europe are the western corn rootworm (Diabrotica virgifera virgifera LeConte, Coleoptera: Chrysomelidae) and the European corn borer (Ostrinia nubilalis Hübner, Lepidoptera: Crambidae), the control of which represents the greatest challenge in European maize production6. Other insects, such as aphids (Aphidoidea) and thrips (Thysanoptera), also occur in all maize croplands and may occasionally contribute to crop losses7–10. Several arthropod predators preying on these and other arthropod herbivores are also found in maize croplands, the most important of which are ground beetles (Coleoptera: Carabidae), ladybird beetles (Coleoptera: Coccinellidae), rove beetles (Coleoptera: Staphylinidae), predatory thrips (Aleolothripidae), wolf spiders (Areneae: Lycosidae), Syrphid larvae (Diptera: Syrphidae) and minute pirate bugs (Orius spp.)5. While several previous studies have assessed the effects of GM maize toxins (especially Bt toxins) on non-target (particularly predatory) arthropods3,11–16, few studies have focussed on the strength of species’ trophic interactions and associated parameters among GM maize systems. This lack of knowledge represents a major issue in the analyses of arthropod communities associated with different GM plant species17.
Arthropod community studies in GM crop systems must involve a comprehensive and multi-methodological field assessment based on different collection methods of all arthropods and weeds from several different GM and non-GM control crops, over several years and throughout the maize growing season. Such datasets may allow us to assess the functional diversity of arthropod communities in GM and non-GM crops to be analysed at the same time, providing consistent species abundance and prey preference data1,3,18. However, datasets with hundreds of thousands of arthropod individuals from GM maize and its non-GM controls are rare, with no such datasets being freely available until now. The analysis of such data may demonstrate that interactions between species are highly correlated, and can thus be considered a useful tool in assessing the deleterious effects of Bt, and other toxins on a wide range of non-target organisms in GM croplands1,3. These data can also be a useful tool to develop biosafety risk hypotheses for invertebrates exposed to GM plants18–20. During a two-year intensive field assessment, detailed arthropod collections were made on different GM maize varieties (some are novel with no previous field test such as the coleopteran+lepidopteran resistant+glyphosate tolerant maize) and on their non-GM controls (Table 1).
Methods
Site characterisation and sampling procedure
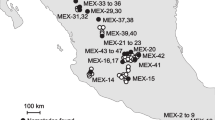
This section is an expanded versions of descriptions in our previous works1–3. The field sites were set up in 2007 in Central Europe, near Budapest, Hungary on chernozem soil in a completely randomised block design with each of the four different GM maize varieties and two non-GM controls. Peach and apricot orchards of about 200 ha dominated the area around the field sites, and no previous maize cultures were present in the immediate area, thus reducing the risk of cross-pollination by GM pollen as much as possible. Both GM and non-GM maize plots were established in 625 m2 (25 m×25 m) blocks, each spaced 3 m apart within each block (Table 1, Figure 1). The GM maize varieties tested contained proteins conferring resistance against two worldwide important maize insect pests (Western corn rootworm D. virgifera virgifera and European corn borer O. nubilalis), or an enzyme conferring glyphosate-resistance, or a combination of these. Tolerance of different GM maize varieties was conferred through different Bt insecticidal crystal (Cry) proteins in different forms that can specially target the above-mentioned two insect pests or the combinations of these. Two other GM maize varieties, in addition to insecticidal proteins, comprised of enzyme (C4 EPSPS) conferring tolerance to glyphosate herbicides (Table 1). Those GM maize varieties that did not contain glyphosate tolerances and the two non-GM controls were seeded in four replicates each. All glyphosate tolerant varieties were replicated eight times from which four blocks were subject to an extra glyphosate treatment in order to test and distinguish the effects of GM varieties vs. effects of glyphosate chemical application on arthropods (Table 1, Figure 1). The extra glyphosate in these blocks was applied in a total amount of 1060 g/ha each year at the four (V4) and eight leaf stages (V8) of the plants (according to the normal application procedures used in maize production). To prevent possible GM pollen contamination of non-GM fields in the wider area, non-GM maize plants of similar maturity were established in the retention zone surrounding the entire experimental fields as a measure to capture pollen. The same planting method, replication units, and glyphosate application were applied in the following year (2008). Maize was planted in late April 2007 and early May 2008, and harvested in late October 2007 and early November 2008. For two years (2007 and 2008), arthropods and weeds were sampled weekly from May, when plants reached the eight leaf stages (V8) until the end of the harvest period. Due to the relatively small block sizes, arthropods are likely to have moved between blocks, thereby reducing the impacts of the different treatments. As such, arthropod samples by pitfall and yellow trapping were taken only from a 10 m×10 m area in the centre of each 625 m2 block. In this way there was a distance of approximately 18–20 m between traps of different blocks. Three standard sampling methods were used to assess and collect arthropods:
-
1
Ground-dwelling arthropods were sampled using pitfall traps. Three traps were placed in the middle of each block (a total of 96 traps in the 32 blocks) and arthropods caught in traps were kept in absolute (99.8%) ethanol until identification. The whole procedure was repeated and traps emptied weekly from May until October. All individuals collected were counted and identified to the lowest taxonomic level possible.
-
2
Plant canopy dwelling arthropods were collected weekly using standard 30 cm×20 cm Pherocon™ yellow sticky traps. Three traps per block (a total of 96 in the 32 blocks) were placed in the centre of each block. Traps were changed once weekly from May to October each year. Arthropods were counted and identified to the lowest taxonomic level possible.
-
3
Arthropods were also assessed by visual observations. Fifteen plants per block were randomly selected (other than those containing yellow traps) from the middle of each block (480 plants per one assessment) and all arthropods found were counted and identified to the lowest taxonomic level possible.
-
4
Weed plant species and their coverage were assessed in all blocks by visual surveys on a random 3×1 m2 area from the middle of each plot as 0-100% area of coverage/species. Weed assessment was made at four different growth stages of maize (eight leaves stage (V8), twelve leaves stage (V12), vegetative stage, tasseling (VT) and reproductive stage, milk (R3)).
Each block was planted in 625 m2 (25 m×25 m) plots spaced 3 m apart. Grey blocks represents extra glyphosate treatments applied. Treatments and replicates are presented in Table 1. Maize plants of similar maturity were established in the retention zone surrounding the experimental fields to capture pollen surrounding the entire experimental field site and prevent pollen contamination of non-GM fields in the wider area around the experimental fields. Part of the figure (field aerial photo only) has been published as supplementary online material in ref. 3 (ece32848-sup-0001-SupInfo.docx).
Data adjustments
This section represents a simplified version of descriptions from our previous works1,3 and describes data adjustments before food web analyses. Arthropod food webs were built using several thousand individuals (lowest=24 972, highest=32 237) per GM maize type (sum of four replications/treatment). First, all arthropods collected were assigned to trophic groups, a widely accepted method in food web studies as it reduces methodological biases related to uneven resolution of taxa within and among species trophic relations21. Trophic groups were defined as taxa that share the same set of predators and/or prey. Before food web constructions, all possible predator-prey interactions of species identified in the GM and non-GM plots were carefully searched in previously published scientific literature. Trophic relations between species (or the next highest level of resolution available, usually genus) were checked using a total of 62 scientific references (Supplementary Table 1). The scientific nomenclature and taxonomy of every resource and consumer were standardised using the Global Names Resolver using the Global Biodiversity Information Facility dataset (http://resolver.globalnames.biodinfo.org/). Designation of trophic links followed methods presented by Gagic et al. 2011 and Jordán et al. 201221,22. To increase the sensibility of the method, arthropod data was further adjusted according to the following parameters:
-
i
Because maize pollen can move long distances by several ways (wind, mechanical, etc.), thereby posing a risk that non-GM maize fields might produce kernels with GM toxins (production of Bt toxins is a dominant trait), food webs of each GM and non-GM maize varieties were built using information on the trophic groups prior tasselling stage (VT) of maize.
-
ii
The presence and abundance of some, or all of the arthropods and weeds investigated varied significantly during the vegetation period. For example, the western corn rootworms adults were absent in April, May, June, present in July and August, and absent again in September, October and November. Therefore, food webs in each GM and non-GM maize varieties were constructed from arthropod and weed data when the abundance of the most frequent species was highest, but prior to pollen spreading (end of June, mid-July).
After adjustments, the food web parameters number of nodes (species), number of edges (trophic relations between species) and K indexes (keystone index) were calculated for each entry following Jordán et al. 201221.
Data Records
The data obtained is available as Data Citation 1. Sample 1_Pitfall Traps file contains arthropod data collected with pitfall traps; Sample 2_Yellow Traps contains data collected with yellow sticky traps; Sample 3_Plant Assessments contains data collected by plant surveys from genetically modified maize events expressing Cry34Ab1, Cry35Ab1, Cry1F and CP4 EPSPS proteins and controls. Values represent number of individuals, except for Sample 3_Plant assessment column, Spider Mites damages %, where the percentage of damages per single plant is given. All arthropod data (Sample 1, 2 and 3) were recorded during the maize developing stages (eight leaves stage (V8), twelve leaves stage (V12), vegetative stage, tasseling (VT) and reproductive stage, milk (R3)) and presented in second column in each dataset). Entry (column 3) describes treatment ID or different GM and non-GM maize varieties (i.e. Entry 1 means Coleopteran resistant GM maize; see Table 1). Replicates (Blocks) (column 4) describes replicates in each entry, along with the code in numbers (as in Table 1). Trap number (column 5) donates the number of traps reported in each entry and its replicates. In Sample 3_Plant Assessment dataset, instead of trap numbers the plant numbers in which arthropods were assessed are given. Sample 4_Weed coverage dataset contains weed coverage (%) in different maize growing stage periods. For weed data the first columns, ‘Sampling’ donates the sampling data made in each year. Columns 2, 3 and 4 contains the same data as described for the previous datasets (2 is Maize growing stage, 3 is Entries and 4 is Blocks). Column 5 (Sample (m2)) describes the number of visual sampling made on weed coverage in each block. Values for each weed species represents soil coverage from 0–100% in 1 m2 area. In food web graph indexes dataset, the number of nodes (species) and the number of edges (trophic relations between species) are presented for each entry. Values ‘in degree’ represents the number of trophic links with the species in the food web (number of other species feeding on this species). Values ‘out degree’ represents the number of trophic links between the species (number of other species eaten by the species). K indices are quantitative rank of species by their topological importance. As the K index increases, the species importance is greater in the functioning of the food web.
Technical Validation
The results of food web analyses using the datasets reported were published in peer-reviewed journals1–3. The arthropod and weed data presented here and in online datasets have also been statistically analysed using standard statistical tools in the associated published papers. The datasets have been carefully checked for possible typing errors in the various published papers. If any discrepancies were found they were corrected before being placed on the web-resource. Weed data were identified to species-level. Arthropod data were identified to the lowest taxonomic level possible, as was the case for most of the predator species. All identified species (weeds and arthropods) were checked by the authors of the paper and by researchers from the Department of Plant Protection and Department of Entomology of the Szent István University, Hungary and by scientists from the Plant Protection Institute of the Hungarian Academy of Science. All datasets were cross validated by carefully checking all data from field sheets and any inconsistencies that might be introduced into the database by different team members were corrected. Additional consistency checks were carried out by carefully analysing the most appropriate scientific references and datasets published in order to establish species similarities found in the present assessment and previously published papers. In addition to these checks, each species identified to this level was checked according to its distribution map. This was done in order to ensure that there were no mistakes in species distribution (i.e. if certain species were indeed previously present in Central European agricultural crops). Necessary corrections and changes were made in the database.
Usage Notes
The database contains information on the functional groups of arthropods from four different GM maize and its isogenic control. To our knowledge, this is the most comprehensive food web/weed/arthropod data assessment from GM maize to date. Future data analyses of the present data that needs further attention include:
-
a
Weed data has not been analysed in detail in comparison with specific trophic groups or species in different GM crops and its control. As weed data has been collected in different maize growth stage periods (V8, V12, VT and R3), detailed arthropod abundance and/or analyses in different growth stages can be made and analysed.
-
b
New methods of species trophic relations and associated metrics analyses can be made.
-
c
Comparison with other arthropod abundances from other GM maize crops can be computed.
-
d
Importance of specific non-target groups and their relative abundance on different GM maize has not been analysed.
-
e
Data can be analysed using several other software applications’ (e.g., R, Matlab, SPSS).
Additional information
How to cite this work: Pálinkás, Z. et al. Arthropods dataset from different genetically modified maize events and associated controls. Sci. Data 5:180019 doi: 10.1038/sdata.2018.19 (2018).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
References
Szénási, Á., Pálinkás, Z., Zalai, M., Schmitz, O. J. & Balog, A. Short-term effects of different genetically modified maize varieties on arthropod food web properties: an experimental field assessment. Sci Rep 4, 5315 (2014).
Pálinkás, Z. et al. Rove beetles (Coleoptera Staphylinidae) – Their abundance and competition with other predatory groups in Bt maize expressing Cry34Ab1, Cry35Ab1, Cry1F and CP4 EPSPS proteins. Crop Prot 80, 87–93 (2016).
Pálinkás, Z. et al. Effects of genetically modified maize events expressing Cry34Ab1, Cry35Ab1, Cry1F, and CP4 EPSPS proteins on arthropod complex food webs. Ecol Evol 7, 2286–2293 (2017).
ISAAA. Global Status of Commercialized Biotech/GM Crops: 2016. ISAAA Brief No. 52. ISAAA, (2016).
Carpenter, J. E. Impact of GM crops on biodiversity. GM Crops 2, 7–23 (2011).
Gassmann, A. J. et al. Field-evolved resistance by western corn rootworm to multiple Bacillus thuringiensis toxins in transgenic maize. PNAS 111, 5141–5146 (2014).
Louis, J. et al. Ethylene contributes to maize insect resistance-mediated maize defense against the phloem sap-sucking corn leaf aphid. Plant Phys 169, 313–324 (2015).
Tzin, V. et al. Dynamic Maize responses to aphid feeding are revealed by a time series of transcriptomic and metabolomic assays. Plant Phys 169, 1727–1743 (2015).
Zhao, M., Ho, H., Wu, Y., He, Y. & Li, M. Western flower thrips (Frankliniella occidentalis) transmits maize chlorotic mottle virus. J Phytopat 162, 532–536 (2014).
Lundgren, J. G., McDonald, T., Rand, T. A. & Fausti, S. W. Spatial and numerical relationships of arthropod communities associated with key pests of maize. J Appl Entomol 139, 446–456 (2015).
Romeis, J., Meissle, M. & Bigler, F. Transgenic crops expressing Bacillus thuringiensis toxins and biological control. Nat Biotech 24, 63–71 (2006).
Raybould, A. Problem formulation and hypothesis testing for environmental risk assessments of genetically modified crops. Environ Biosafety Res 5, 119–125 (2006).
Raybould, A. et al. Non-target organism risk assessment of MIR604 maize expressing mCry3A for control of corn rootworm. J Appl Ent 131, 391–399 (2007).
Naranjo, S. E. Impacts of Bt crops on non-target invertebrates and insecticide use patterns. CAB reviews: Perspectives in agriculture, veterinary science, nutrition and natural resources 4 (2009).
Balog, A., Kiss, J., Szekeres, D., Szénási, Á. & Markó, V. Rove beetle (Coleoptera: Staphylinidae) communities in transgenic Bt (MON810) and near isogenic maize. Crop Prot 6, 567–571 (2010).
Balog, A., Szenasi, A., Szekeres, D. & Palinkas, Z. Analysis of soil dwelling rove beetles (Coleoptera: Staphylinidae) in cultivated maize fields containing the Bt toxins, Cry34/35Ab1 and Cry1F x Cry34/35Ab1. Biocont Sci Tech 21, 293–297 (2011).
Squire, G. R., Hawes, C., Begg, G. S. & Young, M. W. Cumulative impact of GM herbicide-tolerant cropping on arable plants assessed through species-based and functional taxonomies. Environ Sci Pollut Res 16, 85–94 (2009).
Barratt, B. I. P., Todd, J. H., Burgess, E. P. J. & Malone, L. A. Developing biosafety risk hypotheses for invertebrates exposed to GM plants using conceptual food webs: a case study with elevated triacylglyceride levels in ryegrass. Environ Biosafety Res 9, 163–179 (2010).
Powell, J. R. Linking soil organisms within food webs to ecosystem functioning and environmental change. Adv Agr 96, 307–350 (2007).
Powell, J. R. et al. Effects of genetically modified, herbicide-tolerant crops and their management on soil food web properties and crop litter decomposition. J Appl Ecol 46, 388–396 (2009).
Jordán, F., Gjata, N., Mei, S. & Yule, C. M. Simulating food web dynamics along a gradient: quantifying human influence. PLoS ONE 7, e40280 (2012).
Gagic, V. et al. Food web structure and biocontrol in a four-trophic level system across a landscape complexity gradient. Proc R Soc B 278, 2946–2953 (2011).
Data Citations
Pálinkás, Z. Figshare https://doi.org/10.6084/m9.figshare.5501248 (2017)
Acknowledgements
The work/publication is supported by the EFOP-3.6.3-VEKOP-16-2017-00008 project. The project is co-financed by the European Union and the European Social Fund.
Author information
Authors and Affiliations
Contributions
Á.Sz., Z.P., Z.D., M.Z., J.K. and M.Z. performed the experiment, A.B., G.W. and N.S. analysed the data and wrote the main manuscript text and A.B. prepared figures and tables. All authors reviewed the manuscript and were in agreement with submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Additional information
Supplementary information accompanies this paper at
ISA-Tab metadata
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/ The Creative Commons Public Domain Dedication waiver http://creativecommons.org/publicdomain/zero/1.0/ applies to the metadata files made available in this article.
About this article
Cite this article
Pálinkás, Z., Zalai, M., Szénási, Á. et al. Arthropods dataset from different genetically modified maize events and associated controls. Sci Data 5, 180019 (2018). https://doi.org/10.1038/sdata.2018.19
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/sdata.2018.19
This article is cited by
-
Physiological responses in genetically modified cotton and its isohybrid attacked by Aphis gossypii Glover (Hemiptera: Aphididae)
Arthropod-Plant Interactions (2023)