Abstract
How temperature influences development has direct relevance to ascertaining the impact of climate change on natural populations. Reptiles have served as empirical models for understanding how the environment experienced by embryos can influence phenotypic variation, including sex ratio, phenology and survival. Such an understanding has important implications for basic eco-evolutionary theory and conservation efforts worldwide. While there is a burgeoning empirical literature of experimental manipulations of embryonic thermal environments, addressing widespread patterns at a comparative level has been hampered by the lack of accessible data in a format that is amendable to updates as new studies emerge. Here, we describe a database with nearly 10, 000 phenotypic estimates from 155 species of reptile, collected from 300 studies manipulating incubation temperature (published between 1974–2016). The data encompass various morphological, physiological, behavioural and performance traits along with growth rates, developmental timing, sex ratio and survival (e.g., hatching success). This resource will serve as an important data repository for addressing overarching questions about thermal plasticity of reptile embryos.
Design Type(s) | data integration objective • database creation objective |
Measurement Type(s) | developmental plasticity |
Technology Type(s) | data item extraction from journal article |
Factor Type(s) | |
Sample Characteristic(s) | Testudines • Squamata • Sphenodontia • Crocodylia • temperature |
Machine-accessible metadata file describing the reported data (ISA-Tab format)
Similar content being viewed by others
Background & Summary
Conditions experienced early in life are known to impact phenotypes in profound ways that can have long-lasting effects on fitness1,2. Understanding the extent to which developmental environments impact phenotypes is important for addressing many fundamental questions in ecology and evolutionary biology3–5, as well as predicting the effect environmental change will have on populations both locally and globally1,6. Ectothermic vertebrates in particular are sensitive to variation in early developmental temperature, which often is mediated by local climatic conditions, landscape features as well as maternal nest site choice or maternal basing behaviour. In reptiles (turtles and tortoises, tuatara, lizards, snakes and crocodilians), there is a growing empirical literature testing the effects of early thermal environments (i.e., incubation temperatures) on the phenotypic development of a broad range of physiological, morphological and performance traits7–11. Nonetheless, there is currently no database collating and summarising this vast literature in a way that is amenable to updates as the literature grows or that can be expanded to address not only questions on the impacts of temperature, but also other environmental conditions (e.g., moisture, pH) that may be relevant to phenotypic development and survival.
Here, we describe a large database on the effects of incubation temperature on phenotypic traits in oviparous reptiles. Our database differs from others (e.g., BioTraits12), in that it focuses primarily on thermal developmental plasticity by collating studies manipulating temperatures experienced during pre-hatching developmental periods only. Furthermore, ours is the first database on thermal developmental plasticity to provide an updatable platform summarising phenotypic effects of incubation temperatures. A smaller, preliminary version, of the dataset was thoroughly analysed in a related manuscript1 and future plans are to expand the data to capture other environmental drivers of phenotypic variation, such as moisture, pH, and oxygen concentrations. In addition, although the database focuses on oviparous species, it can be expanded to include environmental effects on embryos of viviparous species.
As the database grows, we believe that it can be used to address a wide variety of questions. For example, some of the questions that are currently or have previously been addressed with the database include:
-
What are the overall magnitude of effects of incubation duration on phenotypic development? Qualitative syntheses of this research area have provided an unclear picture of both the magnitude of effect temperature has on phenotypic development, and whether complex patterns alluded to in these reviews13–15 can be explained by species-specific or study-level attributes. Using aspects of these data Noble et al.1 have shown strong overall effects, independent of temperature differences between studies, but found little support for the hypothesis that much of the variation in effects could be explained by phylogeny. Nonetheless, more robust phylogenetic analyses may provide greater insight.
-
Do the effects of early thermal conditions persist late in life? The life-long consequences of adverse early environmental conditions are topics of both theoretical and applied interest, and while immediate effects are well documented2,16, we still know little about whether long-term effects exist, and if so, what the consequences are for population dynamics and life-history evolution16. Noble et al.’s1 analysis suggests that thermal environments can affect the phenotype later in life, however, a more detailed analysis of what predicts variation among species will be worthwhile.
-
Do extreme developmental temperatures elicit developmental stress? Novel environmental conditions, including extreme temperatures, are predicted to affect phenotypic variation17 and can lead to compromised survival1 through developmental stress. We are currently using the database on a more targeted set of traits explore the generality of this prediction.
-
What are the shapes of developmental reaction norms? While previous analyses suggest that many thermal reaction norms for traits exhibit the expected ‘thermal performance curve’ shape1, we are currently exploring the database in more detail on sub-samples of the data to understand the shapes of reaction norms for specific traits that are highly represented in the data (e.g., body size and mass).
-
How realistic thermal fluctuations change the impact thermal developmental environments have on phenotypic development? Previous analyses suggest that more natural, fluctuating conditions decrease the magnitude of phenotypic effects. However, as the database grows and more detailed and realistic thermal conditions are applied experimentally, resolving uncertainty surrounding this question will be possible. It will also be important in establishing the impact thermal conditions have in nature, where temperatures typically vary on daily and longer time scales.
-
How does thermal developmental plasticity facilitate or impede invasion success and adaptation to changing climatic conditions? Phenotypic plasticity is predicted to play an important role in early stages of adaptation to changing environmental conditions17,18 or novel environments (as encountered by invasive species19) and thermal plasticity is expected to feature strongly in this process for ectotherms. As the database grows and thermal reaction norms are more thoroughly characterized in more species these questions maybe feasibly addressed.
The Reptile Development Database can be accessed freely online via a user-friendly webpage (www.repdevo.com) that stores and lodges all versions of past databases in addition to the most up-to-date version. This ensures reproducibility of analyses as the database is updated and evolves to include new types of data. Unpublished data can be submitted through downloaded data templates, and queries can be sought by emailing the team (contact details are on the webpage).
Methods
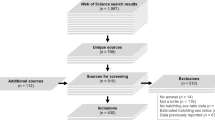
We searched for published literature (1974–2016) describing experiments that manipulated incubation temperature (i.e., pre-hatching developmental period only) in reptiles in Web of Science (v5.13.2) using the following ‘title’ or ‘abstract’ search terms: temperature* AND incubat*, along with one of the following: reptil*, lizard*, squamat*, snake*, turtle*, chelon*, testudin*, crocodil*, alligator*, tuatara*, sphenodon*. In addition, we considered all citations in three major reviews of the topic13–15, and included any additional papers from these sources not identified in our searches. For data currently included in this database, the studies had the following attributes: 1) research on an oviparous reptile (Class Reptilia; excluding birds); 2) employ an experimental manipulation of incubation temperature of eggs; 3) present data on hatching success, incubation duration, or post-hatching phenotypes; 4) consist of eggs that did not receive exogenous hormone application and yolk removal; 6) there was not a substantial delay between oviposition and experimental incubation (e.g., >48-hours). In some cases, papers could not be accessed and/or were in languages that were non-translatable. While we attempted to translate where possible, those that could not be were excluded from the database (approximately 8 studies). We included 684 publications from the primary scientific literature that were relevant based on the title of the paper. If exclusion of papers was not possible based on the title of the publication, we assigned a unique identification number to each publication and considered the abstract and full text for exclusion/inclusion criteria. ‘Citations.csv’ (Table 1; [Data Citation 1]) details the publications considered based on their full-text, and we provide reasons for their exclusion (if excluded) along with full reference information.
Following exclusion / inclusion based on the criteria we had a dataset of 300 publications from which we extracted complete or partial data. From each publication, we extracted data into ‘Database.csv’ as outlined in Table 2 (available online only) [Data Citation 1]. This included the focal information of incubation temperature regime, phenotypes of study, and summary results [mean, error, sample size – type of error (e.g., standard error, standard deviation or 95% confidence interval) is indicated using a separate column – see Table 2 (available online only)] for each phenotype and incubation treatment. We also noted the age of the specimens (assumes to be age 0, hatchlings, if not indicated) in the sample, the temperature fluctuation of treatments, the sex of the sample (assumes to be mixed if not indicated), whether the data were raw or adjusted (e.g., least square means) phenotypic means and the location of the population sample. Data were taken from text or tables from each manuscript, however, when data were provided in figures we extracted key information from these figures (assuming they were clear and readable) using DataThief20. In addition, we recorded contextual information regarding species, study design, cofactors included, and comments. Taxonomic naming was standardised using TimeTree.org21. Genus and species names not identified (N=15 species) in TimeTree were cross checked with the EMBL Reptile Database22. Missing data were coded as ‘NA’. All traits measured were classified into one of eight Trait Categories (see Table 2, (available online only)). Taxonomic representation of data was highest in the Orders Squamata (lizards and snakes – N=75 species) and Testudines (turtles and tortoises – N=69 species) in terms of raw numbers of studies (Fig. 1), although representation relative to species richness was highest in Orders Rhynchocephalia (tuatara, 1 species) and Crocodilia (alligators and crocodiles, – N=10 species). Traits classified under the category morphology were the most-commonly collected data (Fig. 2).
Tree was derived from TimeTree.org 21. Each of the four major orders are represented in the database (‘blue’ – Crocodilia; ‘red’ – Tuatara; ‘yellow’ – Squamates; and ‘green’ – Testudines). Bars above taxa indicate the number of studies (scaled by a factor of 10) for each species. Note that 15 taxa were excluded because of ambiguity surrounding their taxonomic position.
For definitions of the specific trait types included in each of these categories refer to Table 2 (available online only). N above the bars are the number of studies.
These methods, as well as further exclusion criteria, were used for and described in a meta-analysis of a subset of the data1.
Code Availability
Code for technical validation (see below) can be found on the Zenodo archived repository [Data Citation 1].
Data Records
Data Record 1
RepDevo is hosted by GitHub (https://github.com/RepDevo/ReptileDevelopmentDatabase) and provided a unique, stable DOI on Zenodo [Data Citation 1]. This zip file contains csv files of the: 1) citation information, 2) extracted data and 3) metadata.
Technical Validation
We have implemented a number of tests that check the database prior to new releases and major updates by using the testthat23 package in R. The tests check the database for structural integrity (i.e., its internal organization), variable consistency (i.e., correct naming rules) as well as data integrity checks (i.e., outliers, correct data types). Additionally, sub-samples of the data have been thoroughly checked by multiple collectors prior to the release of v1.0.0. The online database can also easily be expanded and corrected if errors are identified. Data will be updated annually with new studies and any errors identified corrected.
Usage Notes
Regularly-updated versions of the Reptile Development Database (RepDevo) can be found at, and downloaded from: www.repdevo.com. The Database.csv, Citations.csv and Metadata.csv files are located on external servers hosted by GitHub and are fully version controlled. Releases will be lodged on GitHub and version releases will be provided with a unique, stable DOI and permanently stored for improved reproducibility as new updates and errors are documented. This is achieved using Zenodo’s DOI and versioning capabilities (https://doi.org/10.5281/zenodo.1188482). Users can cite within the manuscript the specific version used along with its DOI if necessary, which will be provided with the version downloaded from the webpage (www.repdevo.com). However, minimally the specific version should be specified when the data are used to ensure reproducibility of any resulting analyses.
We have been fairly inclusive in our database and many studies report or manipulate a multitude of factors at once, including moisture conditions, oxygen concentrations or measure phenotypes at different ages, temperatures or post-incubation treatments. Our database contains all these data, and so, any future work should take care to extract relevant data based on the question of interest. We have been careful to indicate the population, species, moisture conditions and any post hatching manipulations (e.g., temperature) or measurements (e.g., age) that data are derived from. Some studies confound incubation treatment with year and we have tried to be careful in identifying the specific year in which the experimental manipulation took place to ensure that comparisons of temperatures across different years is not undertaken. In addition, experimental designs can vary substantially among studies (e.g., random allocation of eggs to treatment, split-clutch designs, etc.). This variation could influence results and care should be taken when calculating effect sizes and comparing results of studies with different designs. To account for this, we have also categorized experimental designs that can be considered by users.
Additional information
How to cite this article: Noble, D. W. A. et al. A comprehensive database of thermal developmental plasticity in reptiles. Sci. Data 5:180138 doi: 10.1038/sdata.2018.138 (2018).
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
References
Noble, D. W. A., Stenhouse, V. & Schwanz, L. E. Developmental temperatures and phenotypic plasticity in reptiles: a systematic review and meta-analysis. Biological Reviews, 93, 72–79. 10.1111/brv.12333 (2018).
Monaghan, P. Early growth conditions, phenotypic development and environmental change. Philosophical Transactions of the Royal Society of London B Biological Sciences 363, 1635–1645 (2008).
Schwarzkopf, L. & Andrews, R. M. Are moms manipulative or just selfish? Evaluating the "maternal manipulation hypothesis" and implications for life-history studies of reptiles. Herpetologica 68, 147–159 (2012).
Shine, R. A new hypothesis for the evolution of viviparity in reptiles. The American Naturalist 145, 809–823 (1995).
While, G. M., Uller, T. & Wapstra, E. Offspring performance and the adaptive benefits of prolonged pregnancy: experimental tests in a viviparous lizard. Functional Ecology 23, 818–825 (2009).
Schwanz, L. E. & Janzen, F. J. Climate change and temperature-dependent sex determination: can plasticity in maternal nesting behavior prevent extreme sex ratios? Physiological and Biochemical Zoology 81, 826–834 (2008).
Du, W., Zheng, R. & Shu, L. The influence of incubation temperature on morphology, locomotor performance, and cold tolerance of hatchling Chinese Three- Keeled pond turtles, Chinemys reevesii. Chelonian Conservation and Biology 5, 294–299 (2006).
Du, W.-G., Warner, D. A., Langkilde, T., Robbins, T. & Shine, R. The physiological basis of geographic variation in rates of embryonic development within a widespread lizard species. The American Naturalist 176, 522–528 (2010).
Warner, D. A., Moody, M. A., Telemeco, R. S. & Kolbe, J. J. Egg environments have large effects on embryonic development, but have minimal consequences for hatchling phenotypes in an invasive lizard. Biological Journal of the Linnean Society 105, 25–41 (2012).
Warner, D. A. & Shine, R. Fitness of juvenile lizards depends on seasonal timing of hatching not offspring body size. Oecologia 154, 65–73 (2007).
Warner, D. A. & Shine, R. Maternal nest-site choice in a lizard with temperature-dependent sex determination. Animal Behaviour 75, 861–870 (2008).
Dell, A. I., Pawar, S. & Savage, V. M. The thermal dependence of biological traits. Ecology 94, 1205 (2013).
Birchard, G. F. in Reptilian incubation: Environment, evolution and behaviour 103–123 (Nottingham University Press, 2004).
Deeming, D. C. in Reptilian incubation: Environment, evolution and behaviour 229–252 (Nottingham University Press, 2004).
Deeming, D. C. & Ferguson, M. W. J in Egg incubation: Its effects on embryonic development in birds and reptiles (eds Deeming, D. C. & Ferguson, M. W. J. ) Ch. 10, 147–171 (Cambridge University Press, 1991).
Lindström, J. Early development and fitness in birds and mammals. Trends in Ecology & Evolution 14, 343–347 (1999).
O'Dea, R. E., Noble, D. W. A., Johnson, S. L., Hesselson, D. & Nakagawa, S. The role of non-genetic inheritance in evolutionary rescue: epigenetic buffering, heritable bet hedging and epigenetic traps. Environmental Epigenetics 2, 1–12 (2016).
Chevin, L. M. & Hoffman, A. A. Evolution of phenotypic plasticity in extreme environments. Philosophical Transactions of the Royal Society of London B Biological Sciences 372, 20160138 (2017).
Lande, R. Evolution of phenotypic plasticty in colonizing species. Molecular Ecology 24, 2038–2045 (2015).
Tummers, B. DataThief software, version 3.0 http://datathief.org/ (2006).
Kumar, S., Stecher, G., Suleski, M. & Hedges, S. B. TimeTree: A Resource for Timelines, Timetrees, and Divergence Times. Molecular Biology and Evolution 34, 1812–1819 (2017).
Uetz, P., Freed, P. & Hošek, J. The Reptile Databasehttp://www.reptile-database.org/ (2018).
Wickham, H. testhat: Get started with testing. The R Journal 3, 5–10 (2011).
Data Citations
Noble, D. et al. Zenodo https://doi.org/10.5281/zenodo.1188482 (2018)
Acknowledgements
We thank Samraat Pawar, Joe Salter, Walter Jetz and an anonymous reviewer for constructive feedback on our Data Descriptor. We also thank a host of colleagues who provided important information and clarification on their papers or who provided raw data including: R. Andrews, G. Birchard, M. Blumberg, G. Blouin-Demers, D. Booth, D. Crews, S. Downes, S. Doody, L. Fisher, S. Freedberg, M. Hagman, D. Irschick, X. Ji, D. Ligon, N. Mitchell, G. Packard, R. Paitz, T. Rhen, T. Rodriguez-Diaz, R. Shine, T. Steyermark, R. Telemeco, N. Valenzuela, and G. Webb. Thanks also to R. O’Dea for checking the database. Compilation of the database was supported by the Australian Research Council (L.E.S.: DE120101446; D.W.A.N.: DE150101774), the School of Biological, Earth and Environmental Sciences, UNSW (L.E.S.), the UNSW Faculty of Science (Faculty Research Grant Program, L.E.S.), a UNSW VC Fellowship (D.W.A.N.), a UNSW VC Visiting Researcher Fellowship (D.A.W.), and as an initiative of the UNSW Grand Challenge on Climate Change (L.E.S.).
Author information
Authors and Affiliations
Contributions
Working group organisers: L.S. and D.W.A.N. All authors contributed to paper screening, literature searches, data collection and organisation, and updating the database. D.W.A.N. and J.R. wrote tests for technical validation of the database. D.W.A.N. and L.S. wrote the data descriptor and all authors contributed to its writing and editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
ISA-Tab metadata
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/ The Creative Commons Public Domain Dedication waiver http://creativecommons.org/publicdomain/zero/1.0/ applies to the metadata files made available in this article.
About this article
Cite this article
Noble, D., Stenhouse, V., Riley, J. et al. A comprehensive database of thermal developmental plasticity in reptiles. Sci Data 5, 180138 (2018). https://doi.org/10.1038/sdata.2018.138
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/sdata.2018.138
This article is cited by
-
ReptTraits: a comprehensive dataset of ecological traits in reptiles
Scientific Data (2024)
-
ROSIE, a database of reptilian offspring sex ratios and sex-determining mechanisms, beginning with Testudines
Scientific Data (2022)
-
Effects of incubation temperature on development, morphology, and thermal physiology of the emerging Neotropical lizard model organism Tropidurus torquatus
Scientific Reports (2022)
-
A review of the effects of incubation conditions on hatchling phenotypes in non-squamate reptiles
Journal of Comparative Physiology B (2022)
-
The odonate phenotypic database, a new open data resource for comparative studies of an old insect order
Scientific Data (2019)