Abstract
Structural studies on living cells by conventional methods are limited to low resolution because radiation damage kills cells long before the necessary dose for high resolution can be delivered. X-ray free-electron lasers circumvent this problem by outrunning key damage processes with an ultra-short and extremely bright coherent X-ray pulse. Diffraction-before-destruction experiments provide high-resolution data from cells that are alive when the femtosecond X-ray pulse traverses the sample. This paper presents two data sets from micron-sized cyanobacteria obtained at the Linac Coherent Light Source, containing a total of 199,000 diffraction patterns. Utilizing this type of diffraction data will require the development of new analysis methods and algorithms for studying structure and structural variability in large populations of cells and to create abstract models. Such studies will allow us to understand living cells and populations of cells in new ways. New X-ray lasers, like the European XFEL, will produce billions of pulses per day, and could open new areas in structural sciences.
Design Type(s) | live cell imaging objective |
Measurement Type(s) | X-ray diffraction data |
Technology Type(s) | X-ray free electron laser |
Factor Type(s) | |
Sample Characteristic(s) | Cyanobium gracile • Synechococcus elongatus |
Machine-accessible metadata file describing the reported data (ISA-Tab format)
Similar content being viewed by others
Background & Summary
Imaging living cells at resolutions higher than the resolution of optical microscopy is challenging. A dose in excess of hundred million Grays (Gy: Jkg−1) is required to reach sub-nanometer resolution on a micron-sized cell, using X-rays or electrons, and no cell can survive this level of irradiation; a dose of only hundred Grays kills most cells1,2. What is known about cells today at high resolution comes from dead cells.
Ultra-short and extremely intense coherent X-ray pulses from X-ray lasers offer the possibility to outrun key damage processes3 and deliver a molecular-level snapshot of a cell that is alive at the time of image formation4 but explodes a few picosecond later5. ‘Diffraction-before-destruction’3,4,6 has been successfully demonstrated on a wide range of biological samples, including protein nanocrystals7, living cells8, cell organelles9 and virus particles10. Recent results also show 3D reconstruction of reproducible virus particles11. The data sets described in this paper are from similar ultra-fast imaging experiments.
The ability to measure millions of diffraction patterns in a day at X-ray free-electron lasers (XFELs) offers new avenues for experiments on cells. The femtosecond illumination ‘freezes’ all motion in the sample on the time scale of atomic vibrations. The massive amount of data emerging from XFELs will represent more than just individual projection images of cells. There is a need to develop algorithms to create abstract models of cells from the data, where no individual image gives us complete information but rather nudges the abstraction to describe common features and common internal interactions. Conversely, the data can be used to describe structural variability in populations. With so many images per day, even statistically rare events could be pinpointed and studied. The data sets also offer innovative avenues for data-driven discovery, and helping this effort was one of our motivations in releasing the data sets.
Cyanobacteria were used in this experiment because of their small size and for being remarkably robust. Solitary C. gracile cells are between 0.25–0.4 μm in diameter and 0.4–2.4 μm long12. The S. elongatus cells are similar in diameter but are longer on average by up to a micron. Both species divide symmetrically by binary fission. The two daughter cells separate from each other after reaching the size and shape of the mother cell13. We used non-synchronized cell cultures in our studies, undergoing active growth and providing cells in various stages of their cell cycle.
The live cells were delivered into the pulse train of the Linac Coherent Light Source (LCLS) in an aerosol at a reduced pressure using methods developed for studies on giant viruses10. This type of sample injection delivers truly isolated samples into the X-ray beam and gives diffraction patterns with practically no background noise. In addition, the contrast between the sample and its surrounding (wet helium gas expanding into a vacuum chamber) is also exceptionally high. Injected cells arrive in random order and are imaged in random orientations. The data sets include images with signal extending beyond 4 nm resolution.
At these wavelengths and the scattering angles of the strongest patterns, a single diffraction pattern contains limited depth information, and this information may be retrieved by a numerical propagation of the complex-valued wave front4,6,14,15. There is a need to explore possibilities to extract depth information from the patterns, and a community effort would speed up progress here.
In order to facilitate developments, we present two data records containing a total of 199,000 diffraction patterns from living cells (Data Citation 1), making it the largest freely available X-ray diffraction data set on cells collected at an X-ray FEL. A subset of 11 diffraction patterns from these data sets was used in a recent publication on imaging live cells8. We hope the release of these very large data sets will stimulate interest and help software development.
Methods
Experimental setup
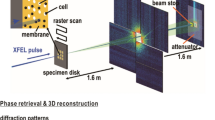
The experiment was executed using the CFEL-ASG Multi-Purpose (CAMP) instrument16, at the AMO end station17 of the Linac Coherent Light Source (LCLS)18, using an experimental configuration identical to that used in ref. 10. The bandwidth of the LCLS is approximately 0.5%. The length of the electron bunch was ~70 fs (full-duration at half-maximum) and the length of the photon bunch is believed to be shorter. The size of the focal spot was 3 μm×7 μm (full width at half maximum).
The two data records presented in this paper come from two experiments, using different experimental parameters (Table 1). In Experiment 1 we collected diffraction patterns from C. gracile cells. The patterns are presented in data record 1. The photon energy of experiment 1 was 517 eV (2.40 nm wavelength). In Experiment 2 we collected diffraction patterns from S. elongatus, presented in data record 2. The photon energy of experiment 2 was 1,100 eV (1.13 nm).
The interaction chamber was equipped with two pairs of pnCCD16 X-ray area detectors (front and back detectors), each consisting of two movable detector panels (Fig. 1). The front detector assembly was placed 220 mm from the interaction point, and the back detector assembly at 741 mm in both experiments. The gap between the two front detector panels was 55.6 mm for experiment 1 and 22.8 mm for experiment 2. The gap between the back detector panels was closed in both experiments. The direct beam exited through openings between the two detector halves and was absorbed in a beam dump behind the back detectors. Each detector panel contained 512×1,024 pixels with 75 μm edge lengths and a full-well capacity of 500,000 electrons/pixel, corresponding to 3,500 photons in experiment 1 and 1,600 photons in experiment 2. The read-out rate matched the 120 Hz repetition rate of the LCLS.
C. gracile and S. elongatus cells were injected into the pulse train of the LCLS18 at 10−6 mbar pressure, using an aerosol sample injector built in Uppsala. The cells are in different stages of division, and arrive in random order and are imaged in random orientations. The diffracted signal is recorded on two detector pairs (front detector and back detector). The direct beam passes through an opening between the two detector halves of each detector pair16. The opening or gap between the front detector halves is 55.6 mm for experiment 1 (C. gracile cells), and 22.8 mm for experiment 2 (S. elongatus cells). The gap between the back detector halves is closed for both experiments.
Cells
Cyanobium gracile PCC 6307 and Synechococcus elongatus PCC 7942 cells were grown in the standard Bg11 medium in batch cultures under constant light. The cell cultures were non-synchronized providing cells in various stages of division. Before the imaging experiments, cells were centrifuged at 6,500 g for 10 min, creating a soft pellet. The pellet was resuspended in 25 mM ammonium acetate, and this buffer exchange was repeated twice to remove salt and contaminants.
Sample injection
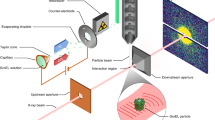
The suspension of live cells was aerosolized with helium in a gas dynamic nebulizer19. The aerosols were delivered into the pulse train of the X-ray laser through an aerodynamic lens20. This method delivers cells in free flight without substrate or other supporting medium, thereby minimizing background scattering, and can produce millions of exposures per day. Most of the nebulizing gas, and vapours of the volatile buffer were pumped away through a differential pumping stage.
Data recording
We recorded a variety of diffraction patterns, originating from single cells, clusters of cells, droplets of buffer or contaminants. The patterns have a large variation in recorded intensity, depending on X-ray pulse-intensity, where in the pulse the particle was hit, and the size of the particle. Figure 2 shows a representative set of diffraction patterns from both experiments.
Ten representative diffraction patterns from each data record were selected. Both data sets contain diffraction patterns from single living cells, multiple cells, large clusters of cells, as well as from contaminants such as spherical droplets, or virus-like particles (possibly being an earlier injected sample). All patterns are normalized individually; dark blue is no scattered signal, dark red is most intense signal in the pattern.
Data preprocessing
We also supply a minimally preprocessed dataset, which includes only diffraction patterns with significant scattered signal (199,000 out of 540,000 patterns). The preprocessing included generation and subtraction of calibration levels, the masking of faulty pixels, the application of the experimental geometry, and a background subtraction. Frames were considered hits if more than 300 pixels record a value above 45 arbitrary detector units (ADU) for experiment 1, and 4,000 pixels recording a value above 45 ADU for experiment 2. The increased threshold in experiment 2 compensates for a stronger background scattering present in experiment 2. All preprocessing steps were done automatically using the Cheetah software package21. The Cheetah configuration files, calibration data, the bad pixel masks, and the respective geometry files are also included into the Data records (Tables 2 and 3).
Data Records
Data record 1
Data record 1 contains the raw and preprocessed data of 473,447 snapshots from C. gracile measured during 77 min of beam-time. The X-ray photon energy was 512 eV and both front and back detector panels were included. The snapshots include blank shots, hits of contaminants, and hits of single and multiple C. gracile cells in random orientation and in random stages of the cell cycle, exposed to different pulse intensities. We estimate the hit ratio of C. gracile cells to be 41% (192,370 diffraction patterns).
The raw data is provided in extended tagged container (XTC) format, and the preprocessed data is provided in CXI format. Both are available for download from the Coherent X-ray Imaging Data Bank (CXIDB)22.
Data record 2
Data record 2 contains the raw and preprocessed data of 66,442 snapshots from S. elongatus measured during 9 min of beam-time. The X-ray wavelength was 1,100 eV and both front and back detector panels were included. The snapshots include blank shots, hits of contaminants, and hits of single and multiple S. elongatus cells in random orientation and in random stages of the cell cycle, exposed to different pulse intensities. We estimate the hit ratio of S. elongatus cells to be 10% (6394 diffraction patterns).
The raw data is provided in XTC format, and the preprocessed data is provided in CXI format, both available at the CXIDB (Data Citation 1).
Technical Validation
Viability of cells
In ref. 8 we show that the injection method was not disruptive to the cells and that the cells were alive at the moment of exposure to the X-ray pulse.
Contamination
We have observed contaminants to be present in each data set, i.e., spherical droplets, and virus-like particles. The former is a common artifact from the injection method, and the latter is most likely samples injected earlier that remained in the pipeline. Both contaminants are easily distinguished by their diffraction patterns (see Fig. 2).
Reconstruction validation
It is shown in ref. 8 that diffraction patterns from this data set can be phased and that the resulting electron densities are matching expectations.
Additional Information
How to cite this article: van der Schot, G. et al. Open data set of live cyanobacterial cells imaged using an X-ray laser. Sci. Data 3:160058 doi: 10.1038/sdata.2016.58 (2016).
References
References
Fairand, B. P. Radiation Sterilization for Health Care Products: X-Ray, Gamma, and Electron Beam. 6, 6–7 (2001).
Game, J. C., Williamson, M. S. & Baccari, C. X-ray survival characteristics and genetic analysis for nine Saccharomyces deletion mutants that show altered radiation sensitivity. Genetics 169, 51–63 (2005).
Neutze, R., Wouts, R., van der Spoel, D., Weckert, E. & Hajdu, J. Potential for biomolecular imaging with femtosecond X-ray pulses. Nature 406, 752–757 (2000).
Bergh, M. et al. Feasibility of imaging living cells at subnanometer resolutions by ultrafast X-ray diffraction. Q. Rev. Biophys. 41, 181–204 (2008).
Rath, A. D. et al. Explosion dynamics of sucrose nanospheres monitored by time of flight spectrometry and coherent diffractive imaging at the split-and-delay beam line of the FLASH soft X-ray laser. Optics Express 22, 28914–28925 (2014).
Chapman, H. N. et al. High-resolution ab initio three-dimensional X-ray diffraction microscopy. J. Opt. Soc. Am. A 23, 1179–1200 (2006).
Chapman, H. N. et al. Femtosecond X-ray protein nanocrystallography. Nature 470, 73–77 (2011).
Van Der Schot, G. et al. Imaging single cells in a beam of live cyanobacteria with an X-ray laser. Nature Communications 6, 5704 (2015).
Hantke, M. F. et al. High-throughput imaging of heterogeneous cell organelles with an X-ray laser. Nature Photonics 8, 943–949 (2014).
Seibert, M. M. et al. Single mimivirus particles intercepted and imaged with an X-ray laser. Nature 470, 78–U86 (2011).
Ekeberg, T. et al. Three-dimensional reconstruction of the giant mimivirus particle with an X-ray free-electron laser. Physical Review Letters 114, 098102 (2015).
Komarek, J., Kopecky, J. & Cepak, V. Generic characters of the simplest cyanoprokaryotes Cyanobium, Cyanobacterium and Synechococcus. Cryptogam. Algol. 20, 209–222 (1999).
Cohen-Bazire, G. S. Fine-Structure of Cyanobacteria. Methods Enzymol. 167, 157–172 (1988).
Spence, J. C. H. et al. Phase recovery and lensless imaging by iterative methods in optical, X-ray and electron diffraction. Phil. Tran.s A Math. Phys. Eng. Sci 360, 875–895 (2002).
Raines, K. S. et al. Three-dimensional structure determination from a single view. Nature 463, 214–217 (2010).
Strüder, L. et al. Large-format, high-speed, X-ray pnCCDs combined with electron and ion imaging spectrometers in a multipurpose chamber for experiments at 4th generation light sources. Nucl. Instrum. Methods Phys. Res., Sect. A 614, 483–496 (2010).
Bostedt, C. et al. Ultra-fast and ultra-intense x-ray sciences: first results from the Linac Coherent Light Source free-electron laser. J. Phys. B 46, 164003 (2013).
Emma, P. et al. First lasing and operation of an angstrom-wavelength free-electron laser. Nat. Photonics 4, 641–647 (2010).
DePonte, D. P. et al. Gas dynamic virtual nozzle for generation of microscopic droplet streams. J. Phys. D: Appl. Phys. 41, 195505 (2008).
Bogan, M. J. et al. Single particle X-ray diffractive imaging. Nano Lett. 8, 310–316 (2008).
Barty, A. et al. A new resource for processing serial X-ray diffraction data. J. Appl. Cryst. 47, 1118–1131 (2014).
Maia, F. R. N. C. The Coherent X-ray Imaging Data Bank. Nat. Methods 9, 854–855 (2012).
The HDF Group. Hierarchical Data Format, version 5, 1997-2016. http://www.hdfgroup.org/HDF5/.
Data Citations
van der Schot, G. Coherent X-ray Imaging Data Bank http://dx.doi.org/10.11577/1245696 (2016)
Acknowledgements
This work was supported by the Swedish Research Council (621-2011-5710, 2015-05007, 628-2008-1109, 828-2012-108, 822-2012-5260, 822-2010-6157, 621-2012-3404, ICA 12-0064, 621-2012-3570), the Knut and Alice Wallenberg Foundation (KAW-2011.081), the European Research Council (ERC-291602), the Röntgen-Ångström Cluster (349-2011-6488, 2015-06107, 349-2011-6497, 349-2013-574, 05K12CH1), Stiftelsen Olle Engkvist Byggmästare, ELI—Extreme Light Infrastructure—phase 2 (CZ.02.1.01/0.0/0.0/15_008/0000162) from European Regional Development Fund, ‘The Hamburg Center for Ultrafast Imaging’ of the Deutsche Forschungsgemeinschaft (CUI, DFG-EXC1074), and the Helmholtz Association through program funds to DESY. Portions of this research were carried out at the Linac Coherent Light Source, a national user facility operated by Stanford University on behalf of the U.S. Department of Energy, Office of Basic Energy Sciences. We are grateful to the scientific and technical staff of the LCLS for support. We thank the CAMP collaboration for giving us access to their experimental setup and for supporting the experiment at the LCLS. We also acknowledge the Max Planck Society for funding the development and operation of the CAMP instrument.
Author information
Authors and Affiliations
Contributions
J.H., F.R.N.C.M. and T.E. developed the imaging concept and conceived the experiment. G.vd.S, F.R.N.C.M., T.E., A.B., N.D.L., A.M., and D.L. developed ideas and software to process the diffraction data. M.S., D.H., G.C., F.N.A. and D.O. prepared and characterised the cells for the study. J.H., B.I., D.P.D., M.S., J.A., J.B., M.M.S., S.B. and D.W. developed and operated the sample injector. J.D.B., C.B., S.C., K.F., S.S., N.T., M.L., F.S., M.S. and M.M.S. operated the beam line at the LCLS R.H. and N.K. operated the pnCCD detectors. M.H., F.R.N.C.M., T.E., D.L., G.vd.S, A.B., J.A., M.M.S., H.N.C., M.S., M.L., F.S., D.R., I.A., A.R., S.E. and J.H. characterized the imaging apparatus and carried out the experiment. G.vd.S, F.R.N.C.M. and T.E. processed the data. G.vd.S, T.E., F.R.N.C.M., and J.H. analysed the results. G.vd.S, J.H. and T.E. wrote the manuscript with input from others.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
ISA-Tab metadata
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0 Metadata associated with this Data Descriptor is available at http://www.nature.com/sdata/ and is released under the CC0 waiver to maximize reuse.
About this article
Cite this article
van der Schot, G., Svenda, M., Maia, F. et al. Open data set of live cyanobacterial cells imaged using an X-ray laser. Sci Data 3, 160058 (2016). https://doi.org/10.1038/sdata.2016.58
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/sdata.2016.58
This article is cited by
-
The trickle before the torrent—diffraction data from X-ray lasers
Scientific Data (2016)