Abstract
Study design:
Spinal cord injury (SCI) is a devastating and common neurologic disorder that has profound influences on modern society from physical, psychosocial and socio-economic perspectives.
Objectives:
To analyze the dynamic changes in protein expression during SCI after ischemia–reperfusion.
Methods:
We used two-dimensional difference gel electrophoresis combined with matrix-assisted laser desorption/ionization time-of-flight/time-of-flight MS to give a global analysis of protein dynamic change during SCI after ischemia–reperfusion. Dynamic changes in protein expression were investigated from 6 to 48 h in SCI after ischemia–reperfusion using a proteomics tool.
Results:
Twenty-one proteins were identified in total, including neuronal proteins, glycometabolism enzymes, stress-related proteins and cytoskeleton-related proteins. These were divided into upregulated and downregulated groups. Results identified 24 h as a key time point when all proteins were changed dramatically. In addition, changes in Fascin expression were discovered in SCI for the first time.
Conclusion:
In conclusion, we observed dynamic proteome change correlated with SCI by ischemia–reperfusion, and provided a clue to this pathological mechanism by protein identification and analysis.
Similar content being viewed by others
Introduction
Traumatic spinal cord injury (SCI) is a severe and costly disease frequently affecting young and healthy individuals. SCI has both primary and secondary mechanisms of injury. In the primary stage, acute SCI causes pathologic changes involving hemorrhage, edema, axonal and neuronal necrosis, and demyelination, followed by cyst formation and infarction. In the secondary stage, damage can be exacerbated by ischemia–reperfusion, which may result in greater injury than ischemia itself.1 Reactive oxidation species have been implicated in this pathogenesis2 and recent proteomics studies of primary stage SCI have identified some important molecules.3, 4 However, the exact mechanism of ischemia–reperfusion-induced SCI remains unclear and the protein differential level of the second stage of injury is seldom reported. High-throughput proteomics tools promise discovery of molecules involved in the pathological processes of ischemia–reperfusion injury.
In this study, we used two-dimensional difference gel electrophoresis (2D DIGE) combined with matrix-assisted laser desorption/ionization time-of-flight/time-of-flight (MALDI-TOF-TOF) MS to perform a global analysis of the dynamic changes in protein expression after ischemia–reperfusion-induced SCI. This study will provide further understanding of the pathological process and identify important molecules involved in SCI after ischemia–reperfusion.
Materials and methods
Animal model and surgical protocol
Rabbits were obtained from the Institute of Zoology, Chinese Academy of Sciences in Beijing, China. New Zealand rabbits were housed in separate cages at 20 °C. The ischemia–reperfusion model was performed according to Zivin and DeGirolami,5 with six randomly assigned groups of six animals each. At 5 months of age, rabbits were anesthetized (10% chloral hydrate, 300 mgkg−1), and a 5-cm cut was made on the skin and peritoneum. Heparin (25 U) was injected and the abdominal aorta was separated and exposed for 25 min without treatment (control group) or blocked with a vascular clamp for 25 min (ischemia group). Four time points were set for ischemia–reperfusion: 6, 12, 24 and 48 h after abdominal aorta blocking. An antibiotic (cidomycin 80 000 U) was injected into the abdominal cavity and the cut was sutured. Rabbits were anesthetized with 10% chloral hydrate. The spinal cord from lumbar 3 to 5 was removed. One half was frozen in liquid nitrogen and the other was fixed with 10% formalin. Then the rabbits were killed by cervical dislocation. All procedures were conducted in accordance with the policies promulgated by the Ethics Committee of the Third Hospital of Jilin University.
The fixed spinal specimens were paraffin embedded, sectioned at 4 μm, de-paraffinized in xylene, rehydrated through a graded series of alcohol, and stained with hematoxylin and eosin. The slides were examined with an Olympus BHTU microscope (New York, NY, USA).
Nerve function evaluation
To evaluate nerve function, a modified Tarlov Score6 was introduced. Scores are described in detail in Table 1.
2D DIGE and image analysis
Protein extraction concentration was determined by the BCA method and pH was adjusted to 8.5. Equal amounts of proteins from the six pairs of samples were pooled together as the internal standard. Control and ischemia–reperfusion groups were randomly labeled with Cy3 or Cy5 (GE Healthcare, Washington, DC, USA), and combined before mixing with Cy2-labeled internal standard (50 μg each), sample buffer and rehydration buffer.
Samples were actively rehydrated into 24 cm pH 3–10 IPG strips (Bio-Rad, Hercules, CA, USA) using a Protean IEF cell (Bio-Rad). Isoelectric focusing was performed at 10 000 V for 8 h, for a total of 80 kV h. The IPG strips were equilibrated, then placed on 12% homogeneous polyacrylamide gels precast with low fluorescence glass plates using an Ettan DALT 12 gel caster. The 2D SDS-PAGE was carried out using the Protean Plus system (Bio-Rad). After 2D DIGE, gels were scanned on the Typhoon 9410 scanner using excitation/emission wavelengths specific for Cy2, Cy3 and Cy5. The intensity was adjusted to ensure that the maximum volume of each image was within 60 000–90 000.
2D DIGE was analyzed using DeCyder 5.02 software (GE Healthcare) according to the manufacturer’s instructions. The DeCyder biological variation analysis module was used to detect spots (estimated number, 2500) and simultaneously match all 49 protein spot maps from three gels. All matches were confirmed manually. A paired t-test was used for statistical analysis of the data. Protein spots with a variation in density between the control and ischemia–reperfusion groups of <2.0 were selected for identification. Spot picking and in-gel digestion were carried out with preparative gels. Two-dimensional electrophoresis was performed as described under ‘2D DIGE and Imaging’, except that IPG strips were loaded with 500–1000 μg of protein, and gels were stained with Coomassie Brilliant Blue. Protein spots of interest were excised and destained. Gels were then dried by centrifugal lyophilization and digested with trypsin. The supernatant was collected, and the tryptic peptides were sequentially extracted from the gel. The extracts were pooled and dried by centrifugal lyophilization.
Protein identification
Peptide–ACN–TFA mixtures were spotted on the target plate. Peptide mass fingerprinting and sequence analysis were carried out on a MALDI-TOF-TOF MS (4800 Proteomics Analyzer, Applied Biosystems, Beijing, China). Peptide mass maps were acquired in positive reflection mode, averaging 1500 laser shots per MALDI-TOF-TOF spectrum and 3000 shots per TOF/TOF spectrum (resolution 20 000). The spectrum was calibrated to a mass tolerance within 0.1 Da using 4800 calibration mixtures (Applied Biosystems, Foster, CA, USA). Parent mass peaks with a mass range of 600–4000 Da and minimum signal to noise ratio of 15 were picked for tandem TOF/TOF analysis. Combined mass and mass/mass spectra were used to interrogate rabbit sequences in the NCBI rabbit protein database and Nr database using MASCOT database search algorithms (version 1.9). Modifications allowed were carbamidomethylation, oxidation and one missed trypsin cleavage site. The tolerance of precursor and fragment ions was 0.2 Da. All automatic data analysis and database searching was performed by GPS Explorer software (version 3.6, Applied Biosystems). Contaminant proteins were excluded manually. Confidently identified proteins had a statistically significant (P⩽0.05) protein score (based on combined mass and mass/mass spectra) and best ion score (based on mass/mass spectra). Redundancy of proteins with several names and accession numbers was eliminated. If multiple proteins were identified in one spot, the single highest scoring protein member was chosen from the protein family. The molecular weight and pI values of most proteins were consistent with the gel regions from which the spots were excised.
Western blot
Proteins from the injured tissues were separated on 12% SDS-PAGE and transferred to PVDF membranes (Amersham Biosciences, Piscataway, NJ, USA). Blots were blocked in a buffer containing TBS-Tween-20 with a 5% concentration (w/v) of skim milk powder and incubated with primary antibody (mouse anti-α-tubulin; Invitrogen (New York, CA, USA), Zymed-180092, 1:50) for 2 h at room temperature. Blots were then washed and incubated with horseradish peroxidase-conjugated secondary antibody (goat anti-mouse antibody, 1:10 000; Santa Cruz Biotechnology, Santa Cruz, CA,USA) for 1 h at room temperature. Immunoreactive complexes were visualized using ECL reagents (Santa Cruz Biotechnology).
Immunohistochemistry
The excised spinal specimens were formalin fixed and paraffin embedded. Slices of 4 μm were then de-paraffinized in xylene, rehydrated through a graded series of alcohol and washed in distilled water. Tissue sections were heated in a microwave oven at 700 W in citrate buffer (pH 6.0) for 10 min, and then processed by the standard streptavidin-biotin-immunoperoxidase method. The endogen peroxidase activity was blocked with 3% H2O2 in methanol for 20 min, then the primary antibody in Tris-HCl buffer (0.05 M, pH 7.6) with 0.1% bovine serum albumin was applied for 45 min at room temperature (20–25 °C). The secondary antibody was then applied for 15 min, followed by treatment with streptavidin-peroxidase complex (1:100 in Tris-HCl buffer) for 15 min at room temperature. Between incubations, the sections were washed three times in Tris-HCl buffer for 3 min. 3,3′-Diaminobenzidine (DAB, Zhongshan Goldenbridge Biotechnology, Beijing, China) was then applied for 10 min, and nuclear counterstaining was done with hematoxylin (Chengdu Kelong Chemical Reagent Factory, Chengdu, China). Negative controls for antibodies involved substituting the primary antibody with normal mouse serum. Slides were examined under an Olympus BX51 light microscope.
Results
Establishment of ischemia–reperfusion model
The average Tarlov score for hind limb movement at different reperfusion times is shown in Table 1. Significance was found between the control group and each reperfusion group (P<0.05). Moreover, we found significant variation between the 6- and 12-h reperfusion groups (P<0.05).
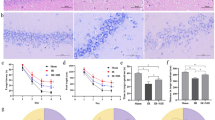
Hematoxylin and eosin staining showed pathological alterations (Figures 1a–j). As shown in Figure 1, some neurons gradually altered in shape with increasing time of ischemia–reperfusion injury. Neurons became elongated, and axons and dendrites were lost after 12 h of ischemia–reperfusion, and spindle-shaped after 48 h. Initially, nuclei were located in the cell center and were swollen and pale with a clear membrane. With increased time after ischemia–reperfusion injury, nuclei became smaller, darker and shifted location. After 12 h, ischemia–reperfusion nuclei were fragmented and the cytoplasm demonstrated acidophilia. Spinal cord stroma showed increasing edema and many microglias.
Pathologic change of spinal cord after ischemia–reperfusion by hematoxylin and eosin staining. (a, b) The control tissue before ischemia. (c–j) Hematoxylin and eosin staining showing tissue injury at 6 h (c, d), 12 h (e, f), 24 h (g, h) and 48 h (i, j) after ischemia–reperfusion. a, c, e, g, and i show neuronal changes, whereas b, d, f, h, and j show mesenchymal changes.
Analysis of differentially expressed proteins
Among 1600 matched protein spots, 21 were significantly up- or downregulated at the analyzed ischemia–reperfusion time points compared with the control group (Figure 1a). Figure 1b shows the comparison between the control group and each ischemia–reperfusion group by 2D DIGE.
Identification of differentially expressed proteins
The mass spectrum analysis was based on available protein databases. Our protein database consisted of proteins from multiple species to ensure all proteins were identified, a strategy previously employed in rabbit myocardium protein profiling because many proteins are highly conserved between mammalian species.7
Forty-nine differentially expressed spots corresponding to 21 gene products were identified (Supplementary Table S1). Two patterns were seen with increased reperfusion times (Figures 2a and b). In the first, four spots corresponding to two proteins, α-tubulin and neurofilament protein M (NEFM), were significantly upregulated after 24 h (Supplementary Table 2). In the second, 26 spots corresponding to 20 proteins were significantly downregulated after 24 h. Seven of these were metabolic enzymes: β-enolase (ENO3); glycogen phosphorylase, brain form (PYGB); glycogen phosphorylase, muscle form (PYGM); pyruvate kinase isozymes M1/M2 (PKM2); glyceraldehyde-3-phosphate dehydrogenase (GAPDH); succinate dehydrogenase complex subunit A (SDHA); and mitochondrial glycerol-3-phosphate dehydrogenase 2 (GPD2) (Supplementary Table 3). Three were nervous system proteins: NEFM, neurofilament-L (Nefl) and glial fibrillary acidic protein. The remaining proteins were associated with the cytoskeleton, transport and stress activity. Over 50% of the differentially expressed proteins were localized to the cytoplasm or mitochondria by Gene Ontology.8 Supplementary Figures 1A–C show the distribution of protein function and localization.
As previously described, multiple spots were identified as products of the same gene. Interestingly, several of these were differentially expressed. For example, NEFM corresponded to six spots: 142, 203, 815, 821, 971 and 1844 (Figures 3a). Five spots showed variable molecular mass and were variably distributed in different pH ladders. PYGM, ENO3, stress-induced-phosphoprotein 1 (STIP1) and SDHA showed similar patterns.
Protein validation by western blot and immunohistochemistry
To verify DIGE results, the ischemia–reperfusion samples were further analyzed by western blot analysis using a α-tubulin antibody. We found α-tubulin was significantly upregulated after 24 h reperfusion compared with other time points (Figure 4a); this result was consistent with its variation curve (Figure 4b). Immunohistochemistry demonstrated that changes in α-tubulin expression correlated with the reperfusion time in the injured spinal cord (Figure 4c).
Validation of changes in α-tubulin expression levels by western blot and immunohistochemistry. (a) The expression curve of α-tubulin. (b) The results of anti-α-tubulin western blot analysis. (c) Immunohistochemistry results after 0 h (I25R0), 6 h (I25R6), 12 h (I25R12), 24 h (I25R24) and 48 h (I25R48) in SCI by ischemia–reperfusion.
Discussion
SCI causes severe neuropathology and limited functional recovery and is a major public health problem. After ischemia–reperfusion, SCI becomes more serious because oxygen-derived free radicals are produced.9 A few studies document proteomics analysis of SCI in animal models,3, 4 but our current study is the first to report dynamic analysis of protein expression during ischemia–reperfusion-induced SCI using 2D DIGE. We identified 21 differentially expressed gene products. Protein differential expression profiles indicated that protein expression changed dramatically 24 h after ischemia–reperfusion.
Our results identified three nervous system-related proteins: Nefl, glial fibrillary acidic protein and NEFM. Nefl is an intermediate filament protein involved in the maintenance of neuronal caliber. A previous study indicated that Nefl is degraded by oxidative modification in SCI, especially in the second stage.10 Our results also showed Nefl downregulation 24 h after ischemia–reperfusion. Glial fibrillary acidic protein (a class-III intermediate filament protein) was also downregulated at this time point, although a previous study demonstrated glial fibrillary acidic protein upregulation in SCI induced by laminectomy.4 NEFM, another neurofilament protein, was distributed at six spots with both up- and downregulated patterns. A previous study reported that NEFM expression is influenced strongly in oxidative stress, and estradiol may protect neurons by inducing NEFM expression.11 In our results, NEFM was upregulated after 24 h of reperfusion, suggesting that this mechanism of protection may occur at 24 h after injury. In addition, we found the pI of NEFM in the acidic terminal of the 2D-gel in two spots (Figure 3a, spots 815 and 821). Considering that the phosphorylated form of NEFM plays a role in protecting the membrane-rich axon,12 NEFM may be phosphorylated for protection in ischemia–reperfusion injury. However, some NEFM protein spots with lower molecular weight than normal (Figure 3a, spot 1844) were observed. After ruling out the possibility of protein degradation in the sample preparation, it is possible that NFM mRNA was spliced and translated into several isoforms in injury, which has not been reported.
Several enzymes involved in carbohydrate and energy metabolism were downregulated, including ENO3, GAPDH, GPD2, PKM2, PYGM, PYGB and SDHA. In the literature, the expression of neuron-specific enolase (γ-enolase) is increased with prolonged time of acute SCI.13 However, enolase was downregulated in our study, possibly representing the pathological differences between acute and ischemia–reperfusion SCI. PKM2, SDHA, PYGM, PYGB and GPD2 are carbon-centric enzymes involved in glycolysis and the tricarboxylic acid cycle. Our study is the first to report changes in these proteins in SCI. The amount of glycogen phosphorylase decreases transiently (24 h) in sectioned sciatic axons,14 and a prior study in cerebral ischemia showed that glucose treatment could reduce the extent of tissue injury.15 Therefore, the downregulation of these enzymes in our study may suggest activation of a similar protective mechanism in SCI after ischemia–reperfusion.
Proteins such as heat shock protein family members are induced by severe tissue and organ stress.16 In our study, three stress-related proteins were downregulated, including heat shock cognate 70 kDa protein (HSC70) and STIP1. Heat shock protein 70 (HSP70) has a protective effect against SCI.17 HSC70 and STIP1 are related to HSP70 in the stress response, and the expression of these proteins was changed after 6 h of ischemia–reperfusion (Supplementary Figures 2A and B). STIP1 may mediate the association between HSP70 and HSP9018 and is downregulated in rat SCI,19 which was consistent with our study. Therefore, STIP1 is a valuable molecule for further functional research in SCI.
Three cytoskeleton proteins were detected in 2D DIGE gels, namely α-tubulin (Tuba), fascin (Fscn1) and dynamin 1 (Dnm1). Tubulin is the major component of microtubules and was upregulated at 24 h in our experiments; this was further validated by western blot analysis (Figure 4). This result was also discovered in rat SCI.19 Dynamin 1 is a microtubule-associated protein involved in bundling microtubules and receptor-mediated endocytosis. Dynamin 1 mRNA levels were decreased at 24 h in SCI made by transection of column nerve fibers,20 and its protein level was also decreased at the same time in our results. Together, these results suggest that the regulation of Dnm1 is determined at the level of transcription. Our study is the first to report changes in Fscn1 expression in SCI.
Conclusion
Our study is the first to use proteomics tools to profile and quantify changes in protein expression in ischemia–reperfusion-induced SCI and indicates that 24 h post injury represents a critical time for changes in protein expression. From these changes, we suggest the following pathological process: SCI causes damage to structural neural proteins and a series of protective mechanisms is activated, including heat shock proteins and related molecules. In addition, glycol metabolism decreases to retain blood glucose concentration and reduce damage. Further investigation is needed to explore the molecular mechanisms of these pathological processes.
Data archiving
There were no data to deposit.
References
Dumont RJ, Okonkwo DO, Verma S, Hurlbert RJ, Boulos PT, Ellegala DB et al. Acute spinal cord injury, part I: pathophysiologic mechanisms. Clin Neuropharmacol 2001; 24: 254–264.
Lu K, Liang CL, Liliang PC, Yang CH, Cho CL, Weng HC et al. Inhibition of extracellular signal-regulated kinases 1/2 provides neuroprotection in spinal cord ischemia/reperfusion injury in rats: relationship with the nuclear factor-kappaB-regulated anti-apoptotic mechanisms. J Neurochem 2010; 114: 237–246.
Ding Q, Wu Z, Guo Y, Zhao C, Jia Y, Kong F et al. Proteome analysis of up-regulated proteins in the rat spinal cord induced by transection injury. Proteomics 2006; 6: 505–518.
Kang SK, So HH, Moon YS, Kim CH . Proteomic analysis of injured spinal cord tissue proteins using 2-DE and MALDI-TOF MS. Proteomic 2006; 6: 2797–2812.
Zivin JA, DeGirolami U . Spinal cord infarction: a highly reproducible stroke model. Stroke 1980; 11: 200–202.
Gale K, Kerasidis H, Wrathall JR . Spinal cord contusion in the rat: behavioral analysis of functional neurologic impairment. Exp Neurol 1985; 132: 123–134.
Vissers JP, Pons S, Hulin A, Tissier R, Berdeaux A, Connolly JB et al. The use of proteome similarity for the qualitative and quantitative profiling of reperfused myocardium. J Chromatogr B Analyt Technol Biomed Life Sci 2009; 877: 1317–1326.
Hill DP, Smith B, McAndrews-Hill MS, Blake JA . Gene Ontology annotations: what they mean and where they come from. BMC Bioinformatics 2008; 9: S2.
Shan LQ, Ma S, Qiu XC, Zhou Y, Zhang Y, Zheng LH et al. Hydroxysafflor Yellow A protects spinal cords from ischemia/reperfusion injury in rabbits. BMC Neurosci 2010; 13: 98.
Leski ML, Bao F, Wu L, Qian H, Sun D, Liu D . Protein and DNA oxidation in spinal injury: neurofilaments – an oxidation target. Free Radic Biol Med 2001; 30: 613–624.
Chiasson K, Lahaie-Collins V, Bournival J, Delapierre B, Gélinas S, Martinoli MG . Oxidative stress and 17-alpha- and 17-beta-estradiol modulate neurofilaments differently. J Mol Neurosci 2006; 30: 297–310.
Wataya T, Nunomura A, Smith MA, Siedlak SL, Harris PL, Shimohama S et al. High molecular weight neurofilament proteins are physiological substrates of adduction by the lipid peroxidation product hydroxynonenal. J Biol Chem 2002; 277: 4644–4648.
Cao F, Yang XF, Liu WG, Hu WW, Li G, Zheng XJ et al. Elevation of neuron-specific enolase and S-100beta protein level in experimental acute spinal cord injury. J Clin Neurosci 2008; 15: 541–544.
Woolf CJ, Chong MS, Ainsworth A . Axotomy increases glycogen phosphorylase activity in motoneurones. Neuroscience 1984; 12: 1261–1269.
Vannucci RC . Cerebral carbohydrate and energy metabolism in perinatal hypoxic-ischemic brain damage. Brain Pathol 1992; 2: 229–234.
Hecker JG, McGarvey M . Heat shock proteins as biomarkers for the rapid detection of brain and spinal cord ischemia: a review and comparison to other methods of detection in thoracic aneurysm repair. Cell Stress Chaperones 2011; 16: 119–131.
Matsumoto M, Ohtake K, Wakamatsu H, Oka S, Kiyoshima T, Nakakimura K et al. The time course of acquisition of ischemic tolerance and induction of heat shock protein 70 after a brief period of ischemia in the spinal cord in rabbits. Anesth Analg 2001; 92: 418–423.
Gonçalves DC, Gava LM, Ramos CH . Human Hsp70/Hsp90 organizing protein (Hop) D456G is a mixture of monomeric and dimeric species. Protein Pept Lett 2010; 17: 492–498.
Afjehi-Sadat L, Brejnikow M, Kang SU, Vishwanath V, Walder N, Herkner K et al. Differential protein levels and post-translational modifications in spinal cord injury of the rat. J Proteome Res 2010; 9: 1591–1597.
Riley LA, Bernstein JJ . Changes in dynamin and actin mRNA expression in the dorsal column-medial lemniscal system following dorsal column lesion. J Neurosci Res 1996; 44: 47–51.
Acknowledgements
This work was supported in part by the National Natural Science Foundation of China for Creative Research Groups (Grant 30872609/H605).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies this paper on the Spinal Cord website
Rights and permissions
About this article
Cite this article
Zhu, B., Li, Y., Li, M. et al. Dynamic proteome analysis of spinal cord injury after ischemia–reperfusion in rabbits by two-dimensional difference gel electrophoresis. Spinal Cord 51, 610–615 (2013). https://doi.org/10.1038/sc.2013.24
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sc.2013.24
Keywords
This article is cited by
-
Inhibition of heat shock protein family A member 8 attenuates spinal cord ischemia–reperfusion injury via astrocyte NF-κB/NLRP3 inflammasome pathway
Journal of Neuroinflammation (2021)
-
Protein Degradome of Spinal Cord Injury: Biomarkers and Potential Therapeutic Targets
Molecular Neurobiology (2020)
-
Clinical proteomics of enervated neurons
Clinical Proteomics (2016)