Abstract
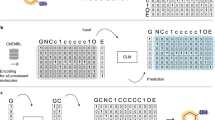
Deep learning has acquired considerable momentum over the past couple of years in the domain of de novo drug design. Here, we propose a simple approach to the task of focused molecular generation for drug design purposes by constructing a conditional recurrent neural network (cRNN). We aggregate selected molecular descriptors and transform them into the initial memory state of the network before starting the generation of alphanumeric strings that describe molecules. We thus tackle the inverse design problem directly, as the cRNNs may generate molecules near the specified conditions. Moreover, we exemplify a novel way of assessing the focus of the conditional output of such a model using negative log-likelihood plots. The output is more focused than traditional unbiased RNNs, yet less focused than autoencoders, thus representing a novel method with intermediate output specificity between well-established methods. Conceptually, our architecture shows promise for the generalized problem of steering of sequential data generation with recurrent neural networks.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The curated datasets used to train all models are available at https://github.com/pcko1/Deep-Drug-Coder/tree/master/datasets.
Code availability
The Python code and the trained neural networks used in this work are available under MIT licence57 in the Deep Drug Coder (DDC) GitHub repository https://github.com/pcko1/Deep-Drug-Coder and https://doi.org/10.5281/zenodo.3739063, which also includes an optional encoding network to constitute a molecular heteroencoder.
References
Lopyrev, K. Generating news headlines with recurrent neural networks. Preprint at https://arxiv.org/pdf/1512.01712.pdf (2015).
Briot, J.-P., Hadjeres, G. & Pachet, F.-D. Deep Learning Techniques for Music Generation (Springer, 2020).
Wang, Z. et al. Chinese poetry generation with planning based neural network. In Proceedings of 26th International Conference of Computing and Linguistics 1051–1060 (COLING 2016 Organizing Committee, 2016).
Elgammal, A., Liu, B., Elhoseiny, M. & Mazzone, M. CAN: Creative adversarial networks, generating ‘art’ by learning about styles and deviating from style norms. Preprint at https://arxiv.org/abs/1706.07068 (2017).
Segler, M. H. S., Preuss, M. & Waller, M. P. Planning chemical syntheses with deep neural networks and symbolic AI. Nature 555, 604–610 (2018).
Ronneberger, O., Fischer, P. & Brox, T. U-Net: convolutional networks for biomedical image segmentation. In Proceedings of Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015 (eds Navab, N., Hornegger, J., Wells, W. M. & Frangi, A. F.) 234–241 (Springer, 2015).
Chen, H., Engkvist, O., Wang, Y., Olivecrona, M. & Blaschke, T. The rise of deep learning in drug discovery. Drug Discov. Today 23, 1241–1250 (2018).
Xu, Y. et al. Deep learning for molecular generation. Future Med. Chem. 11, 567–597 (2019).
Elton, D. C., Boukouvalas, Z., Fuge, M. D. & Chung, P. W. Deep learning for molecular design-a review of the state of the art. Mol. Syst. Des. Eng. 4, 828–849 (2019).
Vamathevan, J. et al. Applications of machine learning in drug discovery and development. Nat. Rev. Drug Discov. 18, 463–477 (2019).
Sanchez-Lengeling, B. & Aspuru-Guzik, A. Inverse molecular design using machine learning: generative models for matter engineering. Science 361, 360–365 (2018).
Weininger, D. SMILES, a chemical language and information system. 1: Introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 28, 31–36 (1988).
Schwalbe-Koda, D. & Gómez-Bombarelli, R. Generative models for automatic chemical design. Preprint at https://arxiv.org/pdf/1907.01632.pdf (2019).
Lipton, Z. C., Berkowitz, J. & Elkan, C. A critical review of recurrent neural networks for sequence learning. Preprint at https://arxiv.org/pdf/1506.00019.pdf (2015).
Arús-Pous, J. et al. Exploring the GDB-13 chemical space using deep generative models. J. Cheminform. 11, 20 (2019).
Arús-Pous, J. et al. Randomized SMILES strings improve the quality of molecular generative models. J. Cheminform. 11, 71 (2019).
Segler, M. H. S., Kogej, T., Tyrchan, C. & Waller, M. P. Generating focused molecule libraries for drug discovery with recurrent neural networks. ACS Cent. Sci. 4, 120–131 (2018).
Olivecrona, M., Blaschke, T., Engkvist, O. & Chen, H. Molecular de-novo design through deep reinforcement learning. J. Cheminform 9, 48 (2017).
Zhou, Z., Kearnes, S., Li, L., Zare, R. N. & Riley, P. Optimization of molecules via deep reinforcement learning. Sci. Rep. 9, 10752 (2019).
Popova, M., Isayev, O. & Tropsha, A. Deep reinforcement learning for de novo drug design. Sci. Adv 4, 7 (2018).
Gómez-Bombarelli, R. et al. Automatic chemical design using a data-driven continuous representation of molecules. ACS Cent. Sci. 4, 268–276 (2018).
Polykovskiy, D., Artamonov, A., Veselov, M., Kadurin, A. & Nikolenko, S. Molecular Sets (MOSES): a benchmarking platform for molecular generation models. Preprint at https://arxiv.org/pdf/1811.12823.pdf (2019).
Brown, N., Fiscato, M., Segler, M. H. S. & Vaucher, A. C. GuacaMol: benchmarking models for de novo molecular design. J. Chem. Inf. Model. 59, 1096–1108 (2019).
Bjerrum, E. J. SMILES enumeration as data augmentation for neural network modeling of molecules. Preprint at https://arxiv.org/pdf/1703.07076.pdf (2017).
Bjerrum, E. J. & Sattarov, B. Improving chemical autoencoder latent space and molecular de novo generation diversity with heteroencoders. Biomolecules 8, 131 (2018).
Winter, R., Montanari, F., Noé, F. & Clevert, D. A. Learning continuous and data-driven molecular descriptors by translating equivalent chemical representations. Chem. Sci. 10, 1692–1701 (2019).
Blaschke, T., Olivecrona, M., Engkvist, O., Bajorath, J. & Chen, H. Application of generative autoencoder in de novo molecular design. Mol. Inform. 37, 1–11 (2018).
Winter, R. et al. Efficient multi-objective molecular optimization in a continuous latent space. Chem. Sci 10, 8016–8024 (2019).
Prykhodko, O. et al. A de novo molecular generation method using latent vector based generative adversarial network. J. Cheminform 11, 74 (2019).
Lim, J., Ryu, S., Kim, J. W. & Kim, W. Y. Molecular generative model based on conditional variational autoencoder for de novo molecular design. J. Cheminform. 10, 31 (2018).
Jin, W., Barzilay, R. & Jaakkola, T. S. Multi-resolution autoregressive graph-to-graph translation for molecules. Preprint at https://chemrxiv.org/articles/Multi-Resolution_Autoregressive_Graph-to-Graph_Translation_for_Molecules/8266745/1 (2019).
Hochreiter, S. & Schmidhuber, J. Long short-term memory. Neural Comput. 9, 1–32 (1997).
Preuer, K., Renz, P., Unterthiner, T., Hochreiter, S. & Klambauer, G. Fréchet ChemNet distance: a metric for generative models for molecules in drug discovery. J. Chem. Inf. Model. 58, 1736–1741 (2018).
Duvenaud, D. K. et al. Convolutional networks on graphs for learning molecular fingerprints. In Advances in Neural Information Processing Systems Vol. 28 (eds Cortes, C., Lawrence, N. D., Lee, D. D., Sugiyama, M. & Garnett, R.) 2224–2232 (Curran Associates, 2015).
Bickerton, G. R., Paolini, G. V., Besnard, J., Muresan, S. & Hopkins, A. L. Quantifying the chemical beauty of drugs. Nat. Chem. 4, 90–98 (2012).
Ester, M., Kriegel, H., Xu, X. & Miinchen, D. A density-based algorithm for discovering clusters in large spatial databases with noise. In Proceedings of the Second International Conference on Knowledge, Discovery and Data Mining 226–231 (AAAI Press, 1996).
Škrlj, B., Džeroski, S., Lavrač, N. & Petkovič, M. Feature importance estimation with self-attention networks. Preprint at https://arxiv.org/pdf/2002.04464.pdf (2020).
Olden, J. D., Joy, M. K. & Death, R. G. An accurate comparison of methods for quantifying variable importance in artificial neural networks using simulated data. Ecol. Model. 178, 389–397 (2004).
Hung, L. & Chung, H. Decoupled control using neural network-based sliding-mode controller for nonlinear systems. Expert Syst. Appl. 32, 1168–1182 (2007).
Gaulton, A. et al. The ChEMBL database in 2017. Nucleic Acids Res. 45, D945–D954 (2017).
Sun, J. et al. ExCAPE-DB: an integrated large scale dataset facilitating big data analysis in chemogenomics. J. Cheminform. 9, 41 (2017).
Swain, M. MolVS: Molecule Validation and Standardization v0.1.1 (2019); https://molvs.readthedocs.io/en/latest/
Sun, J. et al. ExCAPEDB (2019); https://solr.ideaconsult.net/search/excape/
Landrum, G. et al. RDKit: Open-Source Cheminformatics Software (2019); https://www.rdkit.org/
Butina, D. Unsupervised data base clustering based on daylight’s fingerprint and Tanimoto similarity: a fast and automated way to cluster small and large data sets. J. Chem. Inf. Comput. Sci. 39, 747–750 (1999).
Bjerrum, E. J. Molvecgen: Molecular Vectorization and Batch Generation (2019); https://github.com/EBjerrum/molvecgen
Pedregosa, F. et al. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 12, 2825–2830 (2011).
O’Boyle, N. M. & Sayle, R. A. Comparing structural fingerprints using a literature-based similarity benchmark. J. Cheminform. 8, 36 (2016).
Probst, D. & Reymond, J. L. A probabilistic molecular fingerprint for big data settings. J. Cheminform. 10, 66 (2018).
Chollet, F. Keras (2019); https://keras.io/
Abadi, M. et al. TensorFlow: large-scale machine learning on heterogeneous distributed systems. Preprint at https://arxiv.org/pdf/1603.04467.pdf (2016).
Arora, R., Basu, A., Mianjy, P. & Mukherjee, A. Understanding deep neural networks with rectified linear units. Preprint at https://arxiv.org/pdf/1611.01491.pdf (2016).
Williams, R. J. & Zipser, D. A learning algorithm for continually running fully recurrent neural networks. Neural Comput. 1, 270–280 (2008).
Kingma, D. P. & Ba, J. Adam: a method for stochastic optimization. In 3rd International Conference for Learning Representations, (ICLR) 2015, San Diego, CA, USA, May 7-9, 2015, Conference Track Proceedings (eds Bengio, Y. & LeCun, Y.) (2015).
Wildman, S. A. & Crippen, G. M. Prediction of physicochemical parameters by atomic contributions. J. Chem. Inf. Comput. Sci. 39, 868–873 (1999).
Tan, C. et al. A survey on deep transfer learning. In Artificial Neural Networks and Machine Learning — ICANN 2018 (eds Krurková, V., Manolopoulos, Y., Hammer, B., Iliadis, L. & Maglogiannis, I.) 270–279 (Springer, 2018).
MIT Licence; https://opensource.org/licenses/MIT
Acknowledgements
We thank the entire MolecularAI team at AstraZeneca for their invaluable input and the fruitful discussions held during development of the present work. J.A.-P. is supported financially by the European Union’s Horizon 2020 research and innovation programme under a Marie Skłodowska-Curie grant (agreement no. 676434, ‘Big Data in Chemistry’, ‘BIGCHEM’; http://bigchem.eu).
Author information
Authors and Affiliations
Contributions
P.-C.K. and E.J.B. planned the project and jointly performed analysis of the results. P.-C.K. developed the necessary code. E.J.B. supervised the overall project. J.A.-P. assisted with the preprocessing of the datasets. J.A.-P., H.C., O.E. and C.T. provided valuable feedback on the methods used, the experimental set-up and the results at every stage. P.-C.K. wrote the manuscript and all authors reviewed it.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Distribution of physicochemical properties of datasets.
a, Wildman-Crippen coefficient (logP), b, topological polar surface area (TPSA), c, molecular weight (MW), d, drug-likeness score (QED), e, number of hydrogen bond acceptors (HBA) and f, hydrogen bond donors (HBD) with respect to the complete CHEMBL25 and DRD2 datasets before splitting. Subfigures a-d show the continuous histogram density as estimated by the kdeplot method of the seaborn Python library using default parameters.
Extended Data Fig. 2 Tanimoto similarity and predicted activity of generated structures.
a, Distribution of pairwise Tanimoto similarity of uniquely generated Murcko scaffolds to the seeding Murcko scaffold. The physchem-based (PCB) model generates SMILES that correspond to new scaffolds whereas the fingerprint-based (FPB) model generates scaffolds that are more similar or even identical to the seeding scaffold. b, Predicted active probability of all unique structures behind all generated SMILES strings per model. Both models generate SMILES that are predicted to be active with similar probability distributions.
Extended Data Fig. 3 Novelty of uniquely generated underlying molecules with respect to different datasets.
Novelty is assessed with respect to the train and test ChEMBL datasets using the physchem-based (PCB) and fingerprint-based (FPB) models. The first element of every pair on the x-axis corresponds to the dataset the conditions were drawn from. The second element represents the dataset with respect to which novelty was calculated. For any model the difference between datasets is insignificant, reflecting a consistent generation of novel compounds regardless of the seeding conditions. The numbers correspond to the fraction of valid unique novel molecules out of 25,600 generated SMILES strings.
Extended Data Fig. 4 Optimization of properties individually in every direction with the physchem-based model.
The pattern of the molecular properties of the generated valid SMILES (blue dots) seems to follow the set conditions (red lines). The length of a step represents the number of valid SMILES for that setpoint out of 256 sampled SMILES strings. Low molecular weight or high QED setpoints lead to unstable generation of valid SMILES for the given condition. QED displays the largest deviations from the seed conditions and is the hardest property to control as the formula contains a weighted sum of the other five properties. The area annotated by arrows refers to an input combination with a high QED target that caused the output to collapse with respect to the rate of valid SMILES and the fulfillment of the specified conditions. The exact percentage of unique molecules stemming from all valid SMILES sampled at each step is shown in Supplementary Fig. 12.
Supplementary information
Supplementary Information
Likelihood of sampling of canonical SMILES and Figs. 1–12.
Rights and permissions
About this article
Cite this article
Kotsias, PC., Arús-Pous, J., Chen, H. et al. Direct steering of de novo molecular generation with descriptor conditional recurrent neural networks. Nat Mach Intell 2, 254–265 (2020). https://doi.org/10.1038/s42256-020-0174-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-020-0174-5
This article is cited by
-
Integrating QSAR modelling and deep learning in drug discovery: the emergence of deep QSAR
Nature Reviews Drug Discovery (2024)
-
LOGICS: Learning optimal generative distribution for designing de novo chemical structures
Journal of Cheminformatics (2023)
-
Regression Transformer enables concurrent sequence regression and generation for molecular language modelling
Nature Machine Intelligence (2023)
-
Testing the limits of SMILES-based de novo molecular generation with curriculum and deep reinforcement learning
Nature Machine Intelligence (2023)
-
Hierarchical Molecular Graph Self-Supervised Learning for property prediction
Communications Chemistry (2023)