Abstract
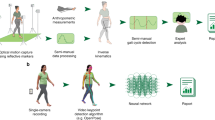
Motion analysis is used in computer vision to understand the behaviour of moving objects in sequences of images. Optimizing the interpretation of dynamic biological systems requires accurate and precise motion tracking as well as efficient representations of high-dimensional motion trajectories so that these can be used for prediction tasks. Here we use image sequences of the heart, acquired using cardiac magnetic resonance imaging, to create time-resolved three-dimensional segmentations using a fully convolutional network trained on anatomical shape priors. This dense motion model formed the input to a supervised denoising autoencoder (4Dsurvival), which is a hybrid network consisting of an autoencoder that learns a task-specific latent code representation trained on observed outcome data, yielding a latent representation optimized for survival prediction. To handle right-censored survival outcomes, our network used a Cox partial likelihood loss function. In a study of 302 patients, the predictive accuracy (quantified by Harrell’s C-index) was significantly higher (P = 0.0012) for our model C = 0.75 (95% CI: 0.70–0.79) than the human benchmark of C = 0.59 (95% CI: 0.53–0.65). This work demonstrates how a complex computer vision task using high-dimensional medical image data can efficiently predict human survival.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data and code availability
Algorithms, motion models and statistical analysis are publicly available on Github under a GNU General Public License (https://github.com/UK-Digital-Heart-Project/4Dsurvival)76. A training simulation is available as a Docker image with an interactive Jupyter notebook hosted on Code Ocean (https://doi.org/10.24433/CO.8519672.v1)77. Personal data are not available due to privacy restrictions.
References
Wang, L., Zhao, G., Cheng, L. & Pietikäinen, M. Machine Learning for Vision-Based Motion Analysis: Theory and Techniques (Springer, London, 2010).
Mei, T. & Zhang, C. Deep learning for intelligent video analysis. Microsoft; https://www.microsoft.com/en-us/research/publication/deep-learning-intelligent-video-analysis/ (2017).
Liang, F., Xie, W. & Yu, Y. Beating heart motion accurate prediction method based on interactive multiple model: an information fusion approach. Biomed. Res. Int. 2017, 1279486 (2017).
Savarese, G. & Lund, L. H. Global public health burden of heart failure. Card. Fail. Rev. 3, 7–11 (2017).
Galie, N. et al. 2015 ESC/ERS guidelines for the diagnosis and treatment of pulmonary hypertension: The Joint Task Force for the Diagnosis and Treatment of Pulmonary Hypertension of the European Society of Cardiology (ESC) and the European Respiratory Society (ERS): Endorsed by: Association for European Paediatric and Congenital Cardiology (AEPC), International Society for Heart and Lung Transplantation (ISHLT). Eur. Heart J. 37, 67–119 (2016).
Puyol-Antón, E. et al. A multimodal spatiotemporal cardiac motion atlas from MR and ultrasound data. Med. Image Anal. 40, 96–110 (2017).
Scatteia, A., Baritussio, A. & Bucciarelli-Ducci, C. Strain imaging using cardiac magnetic resonance. Heart Fail. Rev. 22, 465–476 (2017).
Belkin, M. & Niyogi, P. Laplacian eigenmaps and spectral techniques for embedding and clustering. In Advances in Neural Information Processing Systems 14 (eds Dietterich, T. G. et al.) 585–591 (MIT Press, Cambridge, 2002).
Li, K., Javer, A., Keaveny, E. E. & Brown, A. E. X. Recurrent neural networks with interpretable cells predict and classify worm behaviour. Preprint at https://doi.org/10.1101/222208 (2017).
Walker, J., Doersch, C., Gupta, A. & Hebert, M. An uncertain future: forecasting from static images using variational autoencoders. Preprint at https://arxiv.org/abs/1606.07873 (2016).
Bütepage, J., Black, M., Kragic, D. & Kjellström, H. Deep representation learning for human motion prediction and classification. Preprint at https://arxiv.org/abs/1702.07486 (2017).
Johnson, K. W. et al. Enabling precision cardiology through multiscale biology and systems medicine. JACC Basic Transl. Sci. 2, 311–327 (2017).
Cikes, M. & Solomon, S. D. Beyond ejection fraction: an integrative approach for assessment of cardiac structure and function in heart failure. Eur. Heart J. 37, 1642–1650 (2016).
Ahmad, T. et al. Clinical implications of chronic heart failure phenotypes defined by cluster analysis. J. Am. Coll. Cardiol. 64, 1765–1774 (2014).
Shah, S. J. et al. Phenomapping for novel classification of heart failure with preserved ejection fraction. Circulation 131, 269–279 (2015).
Awan, S. E., Sohel, F., Sanfilippo, F. M., Bennamoun, M. & Dwivedi, G. Machine learning in heart failure: ready for prime time. Curr. Opin. Cardiol. 33, 190–195 (2018).
Tripoliti, E. E., Papadopoulos, T. G., Karanasiou, G. S., Naka, K. K. & Fotiadis, D. I. Heart failure: diagnosis, severity estimation and prediction of adverse events through machine learning techniques. Comput. Struct. Biotechnol. J. 15, 26–47 (2017).
Ambale-Venkatesh, B. et al. Cardiovascular event prediction by machine learning: the multi-ethnic study of atherosclerosis. Circ. Res. 121, 1092–1101 (2017).
Yousefi, S. et al. Predicting clinical outcomes from large scale cancer genomic profiles with deep survival models. Sci. Rep. 7, 11707 (2017).
Ching, T., Zhu, X. & Garmire, L. X. Cox-nnet: an artificial neural network method for prognosis prediction of high-throughput omics data. PLoS Comput. Biol. 14, 1–18 (2018).
Katzman, J. et al. DeepSurv: personalized treatment recommender system using a Cox proportional hazards deep neural network. BMC Med. Res. Methodol. 18, 1–12 (2018).
Esteva, A. et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 542, 115–118 (2017).
Ching, T. et al. Opportunities and obstacles for deep learning in biology and medicine.J. R. Soc. Interface 15, 20170387 (2018).
Litjens, G. et al. A survey on deep learning in medical image analysis. Med. Image Anal. 42, 60–88 (2017).
Shen, D., Wu, G. & Suk, H. I. Deep learning in medical image analysis. Annu. Rev. Biomed. Eng. 19, 221–248 (2017).
Bai, W. et al. Automated cardiovascular magnetic resonance image analysis with fully convolutional networks. J. Cardiovasc. Magn. Reson. 20, 65 (2018).
Piras, P. et al. Morphologically normalized left ventricular motion indicators from MRI feature tracking characterize myocardial infarction. Sci. Rep. 7, 12259 (2017).
Zhang, X. et al. Orthogonal decomposition of left ventricular remodeling in myocardial infarction. Gigascience 6, 1–15 (2017).
Zhang, X. et al. Atlas-based quantification of cardiac remodeling due to myocardial infarction. PLoS ONE 9, e110243 (2014).
Dawes, T. et al. Machine learning of three-dimensional right ventricular motion enables outcome prediction in pulmonary hypertension: a cardiac MR imaging study. Radiology 283, 381–390 (2017).
Rifai, S., Vincent, P., Muller, X., Glorot, X. & Bengio, Y. Contractive auto-encoders: explicit invariance during feature extraction. In Proc. 28th International Conference on Machine Learning, 833–840 (Omnipress, 2011).
Rolfe, J. T. & LeCun, Y. Discriminative recurrent sparse auto-encoders. Preprint at 1301.3775 (2013).
Huang, R., Liu, C., Li, G. & Zhou, J. Adaptive deep supervised autoencoder based image reconstruction for face recognition. Math. Probl. Eng. 2016, 14 (2016).
Du, F., Zhang, J., Ji, N., Hu, J. & Zhang, C. Discriminative representation learning with supervised auto-encoder. Neur. Proc. Lett. https://doi.org/10.1007/s11063-018-9828-2 (2018).
Zaghbani, S., Boujneh, N. & Bouhlel, M. S. Age estimation using deep learning. Comp. Elec. Eng. 68, 337–347 (2018).
Beaulieu-Jones, B. K. & Greene, C. S. Semi-supervised learning of the electronic health record for phenotype stratification. J. Biomed. Inform. 64, 168–178 (2016).
Shakeri, M., Lombaert, H., Tripathi, S. & Kadoury, S. Deep spectral-based shape features for Alzheimer’s disease classification. In International Workshop on Spectral and Shape Analysis in Medical Imaging (eds Reuter, M. et al.) 15–24 (Springer, 2016).
Biffi, C. et al. Learning interpretable anatomical features through deep generative models: Application to cardiac remodeling. In International Conference on Medical Image Computing and Computer-Assisted Intervention Vol. 11071 (eds Frangi, A., Schnabel, J., Davatzikos, C., Alberola-López, C. & Fichtinger, G.) (Springer, 2018).
Dawes, T. J. W., Bello, G. A. & O’Regan, D. P. Multicentre study of machine learning to predict survival in pulmonary hypertension. Open Science Framework https://doi.org/10.17605/OSF.IO/BG6T9 (2018).
Grapsa, J. et al. Echocardiographic and hemodynamic predictors of survival in precapillary pulmonary hypertension: seven-year follow-up. Circ. Cardiovasc. Imaging 8, 45–54 (2015).
Bao, W., Yue, J. & Rao, Y. A deep learning framework for financial time series using stacked autoencoders and long-short term memory. PLoS ONE 12, e0180944 (2017).
Lim, B. & van der Schaar, M. Disease-atlas: navigating disease trajectories with deep learning. Preprint at https://arxiv.org/abs/1803.10254 (2018).
Lee, C., Zame, W. R., Yoon, J. & van der Schaar, M. DeepHit: a deep learning approach to survival analysis with competing risks. In 32nd Association for the Advancement of Artificial Intelligence ( AAAI) Conference (2018).
Gopalan, D., Delcroix, M. & Held, M. Diagnosis of chronic thromboembolic pulmonary hypertension. Eur. Respir. Rev. 26, 160108 (2017).
Kramer, C., Barkhausen, J., Flamm, S., Kim, R. & Nagel, E. Society for cardiovascular magnetic resonance board of trustees task force on standardized protocols. Standardized cardiovascular magnetic resonance (CMR) protocols 2013 update. J. Cardiovasc. Magn. Reson. 15, 91 (2013).
Woodbridge, M., Fagiolo, G. & O’Regan, D. P. MRIdb: medical image management for biobank research. J. Digit. Imaging 26, 886–890 (2013).
Schulz-Menger, J. et al. Standardized image interpretation and post processing in cardiovascular magnetic resonance: society for cardiovascular magnetic resonance (SCMR) board of trustees task force on standardized post processing. J. Cardiovasc. Magn. Reson. 15, 35 (2013).
Baggen, V. J. et al. Cardiac magnetic resonance findings predicting mortality in patients with pulmonary arterial hypertension: a systematic review and meta-analysis. Eur. Radiol. 26, 3771–3780 (2016).
Hulshof, H. G. et al. Prognostic value of right ventricular longitudinal strain in patients with pulmonary hypertension: a systematic review and meta-analysis. Eur. Heart J. Cardiovasc. Imaging https://doi.org/10.1093/ehjci/jey120 (2018).
Duan, J. et al. Automatic 3D bi-ventricular segmentation of cardiac images by a shape-constrained multi-task deep learning approach. Preprint at 1808.08578 (2018).
Bai, W. et al. A bi-ventricular cardiac atlas built from 1000+ high resolution MR images of healthy subjects and an analysis of shape and motion. Med. Image Anal. 26, 133–145 (2015).
Shi, W. et al. Temporal sparse free-form deformations. Med. Image Anal. 17, 779–789 (2013).
Rueckert, D. et al. Nonrigid registration using free-form deformations: application to breast MR images. IEEE Trans. Med. Imaging 18, 712–721 (1999).
Bai, W et al. Learning a global descriptor of cardiac motion from a large cohort of 1000+ normal subjects. In 8th International Conference on Functional Imaging and Modeling of the Heart (FIMH’15) Vol. 9126 (Springer, Cham, 2015).
Vincent, P., Larochelle, H., Lajoie, I., Bengio, Y. & Manzagol, P.-A. Stacked denoising autoencoders: learning useful representations in a deep network with a local denoising criterion. J. Mach. Learn. Res. 11, 3371–3408 (2010).
Cox, D. Regression models and life-tables. J. R. Stat. Soc. B 34, 187–220 (1972).
Srivastava, N., Hinton, G., Krizhevsky, A., Sutskever, I. & Salakhutdinov, R. Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 15, 1929–1958 (2014).
Goodfellow, I., Bengio, Y. & Courville, A. Deep Learning (MIT Press, Cambridge MA, 2016).
Faraggi, D. & Simon, R. A neural network model for survival data. Stat. Med. 14, 73–82 (1995).
Abadi, M. et al. TensorFlow: Large-Scale Machine Learning on Heterogeneous Systems (TensorFlow, 2015); http://download.tensorflow.org/paper/whitepaper2015.pdf
Chollet, F. et al. Keras https://keras.io (2015).
Kennedy, J. & Eberhart, R. Particle swarm optimization. Proc. IEEE Int. Conf. Neural Net. 4, 1942–1948 (1995).
Engelbrecht, A. Fundamentals of Computational Swarm Intelligence (Wiley, Chichester, 2005).
Lorenzo, P. R., Nalepa, J., Kawulok, M., Ramos, L. S. & Pastor, J. R. Particle swarm optimization for hyper-parameter selection in deep neural networks. In Proc. Genetic and Evolutionary Computation Conference, GECCO ‘17, 481–488 (2017).
Claesen, M., Simm, J., Popovic, D. & De Moor, B. Hyperparameter tuning in Python using Optunity.In Proc. International Workshop on Technical Computing for Machine Learning and Mathematical Engineering Vol. 9 (2014).
Harrell, F., Califf, R., Pryor, D., Lee, K. & Rosati, R. Evaluating the yield of medical tests.J. Am. Med. Assoc. 247, 2543–2546 (1982).
Moons, K. et al. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): explanation and elaboration. Ann. Intern. Med. 162, W1–W73 (2015).
Harrell, F., Lee, K. & Mark, D. Tutorial in biostatistics: multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat. Med. 15, 361–387 (1996).
Efron, B. Estimating the error rate of a prediction rule: some improvements on cross-validation. J. Am. Stat. Assoc. 78, 316–331 (1983).
Efron, B. & Tibshirani, R. in An Introduction to the Bootstrap Ch. 17 (Chapman & Hall, New York, 1993).
Smith, G., Seaman, S., Wood, A., Royston, P. & White, I. Correcting for optimistic prediction in small data sets. Am. J. Epidem. 180, 318–324 (2014).
Liu, B. et al. Normal values for myocardial deformation within the right heart measured by feature-tracking cardiovascular magnetic resonance imaging. Int. J. Cardiol. 252, 220–223 (2018).
Gall, H. et al. The Giessen pulmonary hypertension registry: survival in pulmonary hypertension subgroups. J. Heart Lung. Transplant. 36, 957–967 (2017).
Stekhoven, D. J. & Buhlmann, P. missForest–non–parametric missing value imputation for mixed-type data. Bioinformatics 28, 112–118 (2011).
Schroder, M. S., Culhane, A. C., Quackenbush, J. & Haibe-Kains, B. survcomp: an R/Bioconductor package for performance assessment and comparison of survival models. Bioinformatics 27, 3206–3208 (2011).
Bello, G. A. & O’Regan, D. Deep learning cardiac motion analysis for human survival prediction (4Dsurvival) Zenodo https://doi.org/10.5281/zenodo.1451540 (2019).
Bello, G. et al. Deep learning cardiac motion analysis for human survival prediction (4Dsurvival). Code Ocean https://doi.org/10.24433/CO.8519672.v1 (2018).
Acknowledgements
The research was supported by the British Heart Foundation (NH/17/1/32725, RE/13/4/30184); the National Institute for Health Research Biomedical Research Centre based at Imperial College Healthcare NHS Trust and Imperial College London; and the Medical Research Council, UK. The TITAN Xp GPU used for this research was kindly donated by the NVIDIA Corporation.
Author information
Authors and Affiliations
Contributions
G.A.B., C.B. and T.J.W.D. contributed to methodology, software, formal analysis and writing original draft. J.D. contributed to methodology, software and writing original draft; A.d.M. was involved with formal analysis; L.S.G.E.H., J.S.R.G., M.R.W. and S.A.C. were involved in investigation; D.R. contributed to software and supervision; D.P.O. was responsible for conceptualization, supervision, writing (review and editing) and funding acquisition. All authors reviewed the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Notes
Rights and permissions
About this article
Cite this article
Bello, G.A., Dawes, T.J.W., Duan, J. et al. Deep-learning cardiac motion analysis for human survival prediction. Nat Mach Intell 1, 95–104 (2019). https://doi.org/10.1038/s42256-019-0019-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42256-019-0019-2
This article is cited by
-
Deep Learning Based Prediction of Pulmonary Hypertension in Newborns Using Echocardiograms
International Journal of Computer Vision (2024)
-
Automated inversion time selection for late gadolinium–enhanced cardiac magnetic resonance imaging
European Radiology (2024)
-
Automated segmentation of long and short axis DENSE cardiovascular magnetic resonance for myocardial strain analysis using spatio-temporal convolutional neural networks
Journal of Cardiovascular Magnetic Resonance (2023)
-
Biopsy-free AI-aided precision MRI assessment in prediction of prostate cancer biochemical recurrence
British Journal of Cancer (2023)
-
Deep learning-based prognostic model using non-enhanced cardiac cine MRI for outcome prediction in patients with heart failure
European Radiology (2023)