Abstract
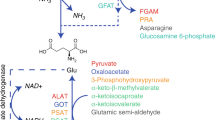
Branched-chain amino acids (BCAAs) supply both carbon and nitrogen in pancreatic cancers, and increased levels of BCAAs have been associated with increased risk of pancreatic ductal adenocarcinomas (PDACs). It remains unclear, however, how stromal cells regulate BCAA metabolism in PDAC cells and how mutualistic determinants control BCAA metabolism in the tumour milieu. Here, we show distinct catabolic, oxidative and protein turnover fluxes between cancer-associated fibroblasts (CAFs) and cancer cells, and a marked reliance on branched-chain α-ketoacid (BCKA) in PDAC cells in stroma-rich tumours. We report that cancer-induced stromal reprogramming fuels this BCKA demand. The TGF-β–SMAD5 axis directly targets BCAT1 in CAFs and dictates internalization of the extracellular matrix from the tumour microenvironment to supply amino-acid precursors for BCKA secretion by CAFs. The in vitro results were corroborated with circulating tumour cells (CTCs) and PDAC tissue slices derived from people with PDAC. Our findings reveal therapeutically actionable targets in pancreatic stromal and cancer cells.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data that support the plots within this paper and other findings of this study are available from the corresponding author upon reasonable request. Source data are provided with this paper.
Code availability
R code used for stromal/epithelial deconvolution, heatmap generation and violin plots and bioinformatics analysis is available from the corresponding author upon request. Source data are provided with this paper.
References
Tonjes, M. et al. BCAT1 promotes cell proliferation through amino acid catabolism in gliomas carrying wild-type IDH1. Nat. Med. 19, 901–908 (2013).
Raffel, S. et al. BCAT1 restricts αKG levels in AML stem cells leading to IDH mut-like DNA hypermethylation. Nature 551, 384 (2017).
Green, C. R. et al. Branched-chain amino acid catabolism fuels adipocyte differentiation and lipogenesis. Nat. Chem. Biol. 12, 15–21 (2016).
Dey, P. et al. Genomic deletion of malic enzyme 2 confers collateral lethality in pancreatic cancer. Nature 542, 119–123 (2017).
Neesse, A. et al. Stromal biology and therapy in pancreatic cancer. Gut 60, 861 (2011).
Schworer, S., Vardhana, S. A. & Thompson, C. B. Cancer metabolism drives a stromal regenerative response. Cell Metab. 29, 576–591 (2019).
Neinast, M. D. et al. Quantitative analysis of the whole-body metabolic fate of branched-chain amino acids. Cell Metab. 29, 417–429 (2019). e414.
Mayers, J. R. et al. Tissue of origin dictates branched-chain amino acid metabolism in mutant Kras-driven cancers. Science 353, 1161–1165 (2016).
Perera, R. M. & Bardeesy, N. Pancreatic cancer metabolism: breaking it down to build it back up. Cancer Discov. 5, 1247–1261 (2015).
Danai, L. V. et al. Altered exocrine function can drive adipose wasting in early pancreatic cancer. Nature 558, 600–604 (2018).
Uhlén, M. et al. Tissue-based map of the human proteome. Science 347, 1260419 (2015).
Pinho, A. V. et al. ROBO2 is a stroma suppressor gene in the pancreas and acts via TGF-β signalling. Nat. Commun. 9, 5083 (2018).
Bailey, P. et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 531, 47–52 (2016).
Janky, R. et al. Prognostic relevance of molecular subtypes and master regulators in pancreatic ductal adenocarcinoma. BMC Cancer 16, 632 (2016).
Stratford, J. K. et al. A six-gene signature predicts survival of patients with localized pancreatic ductal adenocarcinoma. PLoS Med. 7, e1000307 (2010).
Neesse, A., Algul, H., Tuveson, D. A. & Gress, T. M. Stromal biology and therapy in pancreatic cancer: a changing paradigm. Gut 64, 1476–1484 (2015).
Zhao, H. et al. Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. eLife 5, e10250 (2016).
Bernard, V. et al. Single-cell transcriptomics of pancreatic cancer precursors demonstrates epithelial and microenvironmental heterogeneity as an early event in neoplastic progression. Clin. Cancer Res. 25, 2194–2205 (2019).
Peng, J. et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 29, 725–738 (2019).
Sousa, C. M. et al. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature 536, 479–483 (2016).
Lu, G. et al. Protein phosphatase 2Cm is a critical regulator of branched-chain amino acid catabolism in mice and cultured cells. J. Clin. Invest. 119, 1678–1687 (2009).
Grankvist, N., Lagerborg, K. A., Jain, M. & Nilsson, R. Gabapentin can suppress cell proliferation independent of the cytosolic branched-chain amino acid transferase 1 (BCAT1). Biochemistry 57, 6762–6766 (2018).
Sherman, M. H. et al. Vitamin D receptor-mediated stromal reprogramming suppresses pancreatitis and enhances pancreatic cancer therapy. Cell 159, 80–93 (2014).
Sherman, M. H. et al. Stromal cues regulate the pancreatic cancer epigenome and metabolome. Proc. Natl Acad. Sci. USA 114, 1129–1134 (2017).
Commisso, C. et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature 497, 633–637 (2013).
Madsen, D. H. et al. The non-phagocytic route of collagen uptake: a distinct degradation pathway. J. Biol. Chem. 286, 26996–27010 (2011).
Jurgensen, H. J. et al. Immune regulation by fibroblasts in tissue injury depends on uPARAP-mediated uptake of collectins. J. Cell Biol. 218, 333–349 (2019).
Mishra, P. J. et al. Carcinoma-associated fibroblast-like differentiation of human mesenchymal stem cells. Cancer Res. 68, 4331–4339 (2008).
Yeung, T. L. et al. TGF-β modulates ovarian cancer invasion by upregulating CAF-derived versican in the tumor microenvironment. Cancer Res. 73, 5016–5028 (2013).
Franco-Barraza, J. et al. Matrix-regulated integrin αvβ5 maintains α5β1-dependent desmoplastic traits prognostic of neoplastic recurrence. eLife https://doi.org/10.7554/eLife.20600 (2017).
Ashworth, T. A case of cancer in which cells similar to those in the tumours were seen in the blood after death. Aust. Med. J. 14, 146–149 (1869).
Allard, W. J. et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin. Cancer Res. 10, 6897–6904 (2004).
Nagrath, S., Jack, R. M., Sahai, V. & Simeone, D. M. Opportunities and challenges for pancreatic circulating tumor cells. Gastroenterology 151, 412–426 (2016).
Lin, E. et al. High-throughput microfluidic labyrinth for the label-free isolation of circulating tumor cells. Cell Syst. 5, 295–304 (2017). e294.
Rivera-Baez, L. et al. Expansion of circulating tumor cells from patients with locally advanced pancreatic cancer enable patient derived xenografts and functional studies for personalized medicine. Cancers (Basel) https://doi.org/10.3390/cancers12041011 (2020).
van Geer, M. A. et al. Ex-vivo evaluation of gene therapy vectors in human pancreatic (cancer) tissue slices. World J. Gastroenterol. 15, 1359–1366 (2009).
Roife, D. et al. Ex vivo testing of patient-derived xenografts mirrors the clinical outcome of patients with pancreatic ductal adenocarcinoma. Clin. Cancer Res. 22, 6021–6030 (2016).
Auciello, F. R. et al. A stromal lysolipid–autotaxin signaling axis promotes pancreatic tumor progression. Cancer Discov. 9, 617–627 (2019).
Kamphorst, J. J. et al. Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res. 75, 544–553 (2015).
Halbrook, C. J. et al. Macrophage-released pyrimidines inhibit gemcitabine therapy in pancreatic cancer. Cell Metab. 29, 1390–1399.e1396 (2019).
Dalin, S. et al. Deoxycytidine release from pancreatic stellate cells promotes gemcitabine resistance. Cancer Res. 79, 5723–5733 (2019).
Li, J. T. et al. BCAT2-mediated BCAA catabolism is critical for development of pancreatic ductal adenocarcinoma. Nat. Cell Biol. 22, 167–174 (2020).
Lee, J. H. et al. Branched-chain amino acids sustain pancreatic cancer growth by regulating lipid metabolism. Exp. Mol. Med. 51, 1–11 (2019).
Ohlund, D. et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J. Exp. Med. 214, 579–596 (2017).
Biffi, G. et al. IL1-induced JAK–STAT signaling is antagonized by TGF-β to shape CAF heterogeneity in pancreatic ductal adenocarcinoma. Cancer Discov. 9, 282–301 (2019).
Elyada, E. et al. Cross-species single-cell analysis of pancreatic ductal adenocarcinoma reveals antigen-presenting cancer-associated fibroblasts. Cancer Discov. 9, 1102–1123 (2019).
Mishra, R., Haldar, S., Suchanti, S. & Bhowmick, N. A. Epigenetic changes in fibroblasts drive cancer metabolism and differentiation. Endocr. Relat. Cancer 26, R673–R688 (2019).
Bhagat, T. D. et al. Lactate-mediated epigenetic reprogramming regulates formation of human pancreatic cancer-associated fibroblasts. eLife 8, e50663 (2019).
Olivares, O. et al. Collagen-derived proline promotes pancreatic ductal adenocarcinoma cell survival under nutrient limited conditions. Nat. Commun. 8, 16031 (2017).
Sprangers, S., Behrendt, N., Engelholm, L., Cao, Y. & Everts, V. Phagocytosis of collagen fibrils by fibroblasts in vivo Is independent of the uPARAP/Endo180 receptor. J. Cell Biochem 118, 1590–1595 (2017).
Schmidt, E. K., Clavarino, G., Ceppi, M. & Pierre, P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods 6, 275–277 (2009).
Bilan, D. S. et al. Genetically encoded fluorescent indicator for imaging NAD+/NADH ratio changes in different cellular compartments. Biochim. Biophys. Acta 1840, 951–957 (2014).
Cooper, A. J., Conway, M. & Hutson, S. M. A continuous 96-well plate spectrophotometric assay for branched-chain amino acid aminotransferases. Anal. Biochem. 308, 100–105 (2002).
Boj, S. F. et al. Organoid models of human and mouse ductal pancreatic cancer. Cell 160, 324–338 (2015).
Maurer, C. et al. Experimental microdissection enables functional harmonisation of pancreatic cancer subtypes. Gut 68, 1034–1043 (2019).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411 (2018).
Pon, A. et al. Pathways with PathWhiz. Nucleic Acids Res. 43, W552–W559 (2015).
Acknowledgements
We thank and are grateful to three anonymous reviewers and editors for their incisive comments and several excellent suggestions. D.N. is supported by grants from NCI R01CA227622, R01CA222251 and R01CA204969. DN is also supported by Rogel Cancer Center grant. J.S. is supported by grant from NCI K08 CA234222. EC is funded by NCI grants R21CA231252 and R01CA232256, Core Grant CA06927. This work was supported in part by grants from NCI R01CA208335 to S.N. Z.Z. and A.A. are supported by the University of Michigan Precision Health Scholars Awards. Flow cytometry experiments were performed in the Flow Cytometry Research Core at the University of Michigan, Biomedical Research Core Facilities (BRCF). Confocal microscopy was performed in the Microscopy and Image-analysis Laboratory (MIL) at the University of Michigan, Biomedical Research Core Facilities (BRCF). Scanning electron microscopy was performed at the Michigan Center for Materials Characterization (MC2) at University of Michigan which is supported by the University of Michigan College of Engineering and NSF grant DMR-0320740.
Author information
Authors and Affiliations
Contributions
Z.Z., A.A. and D.N. designed the experiments. Z.Z. performed most experiments. A.A. and O.A. developed and performed all of the bioinformatics analysis. S.O. and S.N. collected PDAC samples, and isolated, characterized and developed CTC lines using the microfluidics Labyrinth chip. A.A., O.A., N.M. and A. Mittal developed and performed all metabolic and mass-spectrometry assays. P.P. and T.W.L. assisted with assays. J.F.-B. and E.C. provided CAFs and helped with stromal characterization, J.S. helped with patient tissue collection, pathological interpretation and IHC, V.G. and V.S. collected blood for CTC analysis, M.H.S. provided CAFs and helped with stromal characterization, A.M.P. helped with proteasomal analysis, A. Maitra analysed the data and helped in clinical correlations and M.A.M. and T.S.L. provided tissue slices and helped in designing various experiments. Z.Z., A.A. and D.N. wrote the manuscript with input from coauthors.
Corresponding author
Ethics declarations
Competing interests
A. Maitra receives royalties from Cosmos Wisdom Biotechnologies for a license related to a biomarker test for pancreatic cancer early detection. V.S. is a consultant at Halozyme, QED, Ipsen and Incyte. V.S. receives funding from Celgene, Bristol-Myers Squibb, Agios, Incyte, Clovis Oncology, Debiopharm Group, FibroGen, Halozyme, MedImmune, Rafael Pharmaceuticals and Ipsen. M.A.M. receives honoraria from AstraZeneca. S.N. is the named inventor on a patent for Microfluidic Labyrinth Technology granted to the University of Michigan. S.N. is a cofounders of Labyrinth Biotech. The funders and the company had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Additional information
Peer review information Primary Handling Editor: Elena Bellafante.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Transcriptomic analysis of BCAA metabolic genes in PDAC tumors.
a. Expression of BCAT1 healthy tissue samples from the GTEx database (Brain, n = 2642; Prostate, n = 245; Testis, n = 361; Pancreas, n = 328; Ovary, n = 180). b. Expression of genes involved in BCAA metabolism in samples from GSE21501 (n = 132). Tumor samples with dominant epithelial markers and dominant fibroblast markers are deconvolved to compare expression of metabolic genes between pancreatic cancer cells and stromal cells in the TME. ROBO1 is a marker for validation that has been found to be expressed in stromal cells but not in cancer cells in independent studies. c. Expression of genes involved in BCAA metabolism in samples from GSE36924 (n = 91). d. Expression of genes involved in BCAA metabolism in samples from GSE62165 (n = 118). Samples with dominant epithelial markers and dominant fibroblast markers are deconvolved to compare expression of metabolic genes between epithelial cells and stromal cells (a–d). e. Gene expression of BCAA pathway genes and ROBO1 in paired epithelial and stromal compartments obtained by laser microdissection (GSE 93326, n = 63 paired samples). Violin plot represents all data points in each group (a–d). Boxplot limits represent median and interquartile range (IQR), and notches represent 1.5*IQR (a–e). Data analyzed using multiple, two-tailed, unpaired, Student’s t-test (a–d); multiple, two-tailed, paired, Student’s t-test (e).
Extended Data Fig. 2 Characterization of BCAA metabolism.
a. Representative IHC staining image comparing BCAT1 expression between stromal and tumor compartments. Experiments were repeated independently three times with similar results. b. Representative IF images showing protein expression of stromal αSMA, BCAT1 and Vimentin from paired healthy and PDAC tissue. Experiments were repeated independently twice with similar results. c. Absolute cell numbers of PDAC CAFs were determined in the presence or absence of BCAA. n = 3 biologically independent samples. d. Absolute cell numbers of PDAC cell were determined in the presence or absence of BCAAs. n = 3 biologically independent samples. Data are presented as mean ± s.d.
Extended Data Fig. 3 PDAC cells are BCAT2 dependent for growth.
a. Fluorescence microscopy images merged with brightfield images comparing growth of GFP-labeled Mia Paca-2 and Panc-1 cells in contact co-cultures with CAFs or NOFs under BCAA deprivation. Experiments were repeated independently three times with similar results. b. Relative growth rates of Mia Paca-2 cells co-cultured with CAFs or NOFs at different seeding ratios under BCAA deprivation. n = 3 biologically independent samples. c. Relative growth rates of AsPC1 and BxPC-3 cells co-cultured with CAFs or NOFs under BCAA deprivation. n = 3 biologically independent samples. d. Relative growth rates of Mia Paca-2 cells in various concentrations of BCAAs or BCKAs. n = 6 biologically independent samples. e. Model for the rescue of proliferation in BCAT2 KD cancer cells by BCKAs released from CAFs under BCAA deprivation. f. Relative growth rates of Mia Paca-2 and Panc-1 cells co-cultured with ATG-5/7 knockdown CAFs. n = 3 biologically independent samples. g. Relative growth rates of Mia Paca-2 and Patu 8988t cells cocultured with CAFs treated with autophagy inhibitors (chloroquine, Bafilomycin A1 and LY294002) under BCAA deprivation. n = 3 biologically independent samples. *P < 0.0001. Data are presented as mean ± s.d. Two-tailed, unpaired, Student’s t-test (c).
Extended Data Fig. 4 BCKDH complex is essential for PDAC cells growth and cell biosynthesis.
a. Relative proliferation rates of Mia Paca-2, Panc-1 and Patu 8988t cells expressing control shRNA or two independent shRNAs to DBT. n = 8 biologically independent samples. b. Colony-formation assay of DBT knockdown pancreatic cell lines. n = 3 biologically independent samples. c. Relative growth rates of Patu 8988t, Mia Paca-2 cells, and CAFs treated with BCKDK inhibitor, 3,6- dichlorobenzo[b]thiophene-2-carboxylic acid (BT2). n = 3 biologically independent samples. d. Relative growth rates of MiaPaca-2 and Patu 8988t cells under BCAA deprivation and low glucose and low glutamine conditions after supplementation with BCKAs. n = 3 biologically independent samples. e. Schematic for the loss of rescue in DBT knockdown cancer cells by BCKAs released from CAFs under BCAA deprivation. f. Colocalization of Mitotracker and RexMito fluorescence in Mia Paca-2 cells. Mitotracker (red), RexMito (green), and DAPI (blue). Experiments were repeated independently three times with similar results. g. Substrate-specific oxygen consumption rate (OCR) in permeabilized pancreatic cancer cells. n = 4 biologically independent samples. h. OCR of Panc-1 cells after BCAT2 and DBT knockdown. n = 18 biologically independent samples. i. Substrate-specific OCR of BCAT2 knockdown pancreatic cancer cells. n = 4 biologically independent samples. j. Substrate-specific of DBT knockdown pancreatic cancer cells. n = 4 biologically independent samples. *P < 0.0001. Data are presented as mean ± s.d. One-way ANOVA with Tukey’s post hoc comparison (a, j); two-way ANOVA with Dunnett’s multiple comparison test (i, j).
Extended Data Fig. 5 CAFs have upregulated collagen uptake under BCAA deprivation.
a. BCAT activity in CAFs treated with Gabapentin measured by spectrophotometric assay. n = 6 biologically independent samples. b. Growth rate of Panc-1 cancer cells with Gabapentin, BCKAs, and CAF coculture under BCAA deprivation c. The effect of knockdown of BCAT1 in CAFs on CAF growth rates. n = 4 biologically independent samples. d. Uptake of DQ-Collagen by CAFs after 24 h measured using confocal imaging. Experiments were repeated independently three times with similar results. e. Uptake of DQ-Collagen by PDAC cell lines and CAFs after 24 h measured using confocal imaging. Experiments were repeated independently three times with similar results. f. Flow cytometry assay of MRC2 expression in PDAC cell lines. Experiments were repeated independently three times with similar results. g. Flow cytometry assay of MRC2 expression in CAFs. Experiments were repeated independently three times with similar results. *P < 0.0001. Data are presented as mean ± s.d. One-way ANOVA with Tukey’s post hoc comparison (b).
Extended Data Fig. 6 CAFs uptake collagen through the proteasome.
a. Uptake of DQ-Collagen by CAFs transfected with siControl or siuPARP measured using confocal imaging after 24 h. Experiments were repeated independently three times with similar results. b. CAFs are cultured with 13C-BCAAs for 12 h prior to inducing BCAA deprivation. Spent media and cells are collected after 6, 12, 24, and 48 h under deprivation. Media samples are analyzed for secreted BCKAs using LC-QTOF and intracellular samples are analyzed for BCAAs using GC-MS. c. Intracellular BCAA levels measured after 6, 12, 24 and 48 h under BCAA deprivation. Mole percent enrichment of intracellular BCAAs measured after 6, 12, 24, and 48 h under BCAA deprivation. n = 3 biologically independent samples. d. Influence of TGF-β and BCAA deprivation on the proteasome activity in CAFs (n = 6). e. Relative growth rates of Mia Paca-2 and Panc-1 cells cocultured with CAFs treated with MG-132 under BCAA deprivation conditions. n = 8 biologically independent samples. f. Mass isotopomer distribution of BCAAs after acid hydrolysis of decellularized ECM proteins produced by CAFs cultured with 13C-BCAAs. n = 3 biologically independent samples. g. Fractional enrichment of amino acids after acid hydrolysis of decellularized ECM proteins produced by CAFs cultured with 13C-BCAAs. n = 3 biologically independent samples. Data are presented as mean ± s.d.
Extended Data Fig. 7 Stromal BCAT1 is regulated by cancer-cell derived TGF-β.
a. BCAT2 expression in NOFs treated with pancreatic cancer cell conditioned media (CM). n = 8 biologically independent samples. b. α-smooth muscle actin, (α-SMA) expression in NOFs cultured with pancreatic cancer cell-CM over 4 weeks. n = 8 biologically independent samples. c. Fibroblast specific protein (FSP1) expression in NOFs cultured with pancreatic cancer cell-CM over 4 weeks. n = 8 biologically independent samples. d. Podoplanin (PDPN) expression in NOFs cultured with pancreatic cancer cell-CM over 4 weeks. n = 8 biologically independent samples. e. BCAT2, α-SMA, FSP-1 and PDPN expression in MSCs treated with pancreatic cancer cell CM. n = 6 biologically independent samples. f. Expression of BCAA related genes in CAFs treated with pancreatic cancer cell-CM. n = 8 biologically independent samples. g. BCAT2 expression in CAFs treated with TGF-β and BCAT1 expression in cancer cells treated with TGF-β. n = 8 biologically independent samples. h. BCAT2 and α-SMA expression in NOFs cultured with pancreatic cancer cell CM in presence of anti-TGFB1 antibodies or isotype antibodies for 3 weeks. n = 8 biologically independent samples. Data are presented as mean ± s.d.
Extended Data Fig. 8 Cancer cells regulate stromal BCAT1 through SMAD5.
a. Representative images from IF analysis of BCAT1 and α-SMA expression in NOFs cultured with pancreatic cancer cell CM in presence of anti-TGFB1 antibodies or isotype antibodies for 3 weeks. Experiments were repeated independently twice with similar results. b. BCAT1 and BCAT2 expression in control and integrin αvβ5 KO CAFs cultured with pancreatic cancer cell CM for 3 weeks. n = 4 biologically independent samples. c. ELISA for TGF-β secretion levels from CAFs and PDAC cell lines. n = 8 biologically independent samples. d. SMAD2 expression in NOFs treated with pancreatic cancer cell CM. n = 8 biologically independent samples. e. SMAD3 expression in NOFs treated with pancreatic cancer cell CM. n = 8 biologically independent samples. f. SMAD4 expression in NOFs treated with pancreatic cancer cell CM. n = 8 biologically independent samples. g. SMAD5 binding motif. h. ChIP assays performed with control IgG and anti-SMAD4 antibodies in CAFs treated with PBS control or TGF-β. n = 6 biologically independent samples. *P < 0.0001. Data are presented as mean ± s.d. Multiple, two-tailed, unpaired, Student’s t-test (b); one-way ANOVA with Tukey’s post hoc comparison (c).
Extended Data Fig. 9 Validation of stromal BCAT1 and PDAC DBT in patient-derived CTCs.
a. Representative images of CTCs separated by Labyrinth. Cells are stained with DAPI (blue), cytokeratin (red), CD45 (green) and Vimentin (pink). Experiments were repeated independently three times with similar results. b. The influence of BCAAs and BCKAs on the growth of CTCs. n = 8 biologically independent samples. c. Extracellular concentration of BCKAs secreted by CAFs in monoculture and cocultured with CTCs over 6, 12, 24, and 48 h. n = 3 biologically independent samples. d. Extracellular concentration of BCKAs secreted by CAFs in monoculture or cocultured with CTCs, and CTCs in monoculture for 48 h. n = 4 biologically independent samples. e. Relative growth rate of PDAC cell lines and CTC lines under BCAA deprivation but supplemented with αKG, malate, succinate, acetate, citrate, NEAA mixture, or a combination in BCAA-deprived media. n = 8 biologically independent samples. f. Schematic of the protocol used to generate CTC derived organoid with CAF secreted ECM. g. Representative images from a CTC-derived organoid. Cytokeratin is shown in green and the nuclei stained with DAPI are shown in blue. Experiments were repeated independently three times with similar results. h. Representative FACS data of Pan-Cytokeratin positive tumor cells in CTC derived organoids. Experiments were repeated independently three times with similar results. i. Representative images of CTC derived organoids. Cells are stained with DAPI (blue), cytokeratin (red), CD45 (green) and Vimentin (Pink). Experiments were repeated independently twice with similar results. *P < 0.0001. Data are presented as mean ± s.d. Multiple, two-tailed, unpaired, Student’s t-test (b).
Extended Data Fig. 10 Validation of stromal BCAT1 and PDAC DBT in patient-derived tissue slices.
a. Representative Live Dead assay of tissue slice at Day 0 and Day 14. Live cells fluoresce bright green, whereas dead cells fluoresce red. Negative controls were fixed by methanol. Experiments were repeated independently three times with similar results. b. Efficiency of BCAT1 and DBT siRNAs in the human PDAC tissue slices. Expression of BCAT1, BCAT2, DBT, BCKDHA and BCKDHB in the human PDAC tissue slices treated with BCAT1 and DBT siRNAs. n = 6 biologically independent samples. Data are presented as mean ± s.d.
Supplementary information
Supplementary Information
Supplementary Figures 1–6
Source data
Source Data Fig. 1
Statistical Source Data
Source Data Fig. 1
Unprocessed Western Blots
Source Data Fig. 2
Statistical Source Data
Source Data Fig. 2
Unprocessed Western Blots
Source Data Fig. 3
Statistical Source Data
Source Data Fig. 4
Statistical Source Data
Source Data Fig. 5
Statistical Source Data
Source Data Fig. 5
Unprocessed Western Blots
Source Data Fig. 6
Statistical Source Data
Source Data Fig. 6
Unprocessed Western Blots
Source Data Extended Data Fig. 1
Statistical Source Data
Source Data Extended Data Fig. 2
Statistical Source Data
Source Data Extended Data Fig. 3
Statistical Source Data
Source Data Extended Data Fig. 4
Statistical Source Data
Source Data Extended Data Fig. 5
Statistical Source Data
Source Data Extended Data Fig. 6
Statistical Source Data
Source Data Extended Data Fig. 7
Statistical Source Data
Source Data Extended Data Fig. 8
Statistical Source Data
Source Data Extended Data Fig. 9
Statistical Source Data
Source Data Extended Data Fig. 10
Statistical Source Data
Rights and permissions
About this article
Cite this article
Zhu, Z., Achreja, A., Meurs, N. et al. Tumour-reprogrammed stromal BCAT1 fuels branched-chain ketoacid dependency in stromal-rich PDAC tumours. Nat Metab 2, 775–792 (2020). https://doi.org/10.1038/s42255-020-0226-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42255-020-0226-5
This article is cited by
-
Targeting stromal metabolism in pancreatic ductal adenocarcinoma
Nature Cell Biology (2024)
-
Research progress on the multi-omics and survival status of circulating tumor cells
Clinical and Experimental Medicine (2024)
-
CAFs targeted ultrasound-responsive nanodroplets loaded V9302 and GLULsiRNA to inhibit melanoma growth via glutamine metabolic reprogramming and tumor microenvironment remodeling
Journal of Nanobiotechnology (2023)
-
Cancer-associated fibroblasts and its derived exosomes: a new perspective for reshaping the tumor microenvironment
Molecular Medicine (2023)
-
Extracellular matrix remodeling in tumor progression and immune escape: from mechanisms to treatments
Molecular Cancer (2023)