Abstract
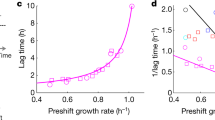
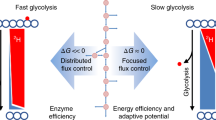
The principles governing cellular metabolic operation are poorly understood. Because diverse organisms show similar metabolic flux patterns, we hypothesized that a fundamental thermodynamic constraint might shape cellular metabolism. Here, we develop a constraint-based model for Saccharomyces cerevisiae with a comprehensive description of biochemical thermodynamics including a Gibbs energy balance. Non-linear regression analyses of quantitative metabolome and physiology data reveal the existence of an upper rate limit for cellular Gibbs energy dissipation. By applying this limit in flux balance analyses with growth maximization as the objective function, our model correctly predicts the physiology and intracellular metabolic fluxes for different glucose uptake rates as well as the maximal growth rate. We find that cells arrange their intracellular metabolic fluxes in such a way that, with increasing glucose uptake rates, they can accomplish optimal growth rates but stay below the critical rate limit on Gibbs energy dissipation. Once all possibilities for intracellular flux redistribution are exhausted, cells reach their maximal growth rate. This principle also holds for Escherichia coli and different carbon sources. Our work proposes that metabolic reaction stoichiometry, a limit on the cellular Gibbs energy dissipation rate, and the objective of growth maximization shape metabolism across organisms and conditions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The data that support the plots within this paper and other findings of this study are available from the corresponding author upon reasonable request. The code is available from the corresponding author upon request and the code to perform the flux balance analyses is deposited on GitHub (https://doi.org/10.5281/zenodo.1401220).
References
Molenaar, D., Van Berlo, R., De Ridder, D. & Teusink, B. Shifts in growth strategies reflect tradeoffs in cellular economics. Mol. Syst. Biol. 5, 323 (2009).
Basan, M. et al. Overflow metabolism in Escherichia coli results from efficient proteome allocation. Nature 528, 99–104 (2015).
Rozpędowska, E. et al. Parallel evolution of the make–accumulate–consume strategy in Saccharomyces and Dekkera yeasts. Nat. Commun. 2, 302 (2011).
Beg, Q. K. et al. Intracellular crowding defines the mode and sequence of substrate uptake by Escherichia coli and constrains its metabolic activity. Proc. Natl Acad. Sci. USA 104, 12663–12668 (2007).
Zhuang, K., Vemuri, G. N. & Mahadevan, R. Economics of membrane occupancy and respiro-fermentation. Mol. Syst. Biol. 7, 500–500 (2014).
Koppenol, W. H., Bounds, P. L. & Dang, C. V. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat. Rev. Cancer 11, 325–337 (2011).
Vander Heiden, M. G., Cantley, L. C. & Thompson, C. B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science 324, 1029–1033 (2009).
Mori, M., Hwa, T., Martin, O. C., De Martino, A. & Marinari, E. Constrained allocation flux balance analysis. PLoS Comput. Biol. 12, e1004913 (2016).
Sánchez, B. J. et al. Improving the phenotype predictions of a yeast genome-scale metabolic model by incorporating enzymatic constraints. Mol. Syst. Biol. 13, 935 (2017).
Zabalza, A. et al. Regulation of respiration and fermentation to control the plant internal oxygen concentration. Plant Physiol. 149, 1087–1098 (2009).
Huberts, D. H. E. W., Niebel, B. & Heinemann, M. A flux-sensing mechanism could regulate the switch between respiration and fermentation. FEMS Yeast. Res. 12, 118–128 (2012).
von Bertalanffy, L. The theory of open systems in physics and biology. Science 111, 23–29 (1950).
von Stockar, U. Biothermodynamics of live cells: a tool for biotechnology and biochemical engineering. J. Non-equilib. Thermodyn. 35, 415–475 (2010).
Lewis, N. E., Nagarajan, H. & Palsson, B. O. Constraining the metabolic genotype–phenotype relationship using a phylogeny of in silico methods. Nat. Rev. Microbiol. 10, 291–305 (2012).
Jol, S. J., Kümmel, A., Hatzimanikatis, V., Beard, D. A. & Heinemann, M. Thermodynamic calculations for biochemical transport and reaction processes in metabolic networks. Biophys. J. 99, 3139–3144 (2010).
Alberty, R. A. et al. Recommendations for terminology and databases for biochemical thermodynamics. Biophys. Chem. 155, 89–103 (2011).
Canelas, A. B., Ras, C., ten Pierick, A., van Gulik, W. M. & Heijnen, J. J. An in vivo data-driven framework for classification and quantification of enzyme kinetics and determination of apparent thermodynamic data. Metab. Eng. 13, 294–306 (2011).
Noor, E., Haraldsdóttir, H. S., Milo, R. & Fleming, R. M. T. Consistent estimation of Gibbs energy using component contributions. PLoS Comput. Biol. 9, e1003098 (2013).
Beard, Da, Liang, S. & Qian, H. Energy balance for analysis of complex metabolic networks. Biophys. J. 83, 79–86 (2002).
Price, N. D., Famili, I., Beard, D. A. & Palsson, B. Ø. Extreme pathways and Kirchhoff’s second law. Biophys. J. 83, 2879–2882 (2002).
Misener, R. & Floudas, C. A. ANTIGONE: Algorithms for coNTinuous / Integer Global Optimization of Nonlinear Equations. J. Glob. Optim. 59, 503–526 (2014).
Schuetz, R., Kuepfer, L. & Sauer, U. Systematic evaluation of objective functions for predicting intracellular fluxes in Escherichia coli. Mol. Syst. Biol. 3, 119 (2007).
Reed, J. L., Vo, T. D., Schilling, C. H. & Palsson, B. O. An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR). Genome. Biol. 4, R54 (2003).
Vemuri, G. N., Altman, E., Sangurdekar, D. P., Khodursky, A. B. & Eiteman, M. A. Overflow metabolism in Escherichia coli during steady-state growth: transcriptional regulation and effect of the redox ratio. Appl. Environ. Microbiol. 72, 3653–3661 (2006).
You, C. et al. Coordination of bacterial proteome with metabolism by cyclic AMP signalling. Nature 500, 301–306 (2013).
Schuetz, R., Zamboni, N., Zampieri, M., Heinemann, M. & Sauer, U. Multidimensional Optimality of Microbial Metabolism. Science 336, 601–604 (2012).
Kümmel, A., Panke, S. & Heinemann, M. Putative regulatory sites unraveled by network-embedded thermodynamic analysis of metabolome data. Mol. Syst. Biol. 2, 2006.0034 (2006).
Henry, C. S., Broadbelt, L. J. & Hatzimanikatis, V. Thermodynamics-based metabolic flux analysis. Biophys. J. 92, 1792–1805 (2007).
Fleming, R. M. T., Thiele, I. & Nasheuer, H. P. Quantitative assignment of reaction directionality in constraint-based models of metabolism: application to Escherichia coli. Biophys. Chem. 145, 47–56 (2009).
Bennett, B. D. et al. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 5, 593–599 (2009).
Bordel, S. & Nielsen, J. Identification of flux control in metabolic networks using non-equilibrium thermodynamics. Metab. Eng. 12, 369–377 (2010).
Noor, E. et al. Pathway thermodynamics highlights kinetic obstacles in central metabolism. PLoS Comput. Biol. 10, e1003483 (2014).
Schrödinger, E. What Is Life? The Physical Aspect of the Living Cell (Cambridge Univ. Press, 1944).
Okabe, K. et al. Intracellular temperature mapping with a fluorescent polymeric thermometer and fluorescence lifetime imaging microscopy. Nat. Commun. 3, 705 (2012).
Lane, N. Hot mitochondria? PLoS Biol. 16, e2005113 (2018).
Baffou, G., Rigneault, H., Marguet, D. & Jullien, L. A critique of methods for temperature imaging in single cells. Nat. Methods 11, 899–901 (2014).
Weber, J. K., Shukla, D. & Pande, V. S. Heat dissipation guides activation in signaling proteins. Proc. Natl. Acad. Sci. USA 112, 10377–10382 (2015).
Slochower, D. R. & Gilson, M. K. Motor-like properties of nonmotor enzymes. Biophys J. 114, 2174–2179 (2018).
Riedel, C. et al. The heat released during catalytic turnover enhances the diffusion of an enzyme. Nature 517, 227–230 (2014).
Golestanian, R. Anomalous diffusion of symmetric and asymmetric active colloids. Phys. Rev. Lett. 102, 188305 (2009).
Gallet, F., Arcizet, D., Bohec, P. & Richert, A. Power spectrum of out-of-equilibrium forces in living cells: amplitude and frequency dependence. Soft Matter 5, 2947 (2009).
Milstein, J. N., Chu, M., Raghunathan, K. & Meiners, J. C. Two-color DNA nanoprobe of intracellular dynamics. Nano. Lett. 12, 2515–2519 (2012).
Weber, S. C., Spakowitz, A. J. & Theriot, J. A. Nonthermal ATP-dependent fluctuations contribute to the in vivo motion of chromosomal loci. Proc. Natl Acad. Sci. USA 109, 7338–7343 (2012).
Chen, Y.-F., Milstein, J. N. & Meiners, J.-C. Protein-mediated DNA loop formation and breakdown in a fluctuating environment. Phys. Rev. Lett. 104, 258103 (2010).
Milstein, J. N. & Meiners, J.-C. On the role of DNA biomechanics in the regulation of gene expression. J. R. Soc. Interface 8, 1673–1681 (2011).
Kochanowski, K. et al. Functioning of a metabolic flux sensor in Escherichia coli. Proc. Natl Acad. Sci. USA 110, 1130–1135 (2013).
Nilsson, A., Nielsen, J. & Palsson, B. O. Metabolic models of protein allocation call for the kinetome. Cell Syst. 5, 538–541 (2017).
Burgard, A. P., Nikolaev, E. V., Schilling, C. H. & Maranas, C. D. Flux coupling analysis of genome-scale metabolic network reconstructions. Genome Res. 14, 301–312 (2004).
Drud, A. S. CONOPT—A large-scale GRG code. ORSA J. Comput. 6, 207–216 (1994).
Hastie, T. J., Tibshirani, R. & Friedman, J. The Elements of Statistical Learning: Data Mining, Inference, and Prediction (Springer-Verlag New York, 2011).
Schellenberger, J., Lewis, N. E. & Palsson, B. Ø. Elimination of thermodynamically infeasible loops in steady-state metabolic models. Biophys. J. 100, 544–553 (2011).
van Hoek, P. et al. Effects of pyruvate decarboxylase overproduction on flux distribution at the pyruvate branch point in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 64, 2133–2140 (1998).
Kümmel, A. et al. Differential glucose repression in common yeast strains in response to HXK2 deletion. FEMS Yeast. Res. 10, 322–332 (2010).
van Winden, W. et al. Metabolic-flux analysis of CEN.PK113-7D based on mass isotopomer measurements of C-labeled primary metabolites. FEMS Yeast. Res. 5, 559–568 (2005).
Fendt, S.-M. & Sauer, U. Transcriptional regulation of respiration in yeast metabolizing differently repressive carbon substrates. BMC Syst. Biol. 4, 12 (2010).
Gombert, A. K., Moreira dos Santos, M., Christensen, B. & Nielsen, J. Network identification and flux quantification in the central metabolism of Saccharomyces cerevisiae under different conditions of glucose repression. J. Bacteriol. 183, 1441–1451 (2001).
Frick, O. & Wittmann, C. Characterization of the metabolic shift between oxidative and fermentative growth in Saccharomyces cerevisiae by comparative 13C flux analysis. Microb. Cell. Fact. 4, 30 (2005).
Perrenoud, A. & Sauer, U. Impact of global transcriptional regulation by ArcA, ArcB, Cra, Crp, Cya, Fnr, and Mlc on glucose catabolism in Escherichia coli. J. Bacteriol. 187, 3171–3179 (2005).
Valgepea, K. et al. Systems biology approach reveals that overflow metabolism of acetate in Escherichia coli is triggered by carbon catabolite repression of acetyl-CoA synthetase. BMC Syst. Biol. 4, 166 (2010).
Nanchen, A., Schicker, A. & Sauer, U. Nonlinear dependency of intracellular fluxes on growth rate in miniaturized continuous cultures of Escherichia coli. Appl. Environ. Microbiol. 72, 1164–1172 (2006).
Peebo, K. et al. Proteome reallocation in Escherichia coli with increasing specific growth rate. Mol. BioSyst. 11, 1184–1193 (1184).
Gerosa, L. et al. Pseudo-transition analysis identifies the key regulators of dynamic metabolic adaptations from steady-state data. Cell Syst. 1, 270–282 (2015).
Schmidt, A. et al. The quantitative and condition-dependent Escherichia coli proteome. Nat. Biotechnol. 34, 104–110 (2015).
Scott, M. et al. Emergence of robust growth laws from optimal regulation of ribosome synthesis. Mol. Syst. Biol. 10, 747 (2014).
Acknowledgements
This work was funded by the Netherlands Organisation for Scientific Research (NWO) through the Systems Biology Centre for Metabolism and Ageing (Groningen), and by the BE-Basic R&D Program, which was granted as FES subsidy from the Dutch Ministry of Economic Affairs, Agriculture and Innovation (EL&I). We thank A. Canelas for sharing raw data, E. Noor for help with the component contribution method, E. Wit for statistics advice, G. Zampar for helpful discussions and B. Bakker, A. Bardow, D. Huberts, A. Ortega, U. Sauer, S. Stratmann and J. Radzikowski for helpful comments on the manuscript.
Author information
Authors and Affiliations
Contributions
B.N., S.L. and M.H. designed the study. B.N. and M.H. developed the concept. B.N. developed and implemented the model for S. cerevisiae. S.L. developed and implemented the model for E. coli. B.N. and S.L. carried out the simulations, analysed the data, and made the figures. B.N., S.L. and M.H. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–18, Supplementary Methods 1–3 and Supplementary Notes 1 and 2
Supplementary Data 1
Model for S. cerevisiae
Supplementary Data 2
Model for E. coli
Rights and permissions
About this article
Cite this article
Niebel, B., Leupold, S. & Heinemann, M. An upper limit on Gibbs energy dissipation governs cellular metabolism. Nat Metab 1, 125–132 (2019). https://doi.org/10.1038/s42255-018-0006-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42255-018-0006-7
Keywords
This article is cited by
-
Predicting microbial interactions with approaches based on flux balance analysis: an evaluation
BMC Bioinformatics (2024)
-
The minimum energy required to build a cell
Scientific Reports (2024)
-
Mitochondrial ATP generation is more proteome efficient than glycolysis
Nature Chemical Biology (2024)
-
Four ways of implementing robustness quantification in strain characterisation
Biotechnology for Biofuels and Bioproducts (2023)
-
Brain mitochondrial diversity and network organization predict anxiety-like behavior in male mice
Nature Communications (2023)