Abstract
Remarkable accomplishments demonstrating the importance of nucleic acids in molecular engineering and computation have been made over the past two decades. However, much of the work in this area so far has been carried out in vitro, utilizing almost exclusively homochiral D-DNAs (or D-RNAs) as chemical building blocks. Such natural building blocks are prone to enzymatic degradation and cross-hybridization with the host’s genetic materials. Here we report the development of an orthogonal nucleic acid system that is made up of a left-handed and a right-handed conformer, and a non-helical peptide nucleic acid analogue. We show that the stereochemical information inherent in the right-handed and left-handed conformers can be interconverted from (R) to (S) and vice versa, along with their helical sense and recognition capability, through strand displacement. The genetic information encoded in these synthetic building blocks can be interfaced with DNA or RNA through a molecular converter.
Similar content being viewed by others
Introduction
In addition to fulfilling their evolutionary roles as a medium for the storage and transmission of genetic information, nucleic acid biopolymers provide a convenient means for organizing molecular self-assembly owing to their specific and predictable nucleobase interactions and defined length scale. Among them, synthetic DNA oligonucleotides have garnered considerable interest as building blocks for molecular engineering because of their chemical stability, ease of synthesis, and low cost of production in comparison to RNA. The seminal work of Chen and Seeman1 more than two decades ago demonstrated its utility in the construction of molecular frameworks for organizing molecular self-assembly. Since then, DNA has been utilized in the bottom-up construction of a wide range of macro-molecular and supra-molecular systems, including polymeric materials2,3, nanoparticles4,5, DNA tiles and origami6,7,8,9,10,11,12,13,14,15, molecular circuits16,17, and autonomous nanoscale machines18,19,20,21, as well as information storage media22,23,24 and molecular computation25,26,27,28,29. However, beyond the feasibility studies that were commonly carried out in test tube, such an approach has not been extensively explored in the biological systems due, in part, to the concerns for enzymatic degradation and inadvertent hybridization of chemical building blocks with the host’s genetic materials, although earnest attempts have been made30,31,32,33,34. Spurious binding is not only counterproductive, but could lead to adverse cellular and cytotoxic effects. Stereo-inverted L-DNA35 and L-RNA (or ‘Spiegelmer’)36, in principle, could be employed to alleviate some of these concerns. However, they are technically challenging to prepare and scale up for basic research consumption in comparison to the natural counterparts. Moreover, because of their recognition orthogonality, they cannot be directly interfaced. This latter issue has recently been addressed by Kabza and Sczepanski37, in which they showed that the stereochemical information inherent in D-nucleic and L-nucleic acids can be interconverted by using peptide nucleic acid (PNA) as an interfacing medium. Such an approach provides another dimension in chirality, in addition to sequence information, for molecular engineering and computation.
A particular interest in many biological applications involves dynamic assembly38, one that relies on the precise spatial and temporal control of the non-equilibrium dynamics to drive the hybridization process. These non-covalent reactions are driven by strand displacement, a biological phenomenon first recognized several decades ago39 and recently pioneered by Yurke18 in nucleic acid programming, in which two strands of partial or full complementary DNA hybridize to each other and in the process displace one or more of the pre-hybridized strands. Strand displacement is initiated upon hybridization of the complementary single-stranded regions, commonly referred to as “toeholds”, where the rate of such a reaction can be controlled by varying the binding strength40. Of a myriad of molecular operations that have been developed, with most geared toward the construction of materials, one that holds considerable promise for molecular diagnostics is hybridization chain reaction (HCR). HCR was developed by Dirks and Pierce41 more than a decade ago as a means for amplifying the transduction signals, in a similar manner to that of polymerase chain reaction (PCR) but without the enzyme. The amplification process relies on the coexistence of two metastable hairpin species with complementary domains in the loops and toeholds. The two hairpins are unable to hybridize to each other because the former is kinetically trapped. HCR is triggered upon the addition of an initiator, causing a cascade reaction between the two hairpins. The importance of HCR in signal amplification has been well-recognized and successfully applied in a number of biosensing applications42,43; however, many of them were directed toward the detection of genetic materials in processed samples, such as fixed cells and tissues, but rarely in their native states44.
Recently, we reported the development of an orthogonal nucleic acid system that is comprised of three components: a right-handed (RH) and a left-handed γPNA conformer (LH), and a non-helical PNA version (NH, Fig. 1a)45. The first two are orthogonal to each other in recognition, while the third is compatible with both the RH and LH conformers, as well as with DNA and RNA. Unlike their natural counterparts, these chemical building blocks are impervious to enzymatic degradation46. Moreover, they are relatively simple to chemically synthesize and structurally modify, as all three components share the same PNA backbone skeleton with the exception of the stereochemistry or the lack thereof at the gamma (γ)-position, which determines whether the resulting oligonucleotide adopts an RH, LH, or NH motif.
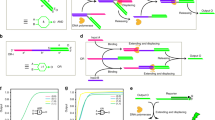
Molecular components for stereochemical conversion. a Chemical structures of the right-handed (RH), left-handed (LH), and non-helical (NH) PNA conformers. B = nucleobases (A, C, G, T). b Strand displacement reactions involving a contiguous template strand (top) and a template strand containing a PEG-spacer (bottom). c The predicted helical sense and conformational state of molecular converters containing a contiguous template strand (left) and a template strand containing a PEG-spacer (right). d Chemical compositions of TS, DS, and IS. DNA oligonucleotides D1, D2, and D3 were included for comparison. TS, template strand (15-nts), DS, displacement strand (10-nts), and IS, invading strand (15-nts). e Chemical structures of glutamic acid residue (Glu), pyrene (Pyr), and PEG-spacer
Here we report the next phase of this investigation, demonstrating that the genetic information encoded in the RH and LH conformers, as well as in natural nucleic acid biopolymers, can be interconverted through strand displacement. Strategic placement of the backbone spacer and systemic evaluation of the experimental conditions provide insight into key parameters for the design of molecular circuits for the detection of genetic materials with improved recognition specificity, detection sensitivity, and conversion rate. The work has implications for the organization and assembly of materials and molecular computation in biological systems.
Results
Design of stereochemical converters
A number of configurationally-inverted nucleic acid systems, including D-DNA and L-DNA35, D-RNA and L-RNA36, and LNA47 and α-L-LNA48, has been developed and has shown to be in discordant with each other in recognition. Likewise, the RH- and LH-γPNA conformers are unable to hybridize to each other45. This aspect suggests that they are relatively rigid in structure. Such a feature may be beneficial for certain biological applications, but could be detrimental for others, such as that involving signal transduction, in the conversion of genetic information from one helical sense to another in order to suppress signal degradation. An increase in the energetic penalty for the conversion of helical sense, as imposed by the structurally rigid γPNA backbone, could hinder or prevent the conversion process altogether. To address this concern, we designed two sets of molecular converters (Fig. 1b), with each comprising a non-helical template strand (TS) and either an RH, LH, or NH displacement strand (DS). The first set contained uninterrupted TS (top), while the second contained TS with a flexible PEG-spacer between the toehold and the stem (bottom). The role of the spacer was to decouple helical induction in the toehold from the stem, making the former conformationally more flexible and, thus, more accommodating to the invading strand (IS) with a mismatched helical sense (Fig. 1c, right). We had decided against poly-T as a spacer49, a common strategy for decoupling base-stacking interactions in DNA, because the RH and LH conformers are relatively rigid, more so than DNA or RNA45; and as such, its inclusion may not be sufficient to prevent helical propagation in the toehold. The chemical compositions of TS, DS, and IS are shown in Fig. 1d, e, along with their HPLC and MALDI-TOF MS spectra (Supplementary Figs. 1–8). Glutamic acid residues were incorporated at the N-terminus of each component to improve water solubility and to enable their separation by gel electrophoresis. Pyrene was chosen as a reporter for monitoring strand displacement50. In its absence, excitation of pyrene monomers at 344 nm results in the fluorescent emissions at 380 and 400 nm. In its presence, excitation of pyrenes at the same wavelength results in the emission of excimers at 485 nm. This ~100 nm red-shift in the fluorescent emission provides a convenient means for monitoring the strand displacement reaction.
Confirmation of recognition orthogonality
To further substantiate the recognition orthogonality of LH conformer with nucleic acid biopolymers, we adopted the HCR system as reported by Dirk and Pierce41 (Fig. 2a, b), and prepared the corresponding DNA (I1), LH-γPNA (I2), and RH-γPNA (I3) initiators (Fig. 2c). The binding affinity of I3 with DNA is too high to be determined accurately by conventional spectroscopic techniques. However, based on the previous findings51, with the 10-mer RH-γPNAs showing dissociation constants (KDs) in the femto-molar range, we estimated the KD value of I3 with DNA (or RNA) to be at least several orders of magnitude lower. We expected I1 and I3 to hybridize to H1A and initiate HCR (Fig. 2b), but not I2, due to the conformational mismatch, unless cross-hybridization takes place between it and H2A. Consistent with this prediction, we observed HCR only upon the addition of I1 (Fig. 2d, lanes 5 and 6) and I3 (lanes 7 and 8), as apparent from the formation of shifted bands and from the inverse molecular weight distribution with respect to initiator concentration (compare lanes 6 to 5, and lanes 8 to 7). No evidence of HCR had taken place following the addition of I2 and incubation of the mixtures at ambient temperature for 16 h (lanes 9 and 10). This result is consistent with the previous report that showed a high degree of recognition orthogonality of LH-γPNA with DNA and, via inference, RNA45.
Confirmation of recognition orthogonality. a Depiction of HCR components. b HCR cascade reactions. c Sequence of HCR components. d Result of HCR following the addition of I1 (lanes 5 and 6), I3 (lanes 7 and 8), and I2 (lanes 9 and 10) to a mixture containing an equimolar ratio of H1A and H1B (lane 4) and incubation at 25 °C for 16 h. The concentrations of H1A and H1B were 1 µM each. The concentrations of initiator were 0.1× and 1× with respect to that of H1A and H1B. The samples were prepared in 1xPBS buffer (137 mM NaCl, 2.7 mM KCl, 10 mM NaPi, pH 7.4), separated on a 10% non-denaturing PAGE, and stained with SYBR-Gold
Evaluation of conformational state
Circular dichroism (CD) was employed to assess the conformations of DS and IS, along with that of the corresponding TS-DS and TS-IS duplexes. Consistent with the published literature52, the individual DS (P2a and P2b) and IS (P3c and P3d) components adopted either an RH or an LH motif as inferred from the CD spectral patterns (Fig. 3a and Supplementary Fig. 9)53. This is in contrast to the weak CD signals observed with the achiral (NH) P1a and P1b templates and P3a invading strand (Supplementary Fig. 10). The difference in CD amplitude between the two sets is due to the difference in probe length. Similar spectral patterns were observed with TS-DS and TS-IS duplexes (Fig. 3b). Interestingly, among the three TS-IS duplexes (P1a-P3a, P1a-P3c, P1a-P3d) with an identical sequence and concentration (Supplementary Fig. 11), P1a-P3a exhibited the weakest CD signals, nearly five-fold lower than that of the latter two. This large difference in the signal strength is reflected in the conformational heterogeneity of the duplex, with P1a-P3a existing as both an RH and an LH conformer in different proportions, whereas the other two were present in a single conformational state. This observation is consistent with the X-ray crystallography data54, along with the findings by Norden55 and Green56, which showed that the helical preference of a PNA–PNA duplex induced by chiral amino acids at the C-terminus does not exist in one predominant state, but rather as a mixture of both RH and LH, with one in slight excess over the other. Incorporation of a PEG-spacer in TS disrupted helical induction in the toehold, as evident from the reduction in CD signals of P1b-P2a and P1b-P2b duplexes as compared to that of the respective P1a-P2a and P1a-P2b (Fig. 3c and Supplementary Fig. 12). However, its inclusion had little effect on the helical induction of the full-length TS-IS duplexes (Fig. 3d), as it is mediated by IS. This result is consistent with our prediction that introduction of a flexible PEG-spacer would disrupt base-stacking interactions, preventing helical induction from propagating into the toehold region and thereby rendering it conformationally more flexible (Fig. 1c, right).
Evaluation of conformational state. CD spectra of a individual DS (P2a and P2b) and IS (P3c and P3d) components, and b the corresponding TS-DS (P1a-P2a and P1a-P2b) and TS-IS (P1a-P3a, P1a-P3c, and P1a-P3d) duplexes. Comparison of the CD spectra of c TS-DS and d TS-IS with and without PEG-spacer. The samples were prepared in a physiologically relevant buffer (137 mM NaCl, 2.7 mM KCl, 10 mM NaPi, 2 mM MgCl2, pH 7.4), and the data were recorded at 25 °C. The concentration of each strand was 5 µM
Assessment of thermal stability
Next, we assessed the prospect of TS-DS as molecular converters by measuring their thermal stability, along with that of the resulting TS-IS products. The data falls in line with the established pattern45, with the conformationally-matched LH-LH or RH-RH duplexes being the most thermally stable, followed by LH-NH or RH-NH, and NH-NH in that respective order (Supplementary Fig. 13). The displacement TS-IS products are generally 10–15 °C higher in melting transition (Tm) than that of the TS-DS converters with the corresponding helical sense. Introduction of a PEG-spacer in TS had little effect on the thermal stability of TS-DS (Supplementary Fig. 14), since backbone disruption is external to the stem region. However, it had a significant destabilizing effect on TS-IS, lowering the Tm by ~15 °C (Supplementary Fig. 15). Nonetheless, it is still higher than that of TS-DS. This finding implies that strand displacement is thermodynamically favorable. This suggestion was further corroborated by gel electrophoresis assay that showed the expected strand displacement reaction upon the addition of IS to TS-DS (Supplementary Fig. 16).
Strand displacement kinetics
Strand displacement data were acquired at a physiologically relevant condition (137 mM NaCl, 2.7 mM KCl, 10 mM NaPi, 2 mM MgCl2, pH 7.4; 37 °C)57, unless otherwise stated. The reaction was monitored at 480 nm following the addition of IS to TS-DS and excitation at 344 nm (Fig. 4a). A DNA counterpart was included for comparison and as a control for validating the mathematical treatment of the kinetics data. The rate constant for strand displacement of DNA was found to be 2.9 × 104 M−1 s−1, which is within the range as reported in the literature (Fig. 4b, black trace)40. Under identical conditions, the rate constants of displacing DSRH by ISNH (red trace) and ISRH (green trace) were similar, 1.9 × 104 and 1.3 × 104 M−1 s−1, respectively, which are slightly lower than that of DNA. We attributed the reduction in the rate to a decrease in water solubility (and possible aggregation of the stock solution) of γPNAs and to an increase in the thermodynamic stability of the stem region of TS-DS, in comparison to that of DNA. Interestingly, strand displacement did not take place with a DNA (D3) invading strand due to the resultant P1a-D3 being thermodynamically less stable than that of P1a-P2b (Fig. 4c). The displacement rate of DSLH by ISRH, or DSRH by ISLH is further reduced by two-fold (Fig. 4d, compare the yellow and blue traces to that of green and red, respectively). Figure 4e provides a compilation of the rate constants. Contrary to our prediction, introduction of a flexible PEG-spacer in TS (P1b) did not improve the rate of strand displacement. In fact, it lowered the rate by roughly three-fold in comparison to that without the spacer (Fig. 5a). Similar trends in the rates were observed with the spacer introduced in TS, IS, or in both (Fig. 5b). We attributed the reduction in the rate to an increase in internal diffusion (or degree of freedom)49. These rate constants, tabulated in Fig. 5c, are similar to those reported by Kabza and Sczepanski37 in the conversion of heterochiral D-DNA and L-DNA using PNA as a template. Temperature-dependent measurements revealed that the rates of strand displacement increased proportionally with temperature (Fig. 5d). An Arrhenius plot of the rates as a function of temperature yielded an activation energy of ~ 73 kJ/mol for strand displacement of a mismatched helical sense (or stereoconversion), which is nearly two-fold lower than that observed with DNA of a similar length but without toehold58.
The kinetics of strand displacement reactions. a Depiction of strand displacement reactions of TS-DS (P1a-P2b) with IS (P3a, P3c, and P3d). b Fluorescent measurements of strand displacement reactions as a function of time. c Result showing that a DNA invading strand (D3) was unable to displace P2b from P1a. d Comparison of strand displacement reactions between the conformationally-matched (green and red traces) and mismatched (yellow and blue traces) IS with TS-DS. e The rate constants of strand displacement reactions as shown in b and d. The concentrations of TS-DS duplexes were 100 nM and those of IS were 1 µM (10-fold excess). The samples were prepared in a physiologically relevant buffer, and the reactions were carried out at 37 °C. Excitation was made 344 nm, and the emissions were recorded at 480 nm
The effects of PEG-spacer and temperature on the rates of strand displacement. Kinetic profiles of strand displacement reactions with and without PEG-spacer performed at 37 °C with a a mismatched helical sense IS and b a non-helical IS. c The tabulated rate constants of the data shown in a and b. d Kinetic profiles as a function of temperature. The samples were prepared in the same way as that mentioned in Fig. 4 caption
Signal transduction
To determine the extent of cross-hybridization of RH-γPNA (or any RH-nucleic acid system for that matter) as a signaling medium with genetic materials, we coupled the molecular converter with a DNA-reporter (Fig. 6a). Despite sequence complementarity, incubation of LH-input (P3c) with the reporter did not yield any fluorescent signals (data not shown), an indication that cross-hybridization between the two did not take place. The fluorescent signals were observed only when all three components, LH-input, converter, and reporter, were present (Fig. 6a, left-side, and Fig. 6b). The fluorescent signals were slightly dampened upon the addition of a DNA quencher (DQ1). The reduction in the fluorescent signals, in this case, is not due to cross-hybridization of P3c with DQ1, but rather due to the inevitable binding of a right-handed output P2b with DQ1 following its successful displacement by Pc3. To minimize this off-target binding, we carried out the experiments at a relatively low temperature—hence, the slow strand displacement kinetics. A similar observation was made upon the addition of a right-handed input (P3d) (Fig. 6a, right-side, and Fig. 6c). However, in this case, the experiment was performed at the usual 37 °C since inadvertent hybridization was not a concern. In contrast to observations made with the left-handed input, addition of a DNA quencher (DQ2) resulted in a complete quenching of the fluorescent signals. This result highlights the potential danger in employing natural or right-handed nucleic acid building blocks in molecular organization and self-assembly in biological systems in the presence of background genetic materials. It should be pointed out that a similar chiral conversion was reported by Kabza and Sczepanski37 using PNA to convert D-DNA to L-DNA and vice versa, but not in the presence of off-target DNA.
The effect of helical sense on signal transduction. a An illustration of molecular conversion in the presence of off-target DNA with an LH-input (left-side) and an RH-input (right-side). Signal transduction generated with b an LH-input and c an RH-input. For LH-input in b, the reactions were carried out at 25 °C to minimize the quenching of RH-output (P2b) by DQ1, whereas those with RH-input in c were performed at 37 °C. The concentration of initiator (input) was 1 µM, the same as that of the converter. DQ1 = D1 + D1′, DQ2 = D1 + P4Q, and REPORTER = P4F + P4Q. D1: 5′-CACGCCGAATCCTAG-3, D1′: 5′-ATTCGGCGTG-3′, P4F: 5′-/5Cy5/CACGCCGAAT-3′, and P4Q: 5′-CGGCGTG/3IAbRQSp/-3′
Discussion
The emerging roles of nucleic acids as engineering materials and as molecular “processors” and signaling mediums, in addition to their natural roles in the storage and transmission of genetic information, lend the possibility for their integration. The interface would provide a more accessible means for controlling life processes. Such an undertaking, however, will require the delivery of these chemical building blocks into live cells and intact organisms, and that they are able to withstand the enzymatic assaults and capable of recognizing their designated partners without cross-interference with the endogenous genetic materials. Natural building blocks, such as DNA oligonucleotides are attractive in this regard and have been extensively utilized in molecular engineering and computation, but they are not well-suited for intracellular or in vivo applications due to the aforementioned concerns. Although synthetic counterparts, such as configurationally-inverted L-DNA and L-RNA, have been employed to alleviate some of these concerns, and as demonstrated by Kabza and Sczepanski37 that the stereochemical information can be interfaced with natural nucleic acid biopolymers by using PNA as a conversion medium, it remains a challenge to reduce such a concept into practice due to the large differences in the hybridization kinetics and thermodynamics of the DNA–DNA and DNA–PNA duplexes. Development of a nucleic acid platform with more closely related components in terms of chemical compositions, binding kinetics and thermodynamics, such as the one reported in this study, might permit greater ease in the design and synthesis of molecular circuits for biological applications.
We have shown that by applying the appropriate chemistry45, a randomly-folded PNA, as originally developed by Nielsen and coworkers59 more than two decades ago, can be preorganized into either an RH or an LH motif. Due to conformational mismatch, the two conformers are unable to hybridize to each other. By employing PNA as an interfacing medium that is capable of recognizing the RH and LH conformers, as well as DNA and RNA, the genetic information encoded in any one of these nucleic acid systems can be interconverted, with rates that are comparable to that of homochiral D-DNA40. Inclusion of conformational chirality provides another dimension in nucleic acid recognition, in addition to sequence information. A distinct advantage of this particular nucleic acid platform is that all three components, RH, LH, and NH, contain the same molecular scaffold, with the exception of the stereochemistry or the lack thereof at the (γ)-backbone that determines the conformations of oligomers. They are relatively simple to chemically synthesize and structurally modify, especially in making nucleobase substitutions and drastic structural modifications in the backbone. They can be made cell-permeable, if necessary, by installing the appropriate chemical groups at the (γ)-backbone51,60,61.
As we have shown one of the major concerns in utilizing natural nucleic acids as chemical building blocks and as signaling molecules in biological systems is off-target binding. One way to mitigate this concern would be to incorporate orthogonal nucleobases62, which can recognize their respective partners but not the natural nucleobases (Fig. 7a). Alternatively, one could envision a design, in which the LH and RH conformers are conjugated through a flexible spacer in such a way that helical induction between the two domains is decoupled, enabling them to hybridize to their respective partners independent of each other (Fig. 7b, c). Such a dual converter-sensor design obviates the need for a separate molecular converter and would produce an output with a matching helical sense as that of the next molecular operator, which would significantly enhance the rate of signal transduction.
A proposed solution to overcome signal degradation. a Conventional molecular operations for the detection of genetic material and for the conversion of an RH-input (T2) to an LH-output (T3). A concern with this design is the potential for inadvertent binding of T2 with background genetic materials. b A solution to the concern in a. Integration of molecular converter with sensor into a single unit bypasses the first step, in the production of T2, and goes directly to producing T3. c Depiction of the various components involved in b. Blue: LH-γPNA, green: RH-γPNA, T1: DNA or RNA target. Orthogonal nucleobases could be incorporated in domain a of “Waste” product to prevent its hybridization to domain a’ of H1B. Circle: PEG-spacer
While it is an attractive method for signal amplification, HCR has limitations. The signal amplification is linear with respect to initiator concentration. As suggested by Dirk and Pierce41, and recently demonstrated by Xuan and Hsing63, higher order growth rates could be envisioned by application of branched or dendritic systems. However, such a design may be challenging to implement due to its high complexity and sheer size for intracellular or in vivo applications. Alternatively, one could envisage an HCR-mediated runaway reaction, whereby the initiator is present in large quantities but in a protected form so that it is unable to trigger HCR until the target is detected. In such an event, the polymerized HCR products catalyze the removal of the protecting group from initiator, causing a runaway HCR. This type of a design is within reach with a chemically facile nucleic acid platform such as γPNA. The development of a rapid and sensitive method for in-situ detection of genetic materials will be essential and an important first step on the road to implementing a global pathogen surveillance system for the monitoring and for the containment of infectious pathogens. Overall, the work provides a foundation for the organization and assembly of materials, and for building molecular circuits and for the execution of molecular processes in biological systems without the concerns for erosion of chemical building blocks or degradation of transduction signals as a result of cross-interference with the host’s genetic materials.
Methods
PNA monomer synthesis
Boc-protected PNA monomers were purchased from PolyOrg Inc. Boc-MiniPEG-RH-γPNA monomers were synthesized from Boc-protected l-serine following the previously reported protocols51. The LH monomers were synthesized by the same protocols but instead starting with Boc-protected d-serine64.
Oligomer synthesis
All oligomers were synthesized on solid-support using Boc-chemistry procedures. The oligomers were cleaved from resin using an m-cresol:thioanisole:trifluoromethanesulfonic acid (TFMSA):trifluoroacetic acid (TFA) (1:1:2:6) cocktail solution and precipitated with ether. PNA oligomers were purified by reverse-phase high performance liquid chromatography (RP-HPLC) in the analytical mode (C18 column with dimensions 4.6 mm × 250 mm, 1 mL/min, 60 °C) or in the semi-prep mode (C18 column with dimensions 19 mm × 100 mm, 5 mL/min, 55 °C) by monitoring UV absorbance at 260 nm. The oligomer structures were characterized by MALDI-TOF MS with CHCA matrix. All PNA stock solutions were prepared using nanopure water, and the concentrations were determined at 90 °C on a Agilent Cary UV–Vis 300 spectrometer using the following extinction coefficients: 13,700 M−1 cm−1 (A), 6,600 M−1 cm−1 (C), 11,700 M−1 cm−1 (G), 8600 M−1 cm−1 (T), and 24,000 M−1 cm−1 (Pyrene). All DNA oligomers were procured from IDT and used without further purification.
UV-melting experiments
All UV-melting samples were prepared by mixing PNA oligomers at the indicated concentrations in a physiologically simulated buffer (137 mM NaCl, 2.7 mM KCl, 10 mM NaPi, 2 mM MgCl2, pH = 7.4), and annealed by incubation at 95 °C for 5 min followed by gradual cooling to room temperature. UV melting curves were collected using Agilent Cary UV–Vis 300 spectrometer equipped with a thermoelectrically controlled multi-cell holder. UV-melting spectra were collected by monitoring UV-absorption at 260 nm from 20 to 95 °C in the heating runs, and from 95 to 20 °C in the cooling runs, both at the rate of 0.2 °C min−1. The cooling and heating curves were nearly identical, indicating that the hybridization process is reversible. The recorded spectra were smoothed using a 20-point adjacent averaging algorithm. The first-order derivatives of the melting curves were taken to determine the melting transitions of the duplexes.
Circular dichroism analyses
CD samples were prepared at 5 μM strand concentrations in a physiologically relevant buffer. The samples were heated to 95 °C and gradually cooled to room temperature. CD experiments were performed on a JASCO J-715 spectropolarimeter using a quartz cuvette with 1 cm path length. The samples were scanned from 320 to 200 nm with a scan rate of 100 nm/min and 15 scan accumulations at 25 °C. All spectra were baseline-subtracted, smoothed using a five-point adjacent averaging algorithm and replotted using Origin software.
Fluorescence measurements
(i) To perform kinetic experiments for nucleic acid conversions, the two components were prepared separately in a physiologically relevant buffer: (1) 216 nM of DS and 180 nM of TS, and (2) 180 nM of IS. The TS-DS mixture was annealed at 95 °C for 5 min before 1 h of gradual cooling to ambient temperature. On Cary Eclipse Fluorescence Spectrometer, the TS-DS duplex was incubated at 37 °C for 5 min followed by addition of IS to reach the final concentrations: [DS] = 72 nM, [TS] = 60 nM, and [IS] = 120 nM. Real-time fluorescence data were collected with wavelengths (λex = 350 nm and λem = 480 nm) and slit sizes (Ex = 5 nm and Em = 10 nm). The first collection point after addition of invading strand was set as t = 0.
The degree of conversion was calculated by Eq. (1):
where Fobs = real-time observed fluorescence intensity; Fi = fluorescence intensity of annealed template and displacement strands before the addition of invading strand; Ff = fluorescence intensity of a pre-annealed sample of invading and template strands, a presumed end-product.
The 2nd order kinetic rate constant k calculated by Eq. (2):
where X = TSo × x.
(ii) To perform kinetic experiment for signal transduction, the four components were prepared separately in a physiologically relevant buffer: (1) 90 nM of P4F and 108 nM of P4Q, (2) 540 nM of P2b and P1a, (3) 2160 nM of DQ1 or DQ2, and (4) 2160 nM of LH or RH input. The four components were annealed separately at 95 °C for 5 min before 1 h of gradual cooling to ambient temperature. On Cary Eclipse Fluorescence Spectrometer, component (1) was incubated at 20 or 37 °C followed by addition of other indicated components to reach the final concentrations. Real-time fluorescence data were collected with wavelengths (λex = 648 nm and λem = 668 nm) and slit sizes (Ex = 5 nm and Em = 5 nm). t = 0 is defined by the first observation after mixing all the indicated components.
The degree of conversion was calculated by Eq. (1), where Fobs = real-time observed fluorescence intensity; Fi = fluorescence intensity before addition of the invading strand; Ff = fluorescence intensity of a pre-annealed sample of P4F and P3d, a presumed end-product.
Gel shift assay
PNA strands were prepared at the indicated concentrations in 1× PBS buffer (137 mM NaCl, 2.7 mM KCl, 10 mM NaPi, pH 7.4). For strand displacement reactions, TS-DS duplexes were annealed at 95 °C for 5 min followed by gradual cooling to ambient temperature in 1 h. The duplexes were then incubated at 37 °C for 5 min before addition of IS oligomers. The reaction mixtures were incubated at the same temperature for another 1 h before gel shift analysis. The samples were then loaded onto 2% agarose-gel containing 1× SYBR-Gold with 1× Tris-borate buffer and electrophoretically separated at 100 V for 15 min. The bands were visualized by UV-Transilluminator. For the results shown in Supplementary Fig. 16b, the samples were separated on a 10% non-denaturing PAGE for 1.5 h at 120 V cm−1.
Data availability
All data are available from the corresponding authors upon request.
References
Chen, J. H. & Seeman, N. C. Synthesis from DNA of a molecule with the connectivity of a cube. Nature 350, 631–633 (1991).
Pianowski, Z. L. & Winssinger, N. Nucleic acid encoding to program self-assembly in chemical biology. Chem. Soc. Rev. 37, 1330–1336 (2008).
Seeman, N. C. DNA in a material world. Nature 421, 427–431 (2003).
Park, S. Y. et al. DNA-programmable nanoparticle crystallization. Nature 451, 553–556 (2008).
Jones, M. R., Seeman, N. C. & Mirkin, C. A. Programmable materials and the nature of the DNA bond. Science 347, 1260901 (2015).
Rothemund, P. W. K. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006).
Pinheiro, A. V., Han, D., Shih, W. M. & Yan, H. Challenges and opportunities for structural DNA nanotechnology. Nat. Biotechnol. 6, 763–772 (2011).
Aldaye, F. A., Palmer, A. L. & Sleiman, H. F. Assembling materials with DNA as the guide. Science 321, 1795–1799 (2008).
Douglas, S. M. et al. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459, 414–418 (2009).
Dietz, H., Douglas, S. W. & Shih, W. M. Folding DNA into twisted and curved nanoscale shapes. Science 325, 725–730 (2011).
Han, D. et al. DNA origami with complex curvatures in three-dimensional space. Science 332, 342–346 (2011).
Wei, B., Dai, M. & Yin, P. Complex shapes self-assembled from single-stranded DNA tiles. Nature 485, 623–626 (2012).
Ke, Y., Ong, L. L., Shih, W. M. & Yin, P. Three-dimensional structures self-assembled from DNA bricks. Science 338, 1177–1183 (2012).
Veneziano, R. et al. Designer nanoscale DNA assemblies programmed from the top down. Science 352, aaf4388–4381 (2016).
Han, D. et al. Single-stranded DNA and RNA origami. Science 358, eaao2648 (2017).
Bath, J. & Turberfield, A. J. DNA nanomachines. Nat. Nanotechnol. 2, 275–284 (2007).
Gerling, T., Wagenbauer, K. F., Neuner, A. M. & Dietz, H. Dynamic DNA devices and assemblies formed by shape-complementarity, non-base pairing 3D components. Science 347, 1446–1452 (2015).
Yurke, B. et al. A DNA-fueled molecular machine made of DNA. Nature 406, 605–608 (2000).
Omabegho, T., Sha, R. & Seeman, N. C. A bipedal DNA brownian motor with coordinated legs. Science 324, 67–71 (2009).
Lund, K. et al. Melecular robots guided by prescriptive landscapes. Nature 465, 206–210 (2010).
Gu, H. Z., Chao, J., Xiao, S. J. & Seeman, N. C. A proximity-based programmable DNA nanoscale assembly line. Nature 465, 202–205 (2011).
Church, G. M., Gao, Y. & Kosuri, S. Next-generation digital information storage in DNA. Science 337, 1628 (2012).
Erlich, Y. & Zielinski, D. DNA Fountain enables a robust and efficient storage architecture. Science 355, 950–954 (2017).
Goldman, N. et al. Towards practical, high-capacity, low-maintenance informaton storage in synthesized DNA. Nature 494, 77–80 (2013).
Adleman, L. M. Molecular computation of solutions to combinatorial problems. Science 266, 1021–1024 (1994).
Benenson, Y. Biomolecular computing systems: principles, progress and potential. Nat. Rev. Gen. 13, 455–468 (2012).
Stojanovic, M. N., Stefanovic, D. & Rudchenko, S. Exercises in molecular computing. Acc. Chem. Res. 47, 1845–1852 (2014).
Qian, L. & Winfree, E. Scaling up digital circuit computation with DNA strand displacement cascades. Science 332, 1196–1201 (2011).
Qian, L., Winfree, E. & Bruck, J. Neural network computation with DNA strand displacement cascades. Nature 475, 368–372 (2011).
Win, M. N. & Smolke, C. D. HIgher-order cellular information processing with synthetic RNA devices. Science 322, 456–460 (2008).
Chen, Y.-J., Groves, B., Muscat, R. A. & Seelig, G. DNA nanotechnology from the test tube to the cell. Nat. Nanotechnol. 10, 748–760 (2015).
Groves, B. et al. Computing in mammalian cells with nucleic acid strand exchange. Nat. Nanotechnol. 11, 287–294 (2016).
Douglas, S. M., Bachelet, I. & Church, G. M. A logic-gated nanorobot for targeted transport of molecular payloads. Science 335, 831–834 (2012).
Delebecque, C. J., Linder, A. B., Silver, P. A. & Aldaye, F. A. Organization of intracellular reactions with rationally designed RNA assemblies. Science 333, 470–474 (2011).
Fujimori, S. & Shudo, K. Enantio-DNA recognizes complementary RNA but not complementary DNA. J. Am. Chem. Soc. 112, 7436–7438 (1990).
Ashley, G. W. Modeling, synthesis, and hybridization properties of (L)-ribonucleic acid. J. Am. Chem. Soc. 114, 9731–9736 (1992).
Kabza, A. M., Young, B. E. & Sczepanski, J. T. Heterochiral DNA strand-displacement circuits. J. Am. Chem. Soc. 139, 17715–17718 (2017).
Zhang, D. Y. & Seelig, G. Dynamic DNA nanotechnology using strand-displacement reactions. Nat. Chem. 3, 103–113 (2011).
Meselson, M. S. & Radding, C. M. A general method for genetic recombination. Proc. Natl Acad. Sci. USA 72, 358–361 (1975).
Zhang, D. Y. & Winfree, E. Control of DNA strand displacement kinetics using toeold exchange. J. Am. Chem. Soc. 131, 17303–17314 (2009).
Dirks, R. M. & Pierce, N. A. Triggered amplification by hybridization chain reaction. Proc. Natl Acad. Sci. USA 101, 15275–15278 (2004).
Choi, H. M. T. et al. Programmable in situ amplification for multiplexed imaging of mRNA expression. Nat. Biotechnol. 28, 1208–1212 (2010).
Jung, C. & Ellington, A. D. Diagnostic applications of nucleic acid circuits. Acc. Chem. Res. 47, 1825–1835 (2014).
Wu, Z., Liu, G.-Q., Yang, X.-L. & Jiang, J.-H. Electrostatic nucleic acid nanoassembly enables hybridization chain reaction in living cells for ultrasensitive mRNA imaging. J. Am. Chem. Soc. 137, 6829–6836 (2015).
Sacui, I. et al. Gamma peptide nucleic acids: as orthogonal nucleic acid recognition codes for organizing molecular self-assembly. J. Am. Chem. Soc. 137, 8603–8610 (2015).
Demidov, V. V. et al. Stability of peptide nucleic acids in human serum and cellular extracts. Biochem. Pharmacol. 48, 1310–1313 (1994).
Wengel, J. Synthesis of 3’-C- and 4’-C-branched oligodeoxynucleotides and the development of locked nucleic acid (LNA). Acc. Chem. Res. 32, 301–310 (1999).
Rajwanshi, V. K., Hakansson, A. E., Dahl, B. M. & Wengel, J. LNA stereoisomers: xylo-LNA (beta-d-xylo configured locked nucleic acid) and alpha-L-LNA (alpha-L-ribo configured locked nucleic acid). Chem. Commun. 1395–1396 (1999).
Genot, A. J., Zhang, D. Y., Bath, J. & Turberfield, A. J. Remote toehold: a mechanism for flexible control of DNA hybridization kinetics. J. Am. Chem. Soc. 133, 2177–2182 (2011).
Yang, C. J. et al. Light-switching excimer probes for rapid protein monitoring in complex biological fluids. Proc. Natl Acad. Sci. USA 102, 17278–17283 (2005).
Sahu, B. et al. Synthesis and characterization of conformationally preorganized, (R)-diethylene glycol-containing g-peptide nucleic acids with superior hybridization properties and water solubility. J. Org. Chem. 76, 5614–5627 (2011).
Dragulescu-Andrasi, A. et al. A simple gamma-backbone modification preorganizes peptide nucleic acid into a helical structure. J. Am. Chem. Soc. 128, 10258–10267 (2006).
Wittung, P. et al. DNA-like double helix formed by peptide nucleic acid. Nature 368, 561–563 (1994).
Rasmussen, H. et al. Crystal structure of a peptide nucleic acid (PNA) duplex at 1.7 A resolution. Nat. Struct. Biol. 4, 98–101 (1997).
Wittung, P. et al. Induced chirality in PNA-PNA duplexes. J. Am. Chem. Soc. 117, 10167–10173 (1995).
Totsingan, F. et al. Conformational heterogeneity in PNA-PNA duplexes. Macromolecules 43, 2692–2703 (2010).
Santoro, S. W. & Joyce, G. F. A general purpose RNA-cleaving DNA enzyme. Proc. Natl Acad. Sci. USA 94, 4262–4266 (1997).
Reynaldo, I. P., Vologodskii, A. V., Neri, B. P. & Lyamichev, V. I. The kinetics of oligonucleotide replacement. J. Mol. Biol. 297, 511–520 (2000).
Nielsen, P. E., Egholm, M., Berg, R. H. & Buchardt, O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science 254, 1497–1500 (1991).
Zhou, P. et al. Novel binding and efficient cellular uptake of guanidine-based peptide nucleic acids (GPNA). J. Am. Chem. Soc. 125, 6878–6879 (2003).
Stanzl, E. G., Trantow, B. M., Vargas, J. R. & Wender, P. A. Fifteen years of cell-penetrating, guanidinium-rich molecular transporters: Basic science, research tools, and clinical applications. Acc. Chem. Res. 46, 2944–2954 (2013).
Wojciechowski, F. & Leumann, C. J. Alternative DNA base-pairs: from efforts to expand the genetic code to potential material applications. Chem. Soc. Rev. 40, 5669–5679 (2011).
Xuan, F. & Shing, I.-M. Triggering hairpin-free chain-branching growth of fluorescent DNA dendrimers for nonlinear hybridization chain reaction. J. Am. Chem. Soc. 136, 9810–9813 (2014).
Yu, Z. et al. Orthogonal yPNA dimerization domains empower DNA binders with cooperativity and versatility mimicking that of transcription factor pairs. Chem. Eur. J. 24, 14183–14188 (2018).
Acknowledgements
The authors would like to thank Dr. James W. Schneider and Yogesh Somasundar for the technical support in fluorescent measurements. Nuclear Magnetic Resonance and MALDI-TOF MS instrumentations at Carnegie Mellon University were partially supported by the National Science Foundation (CHE-9808188, DBI-9729351, CHE-1039870 and CHE-1726525). G.R.M. acknowledged the financial support from Xunta de Galicia. Financial support for D.H.L. and W.-C.H. were provided by the DSF Charitable Foundation.
Author information
Authors and Affiliations
Contributions
W.-C.H., G.R.M., and D.H.L. designed the experiments. W.-C.H. and R.C. synthesized the monomers. W.-C.H., A.W., S.F.W., G.R.M., and D.H.L. prepared PNA oligomers. W.-C.H. and G.R.M. performed spectroscopic studies. W.-C.H. and D.H.L. performed gel analysis. W.-C.H., G.R.M., A.W., S.F.W., and D.H.L. analyzed the data and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hsieh, WC., Martinez, G.R., Wang, A. et al. Stereochemical conversion of nucleic acid circuits via strand displacement. Commun Chem 1, 89 (2018). https://doi.org/10.1038/s42004-018-0089-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42004-018-0089-9
This article is cited by
-
Functionalization of acyclic xenonucleic acid with modified nucleobases
Polymer Journal (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.