Abstract
A growing body of research has shown how important vitamin D is in the prognosis of coronavirus disease 19 (COVID-19). The vitamin D receptor is necessary for vitamin D to perform its effects, and its polymorphisms can help in this regard. Therefore, we aimed to evaluate whether the association of ApaI rs7975232 and BsmI rs1544410 polymorphisms in different severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants were influential in the outcomes of COVID-19. The polymerase chain reaction-restriction fragment length polymorphism method was utilized to determine the different genotypes of ApaI rs7975232 and BsmI rs1544410 in 1734 and 1450 patients who had recovered and deceased, respectively. Our finding revealed that the ApaI rs7975232 AA genotype in the Delta and Omicron BA.5 and the CA genotype in the Delta and Alpha variants were associated with higher mortality rate. Also, the BsmI rs1544410 GG genotype in the Delta and Omicron BA.5 and the GA genotype in the Delta and Alpha variants were related to a higher mortality rate. The A-G haplotype was linked with COVID-19 mortality in both the Alpha and Delta variants. The A-A haplotype for the Omicron BA.5 variants was statistically significant. In conclusion, our research revealed a connection between SARS-CoV-2 variants and the impacts of ApaI rs7975232 and BsmI rs1544410 polymorphisms. However, more research is still needed to substantiate our findings.
Similar content being viewed by others
Introduction
The World Health Organization (WHO) reports demonstrate that severe acute respiratory syndrome associated with coronavirus-2 (SARS-CoV-2) has caused Coronavirus disease 2019 (COVID-19), a worldwide pandemic, in millions of people since December 2019. The symptoms of the COVID-19 can range from mild to moderate and severe to critical, necessitating hospitalization and intensive care unit (ICU) admissions for some patients1. It is appropriate to shift our focus from preventive to therapeutic measures in light of the development of effective vaccines against COVID-19, whose efficacies are unusually high but not absolute, and in light of the prospect that new viral variations may limit its efficacy. Additionally, there are still a significant number of unvaccinated people2.
Vitamin D deficiency has increased the severity of viral illnesses such as influenza3. According to recent research, vitamin D can influence SARS-CoV-2 gene expression and reduce infection rate when it binds to the vitamin D response element4. The angiotensin-converting enzyme 2 (ACE2) mediates the SARS-CoV-2 infection and its receptor, which vitamin D. Additionally regulates, vitamin D is known to increase the generation of antimicrobial proteins, modulate innate and adaptive immune responses, and possibly operate as an anti-inflammatory agent5,6,7.
Vitamin D exerprimarily biological effects through vitamin D receptors (VDRs) are mostly found in the gastrointestinal tract, bones, lungs, and most immune cells. Even though the VDR is abundantly expressed in the lung tissue, it is yet unclear how vitamin D-VDR signaling may contribute to pulmonary immunopathology8.
Regarding the related mechanisms, single nucleotide polymorphisms (SNPs) in the gene encoding the VDR could alter the protective efficacy of vitamin D-mediated host responses. This may be done, for instance, by affecting the structure of the VDR, which will impact the transcription of genes regulated by vitamin D that affect immune function9. VDR polymorphisms have been linked to an increased risk of acute lower respiratory infections in various contexts10,11,12. Several studies have investigated the association between four VDR polymorphisms including, TaqI (rs731236; exon 9; A > G), FokI (rs2228570; exon 2; C > T), ApaI (rs7975232; intron 8; C > A), and BsmI (rs1544410; intron 8; G > A) and the risk of hepatitis B virus infection in different ethnic groups13,14.
It is important to note that the results of genetic studies investigating the function of the ApaI rs7975232 and BsmI rs1544410 polymorphisms in the pathogeneses of COVID-19 remained controversial. Therefore, this study aimed to examine whether these ApaI rs7975232 and BsmI rs1544410 polymorphisms play a role in the susceptibility to the COVID-19 of different variants of SARS-CoV-2.
Materials and methods
Sample collection
We confirm that all experimental protocols were approved by an Ilam University of Medical Science ethical committee. Moreover, all methods were performed in accordance with the relevant guidelines and regulations.
From 14,117 patients who visited a hospital of Ilam University of Medical Sciences between November 2020 to February 2022 during the three peaks (Alpha, Delta, and Omicron BA.5) of the SARS-CoV-2 infection, 3184 patients were selected based on the following criteria: (1) having a positive real-time reverse transcription polymerase chain reaction (rtReal time-PCR) from the pharyngeal swab samples that were selected from a hospital; (2) giving informed consent to participate in the study; (3) having Iranian nationality with the same ethnicity; (4) lack of underlying comorbidities including pulmonary infection (cystic fibrosis, chronic obstructive pulmonary disease, and asthma), liver disease, chronic kidney disease, heart disease (cardiovascular disease, heart failure, and etc.), cancer, immunocompromised disease (transplant patients and human immunodeficiency virus), hypertension, pregnancy, and diabetes.
In this study, we examined two groups. One was patients with mild and moderate symptoms (cough, malaise, loss of taste and smell, fever, muscle pain, sore throat, nausea, diarrhea, vomiting, headache, and oxygen saturation (SpO2) above 94% on room air at sea level), which were considered as the control group (recovered patients), and the second group was patients with severe and critical symptoms (SpO2 of 94% below room air at sea level, PaO2/FiO2 of 300 mm Hg, lung infiltrates less than 50%, septic shock, difficulty breathing during slight movement or even at rest and multiple organ dysfunction) as the case group (deceased patients).
All paraclinical information such as lipid profile, liver enzymes, complete blood count (CBC), real-time PCR cycle threshold (Ct) values, 25-hydroxyvitamin D, C-reactive protein (CRP), uric acid, erythrocyte sedimentation rate (ESR), and creatinine were obtained when visiting the hospital.
ApaI rs7975232 and BsmI rs1544410 genotyping
After DNA extraction of all patients using the High-pure PCR Template Preparation Kit (Roche Diagnostics Deutschland GmbH, Mannheim, Germany), ApaI rs7975232 and BsmI rs1544410 genotyping was performed using polymerase chain reaction-restriction fragment length polymorphism (PCR–RFLP) method.
The forward and reverse sequence primers for ApaI rs7975232 with the PCR product sizes 242 bp included 5'-CTGCCGTTGAGTGTCTGTGT-3' and 5'-TCGGCTAGCTTCTGGATCAT-3', respectively. The forward and reverse sequence primers for BsmI rs1544410 with the PCR product sizes 297 bp were 5'-GGGAGACGTAGCAAAAGGAG-3' and 5'-CCATCTCTCAGGCTCCAAAG-3', respectively. The PCR conditions were following: initial denaturation at 95 °C for 5 min, followed by 35 cycles of 95 °C for 30 s, 57 °C for 30 s, 72 °C for 35 s, and final extension at 72 °C for 10 min was for ApaI rs7975232 and initial denaturation at 95 °C for 5 min, followed by 35 cycles of 95 °C for 30 s, 58 °C for 30 s, 72 °C for 45 s, and final extension at 72 °C for 10 min was for BsmI rs1544410.
The PCR products were digested with ApaI and BsmI, according to the manufacturer's instructions, and were visualized by electrophoresis on 2.5% agarose gel. The product sizes for ApaI rs7975232 after digestion were 191 bp and 51 bp for the CC genotype and 242 bp for the AA genotype, and for BsmI rs1544410, were 192 bp and 105 bp for the GG genotype and 297 bp for the AA genotype15.
For the PCR–RFLP result confirmation, several samples were randomly selected and sequenced on an ABI 3500 DX Genetic Analyzer (ABI, Thermo Fisher Scientific, Waltham, MA, USA) by the Sanger sequencing method. Then raw data were analyzed with ChromasPro software.
Statistical analyses
SPSS version 22.0 (SPSS, Inc, Chicago, IL, USA) was used for analysis. The Chi-square test was used to evaluate the significance of the relationship between the two qualitative groups. The Shapiro–Wilk test was used to determine the distribution's normality, and Mann–Whitney U test was used for quantitative data.
The Chi-square test was used to examine all SNPs for Hardy–Weinberg equilibrium (HWE). Using SNPStats software, the correlation analysis was carried out, including dominant, over-dominant, co-dominant and recessive models. The minor allele frequency (MAF) and linkage disequilibrium (LD) was also determined. The fitting-best model was chosen using the Akaike Information Criterion (AIC) and the Bayesian Information Criterion (BIC). The most effective model was the one with the lowest AIC score (http://bioinfo.iconcologia.net/SNPStats). Logistic regression was used to determine odds ratios (ORs) and their respective 95% confidence intervals (CIs) for each model. P-values lower than 0.05 were deemed significant.
Results
Baseline clinical features and demographics
Table 1 demonstrates the characteristics of the study participants. Three variants of SARS-CoV-2 were included in the study. Among 3184 patients, there were 1022 Alpha variant, 1026 Delta variant, and 1132 Omicron BA.5 variant. The mean age of the patients with the Delta variant (58.0 ± 11.8) was higher than those with Alpha (53.0 ± 12.7) and Omicron BA.5 (53.7 ± 12.9) variants. The number of males and females in the Alpha variant was 479 (46.9%) and 543 (53.1%), respectively. In the Delta variant, these numbers were 546 (53.2%) and 480 (46.8%) and in the Omicron BA.5 variant, they were 546 (53.2%) and 480 (46.8%), respectively.
The 25-hydroxy vitamin D rates in the Alpha, Delta, and Omicron BA.5 variants were (24.2 ± 12.8), (21.8 ± 10.3), and (33.0 ± 13.4), respectively, which was significant between variants (P = 0.029). The mean qPCR Ct values in the Delta variant (17.4 ± 6.1) were higher than the Alpha (20.1 ± 6.4) and Omicron BA.5 (21.9 ± 6.0) variants (P < 0.001).
Relationship between COVID-19 mortality adjusted by SARS-CoV-2 variants and ApaI rs7975232 and BsmI rs1544410 polymorphisms
The COVID-19 death rate was considerably more significant in patients with the ApaI rs7975232 AA genotype than in other genotypes. Patients who have recovered from COVID-19 also had the ApaI rs7975232 CC genotype. Patients with the GG genotype exhibited a higher COVID-19 mortality rate in the BsmI rs1544410 polymorphism.
Table 2 tabulates the inheritance model analysis results for ApaI rs7975232 and BsmI rs1544410 polymorphisms in patients. By comparing the deceased and recovered patients, the codominant and dominant inheritance models with the lowest AIC and BIC values were found to be the best-fitting models for ApaI rs7975232 and BsmI rs1544410. The ApaI rs7975232 AA genotype was linked to a higher risk of COVID-19 mortality (P < 0.0001, OR 1.87, 95% CI 1.49–2.35), whereas the BsmI rs1544410 GA/GG genotype was associated with a higher risk of COVID-19 mortality (P < 0.0001, OR 1.97, 95% CI 1.68–2.30).
The ApaI rs7975232 polymorphism in recovered and deceased patients was compatible with HWE (P > 0.05), while HWE in BsmI rs1544410 was incompatible in both groups (P < 0.001). The MAF for ApaI rs7975232 (A) and BsmI rs1544410 (G) polymorphisms in deceased patients was higher than in recovered patients.
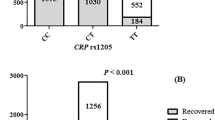
Correlation between the value of vitamin D and ApaI rs7975232 and BsmI rs1544410 polymorphism
The relation between vitamin D values and different genotype frequencies is indicated in Fig. 1. There is significant difference in vitamin D levels between different ApaI rs7975232 (P = 0.041) and BsmI rs1544410 (P = 0.008) genotypes among recovered and deceased patients. The lowest amount of vitamin D was found in ApaI rs7975232 GG and BsmI rs1544410 AA genotypes, while the highest amount in ApaI rs7975232 AA and BsmI rs1544410 CC genotypes.
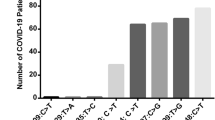
Frequencies of ApaI rs7975232 and BsmI rs1544410 polymorphism between SARS-CoV-2 variants
Our findings showed that the death rate was related to the SARS-CoV-2 variants, much higher in the Delta variant than in the Alpha and Omicron BA.5 variants (P < 0.001).
The frequencies of CC, CA, and AA genotypes in ApaI rs7975232 polymorphism in the Alpha variant were 407 (39.8%), 457 (44.7%), and 158 (15.5%), respectively. These frequencies in the Delta variant was 333 (32.5%), 480 (46.8%), and 213 (20.7%), respectively. In the Omicron BA.5 variant, the frequency of CC was 570 (50.2%), CT was 464 (40.8%), and TT was 102 (9.0%) (Fig. 2A).
When we adjusted the effect of ApaI rs7975232 polymorphism for SARS-CoV-2 variants, the COVID-19 mortality rate was related to ApaI rs7975232 CA (OR 1.32, 95% CI 1.01–1.73) in the Alpha variant and with ApaI rs7975232 AA (OR 3.03, 95% CI 2.09–4.41) and CA (OR 2.97, 95% CI 2.21–3.99) in the Delta variant and with AA in the Omicron BA.5 variant (OR 3.86, 95% CI 2.49–5.99) (Table 3).
The frequencies of AA, GA, and GG genotypes in BsmI rs1544410 polymorphism in the Alpha variant were 430 (42.1%), 507 (49.6%), and 85 (8.3%), respectively. These frequencies in the Delta variant was 426 (41.7%), 479 (46.7%), and 121 (11.6%), respectively. In the Omicron BA.5 variant, the frequency of CC was 420 (37.0%), CT was 646 (56.9%), and TT was 70 (6.1%) (Fig. 2B).
When we adjusted the effect of BsmI rs1544410 polymorphism for SARS-CoV-2 variants, the COVID-19 mortality rate was related to BsmI rs1544410 GA (OR 3.26, 95% CI 2.49–4.27) in the Alpha variant and with BsmI rs1544410 GG (OR 5.08, 95% CI 3.77–6.84) and GA (OR 3.26, 95% CI 2.09–5.10) in the Delta variant and with GG in the Omicron BA.5 variant (OR 2.41, 95% CI 1.44–4.02) (Table 3).
According to our findings, the C-A haplotype was observed to be the predominant form among all SARS-CoV-2 variants. The A-G haplotype was linked with COVID-19 mortality in both the Alpha (OR 1.56, 95% CI 1.27–1.92) and Delta (OR 2.70, 95% CI 2.15–3.38) variants. The A-A haplotype for the Omicron BA.5 (OR 10.37, 95%CI 4.40–24.46) variants was statistically significant (Table 4). There were strong LD between ApaI rs7975232 and BsmI rs1544410 (r2 = 0.91).
Discussion
We investigated how the ApaI rs7975232 and BsmI rs1544410 affected the susceptibility to COVID-19 and showed that they might be used as genetic indicators for infection by different SARS-CoV-2 variants.
Alleles A (0.37) for the ApaI rs7975232 and G (0.34) for the BsmI rs1544410 polymorphisms as MAF were directly related to mortality in patients with COVID-19.
The MAF results in our study for ApaI rs7975232 were almost similar to different Asian populations, including Asian (0.313), East Asian (0.314), and other Asian (0.310), while it was different from South Asian (0.615), European (0.537), Latin American (0.589), and African (0.630) (https://www.ncbi.nlm.nih.gov/snp/rs7975232). The MAF for ApaI rs7975232 in deceased patients (0.42) was slightly higher than recovered patients (0.32) in our study.
The MAF result in our study for BsmI rs1544410 were similar to a study in Iran and South Asian (0.443), Latin American (0.366), European (0.398), and African (0.262), but was different from other regions including Asian (0.060), East Asian (0.056), and other Asian (0.076) (https://www.ncbi.nlm.nih.gov/snp/rs1544410). The MAF for BsmI rs1544410 in deceased patients (0.39) was slightly higher than recovered patients (0.30) in our study.
In this study, the levels of vitamin D in COVID-19 patients, especially those infected with the Delta variant with a higher mortality rate, were lower than the other two variants. It has been found that vitamin D can play an antiviral inhibitory role in nasal epithelial cells in SARS-CoV-2 infection16. This virus enters the host cells after binding to its receptors on the cell’s surface called ACE2 by Spike protein. Type II alveolar cells, in which ACE2 receptors are strongly expressed, are the virus’s primary target17. Calcitriol, a vitamin D agonist, increases the ACE2 expression and soluble ACE2, which may lead to virus trapping and inactivation. The renin–angiotensin–aldosterone system, altered by SARS-CoV-2 infection, is negatively regulated by calcitriol, inhibiting renin expression. This increased availability of angiotensin II leads to tissue damage, inflammation, and multi-organ failure18.
Active forms of vitamin D and lumisterol have been shown to block SARS-CoV-2 replication machinery enzymes (main protease and RNA-dependent RNA polymerase), implying that novel vitamin D and lumisterol metabolites are potential antiviral therapeutic candidates. Moreover, these metabolites may prevent SARS-CoV-2 receptor binding domain from attaching to ACE2 by interacting with transmembrane serine protease 2 (TMPRSS2) and ACE2. The structural and dynamical motion alterations brought on by these interactions could impact TMPRSS2's ability to prime the SARS-CoV-2 spike proteins19. As a result, novel CYP11A1-derived vitamin D3 hydroxyderivative, including 20(OH) vitamin D3 and 20,23(OH)2 vitamin D3, and lumisterol hydroxymetabolites can inhibit COVID-19 via both independent and nuclear receptor-dependent mechanisms, making them excellent candidates for antiviral drug research as well as the informed use of their precursors as nutrients or supplements in the prevention and attenuation of COVID-19 disease20,21.
Vitamin D's active hydroxyl forms have anti-inflammatory and antioxidant effects, and they also boost innate defense to infectious agents. These characteristics are shared by non-calcemic hydroxyderivatives produced by CYP11A1 and calcitriol. They exhibit inverse agonism on the retinoic acid‐related orphan receptors-γ (ROR-γ), suppress the synthesis of pro-inflammatory cytokines, downregulate NF‐κΒ, and combat oxidative stress by activating transcription factor NF‐E2‐related factor 2 (NRF2). As a result, a direct delivery of vitamin D hydroxyderivatives deserves consideration in the therapy of COVID19 of various etiologies22.
Human VDR has more than 14 distinct identified polymorphisms. These polymorphisms may affect how VDR binds to calcitriol to modulate its response. FokI rs2228570, BsmI rs1544410, ApaI rs7975232, and TaqI rs731236 are the four SNPs that are most commonly examined. They were demonstrated independently modifying vitamin D status and in haplotypes23.
The COVID-19 death rate was considerably more significant in patients with the ApaI rs7975232 AA genotype than in other genotypes. The COVID-19 mortality rate was related to ApaI rs7975232 CA in the Alpha variant and with AA and CA in the Delta variant and with AA in the Omicron BA.5 variant. In agreement with our results, Apaydin et al. showed that the AA genotype was common among patients with severe COVID-1924.
Cohorts from Nigeria, Egypt, Ethiopia, Pakistan, Saudi Arabia, Lebanon, Turkey, and Italy were found to frequently have the AA genotype, according to the frequencies of the ApaI rs7975232 polymorphism. In contrast, the cohorts from Iran, the US, Poland, Greece, Mexico, India, the Netherlands, Czechia, Croatia, Russia, Spain, Finland, Brazil, and Tunisia frequently had the AC genotype. The CC genotype of the ApaI rs7975232 gene was most common in deceased patients from Korea, Japan, and China25. Studies with hepatitis B virus demonstrated that CA/AA genotypes of ApaI rs7975232 polymorphism trigger T helper 2 (Th2) cells proliferation, but there are no studies on ApaI rs7975232 and respiratory system viral infection. On the other hand, AA genotypes result in Th1 proliferation and anti-inflammatory cytokine production, which accelerates the progression of liver disease progression to cirrhosis14.
The fact that participants with the AA genotype of the ApaI rs7975232 polymorphism in this study had a greater death rate suggests that Th2 can also release Interleukin-6 (IL-6), which is related to COVID-19 prognosis. IL-6 is one of the key factors in the cytokine storm caused by COVID-19. IL-6 induces endothelial dysfunction with expression of tissue factor and adhesion molecules via upregulation of angiotensin converting enzyme-2 receptor. These negative effects of IL-6 were mitigated by vitamin D and VDR polymorphisms. As a result, it is possible that this is one of the putative mechanism(s) by which vitamin D exerts its positive effects in COVID-19 infection26. Subjects with the severe and moderate disease who had the "CA" genotype compared to "CC and AA" genotypes demonstrated a more severe risk, according to the study by Abdollahzadeh et al. Contrary to "CA and AA" and "CA" genotypes, symptomatic-asymptomatic and moderately-asymptomatic patients with the CC genotype were more likely to have signs and symptoms. In contrast to our findings, none of the deceased participants had the AA genotype15.
This study's patients with the GG genotype in this study exhibited a higher COVID-19 mortality rate in the BsmI rs1544410 polymorphism. The COVID-19 mortality rate was related to BsmI rs1544410 GA in the Alpha variant, BsmI rs1544410 AA and GA in the Delta variant, and GG in the Omicron BA.5 variant. It has been demonstrated that the BsmI rs1544410 G allele can be a risk factor for COVID-19 severity15, while no such relationship was seen in the study of Apaydin et al.24. The BsmI rs1544410 polymorphism's diversity revealed that the cohorts of the US, China, Poland, Turkey, Egypt, Italy, Saudi Arabia, Russia, Czechia, India, Greece, the Netherlands, Croatia, Brazil, Spain, Tunisia, Nigeria, and Lebanon frequently had the BsmI rs1544410 AG genotype. In contrast, the cohorts from Iran, Korea, Japan, Finland, Pakistan, and Mexico frequently had the BsmI rs1544410 GG genotype in deceased patients, but this was not significant25.
The association of BsmI rs1544410 and viral infections such as HIV has been investigated. It has been demonstrated that BsmI rs1544410 A-allele was strongly correlated with the rapid progression of HIV disease. It is unclear exactly how the BsmI rs1544410 G-allele confers protection, while the BsmI rs1544410 A-allele raises the likelihood of disease27. The BsmI rs1544410 G to A polymorphism alteration occurs in the 3' untranslated regions (3' UTRs) of the VDR gene and is hypothesized to affect the VDR messenger RNA stability. This polymorphism has been linked to an increased HIV infection susceptibility and faster rate of HIV disease development28,29.
Strong LD exists between BsmI rs1544410 and another 3′ UTR polymorphism (ApaI rs7975232), which has also been linked to the course of HIV illness. Given that the BsmI rs1544410 polymorphism is a synonymous mutation, the relationships seen may be explained by LD with one or more functional polymorphisms at other locations in the VDR gene30. However, synonymous rather than silent mutations could cause alternations in the protein's expression, conformation, and function. Therefore, BsmI rs1544410 polymorphisms might also directly change the VDR31. The findings that the BsmI rs1544410 A-allele is more influential in disease progression in the Delta variant than the other two may be explained by the difference in the serum vitamin D level, as these levels in patients with the Delta variant were much higher. Also, in this study, there was a strong LD between BsmI rs1544410 and ApaI rs7975232.
According to our findings, the C-A haplotype was more common among all SARS-CoV-2 variations. The A-G haplotype was linked with COVID-19 mortality in both the Alpha and Delta variants. The A-A haplotype for the Omicron variants was statistically significant. These two SNPs may likely function differently in distinct SARS-CoV-2 variants. However, the mechanism underlying this divergence remains unknown.
There were several limitations in our study that should be considered. We did not have any healthy controls who had not previously suffered from COVID-19. Besides, previous vaccination information of all patients was not available. Moreover, this study was conducted in only one population with the same ethnicity. To generalize the relationship between these two polymorphisms to the whole society, more studies should be done on different races in Iran.
In conclusion, our study showed that the serum vitamin D level and BsmI rs1544410 and ApaI rs7975232 polymorphisms were related to the mortality rate of SARS-CoV-2 with different variants. The COVID-19 mortality rate was related to ApaI rs7975232 CA genotype in the Alpha variant and with AA and CA genotypes in the Delta variant and with AA genotype in the Omicron BA.5 variant. Moreover, in BsmI rs1544410 polymorphisms, the mortality rate was correlated with GA genotype in the Alpha variant and with GG and GA genotypes in the Delta variant and with GG genotype in the Omicron BA.5 variant. The A-G haplotype was linked with COVID-19 mortality in both the Alpha and Delta variants. The A-A haplotype for the Omicron BA.5 variants was statistically significant. Further studies in different ethnicities should be done to confirm our results.
Data availability
All data that support all the experimental findings in this article is available in the paper.
References
Varikasuvu, S. R. et al. COVID-19 and vitamin D (Co-VIVID study): A systematic review and meta-analysis of randomized controlled trials. Expert Rev. Anti-infect. Ther. 92, 111356 (2022).
García-Lledó, A. et al. Pharmacological treatment of COVID-19: an opinion paper. Rev. Esp. Quimioter. 35, 115 (2022).
Watkins, R. R., Lemonovich, T. L. & Salata, R. A. An update on the association of vitamin D deficiency with common infectious diseases. Can. J. Physiol. Pharmacol. 93, 363–368 (2015).
Brito, D. T. M., Ribeiro, L. H. C., da Cunha Daltro, C. H. & de Barros Silva, R. The possible benefits of vitamin D in COVID-19. Nutrition 91, 111356 (2021).
Gois, P. H. F., Ferreira, D., Olenski, S. & Seguro, A. C. Vitamin D and infectious diseases: simple bystander or contributing factor?. Nutrients 9, 651 (2017).
AlSafar, H. et al. COVID-19 disease severity and death in relation to vitamin D status among SARS-CoV-2-positive UAE residents. Nutrients 13, 1714 (2021).
Malek Mahdavi, A. A brief review of interplay between vitamin D and angiotensin-converting enzyme 2: Implications for a potential treatment for COVID-19. Rev. Med. Virol. 30, e2119 (2020).
Zeidan, N. et al. Vitamin D deficiency and vitamin D receptor FokI polymorphism as risk factors for COVID-19. Pediatr. Res. 9, 1–8 (2022).
van Etten, E. et al. The vitamin D receptor gene FokI polymorphism: functional impact on the immune system. Eur. J. Immunol. 37, 395–405 (2007).
Kresfelder, T., Janssen, R., Bont, L. & Venter, M. Confirmation of an association between single nucleotide polymorphisms in the VDR gene with respiratory syncytial virus related disease in South African children. J. Med. Virol. 83, 1834–1840 (2011).
Roth, D. E., Jones, A. B., Prosser, C., Robinson, J. L. & Vohra, S. Vitamin D receptor polymorphisms and the risk of acute lower respiratory tract infection in early childhood. J. Infect. Dis. 197, 676–680 (2008).
Albu-Mohammed, W. H. M., Anvari, E. & Fateh, A. Evaluating the role of BglI rs739837 and TaqI rs731236 polymorphisms in Vitamin D receptor with SARS-CoV-2 variants mortality rate. Genes 13, 2346 (2022).
He, Q. et al. Association between vitamin D receptor polymorphisms and hepatitis B virus infection susceptibility: A meta-analysis study. Gene 645, 105–112 (2018).
Hoan, N. X. et al. Vitamin D receptor ApaI polymorphism associated with progression of liver disease in Vietnamese patients chronically infected with hepatitis B virus. BMC Med. Genet. 20, 1–12 (2019).
Abdollahzadeh, R. et al. Association of Vitamin D receptor gene polymorphisms and clinical/severe outcomes of COVID-19 patients. Infect. Genet. Evol. 96, 105098 (2021).
Greiller, C. L. & Martineau, A. R. Modulation of the immune response to respiratory viruses by vitamin D. Nutrients 7, 4240–4270 (2015).
Hoffmann, M. et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280 (2020).
Yuan, W. et al. 1, 25-dihydroxyvitamin D3 suppresses renin gene transcription by blocking the activity of the cyclic AMP response element in the renin gene promoter. J. Biol. Chem. 282, 29821–29830 (2007).
Song, Y. et al. Vitamin D3 and its hydroxyderivatives as promising drugs against COVID-19: A computational study. J. Biomol. Struct. Dyn. 40, 11594–11610 (2022).
Qayyum, S. et al. Vitamin D and lumisterol novel metabolites can inhibit SARS-CoV-2 replication machinery enzymes. Am. J. Physiol.-Endocrinol. Metab. 321, 246–251 (2021).
Qayyum, S., Slominski, R. M., Raman, C. & Slominski, A. T. Novel CYP11A1-derived vitamin D and lumisterol biometabolites for the management of COVID-19. Nutrients 14, 4779 (2022).
Slominski, R. M. et al. COVID-19 and Vitamin D: A lesson from the skin. Exp. Dermatol. 29, 885–890 (2020).
Hasan, H. A., Raed, O. A., Muda, W. A. M. B. W., Mohamed, H. J. B. J. & Samsudin, A. R. Association of Vitamin D receptor gene polymorphisms with metabolic syndrome and its components among adult Arabs from the United Arab Emirates. Diabetes Metab. Syndr. 11, S531–S537 (2017).
Apaydin, T. et al. Effects of vitamin D receptor gene polymorphisms on the prognosis of COVID-19. Clin. Endocrinol. 96, 819–830 (2022).
Karcioğlu, L. Correlation of the variations in prevalence of coronavirus disease 2019 and vitamin D receptor gene polymorphisms in cohorts from 26 countries. Anatolian Clin. J. Med. Sci. 27, 60–70 (2022).
Cimmino, G. et al. Vitamin D Inhibits IL-6 pro-atherothrombotic effects in human endothelial cells: A potential mechanism for protection against COVID-19 infection?. J. Cardiovasc. Dev. Dis. 9, 27 (2022).
Moodley, A., Qin, M., Singh, K. K. & Spector, S. A. Vitamin D related host genetic variants alter HIV disease progression in children. Pediatr. Infect. Dis. J. 32, 1230–1236 (2013).
Alagarasu, K., Selvaraj, P., Swaminathan, S., Narendran, G. & Narayanan, P. 5′ regulatory and 3′ untranslated region polymorphisms of vitamin D receptor gene in South Indian HIV and HIV–TB patients. J. Clin. Immunol. 29, 196–204 (2009).
Sánchez de laTorre, M. et al. Vitamin D receptor gene haplotypes and susceptibility to HIV-1 infection in injection drug users. J. Infect. Dis. 197, 405–410 (2008).
Uitterlinden, A. G., Fang, Y., Van Meurs, J. B., Pols, H. A. & Van Leeuwen, J. P. Genetics and biology of vitamin D receptor polymorphisms. Gene 338, 143–156 (2004).
Sauna, Z. E. & Kimchi-Sarfaty, C. Understanding the contribution of synonymous mutations to human disease. Nat. Rev. Genet. 12, 683–691 (2011).
Acknowledgements
We would like to thank all of the patients who participated in the study.
Author information
Authors and Affiliations
Contributions
A.A and E.A.: Performed the experiments and manuscript preparation, clinical sample and data acquisition; A.F. and E.A.: analyzed and interpreted data; A.F.: designed and supervised clinical study, interpreted data, read and approved manuscript. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Al-Gharrawi, A., Anvari, E. & Fateh, A. Association of ApaI rs7975232 and BsmI rs1544410 in clinical outcomes of COVID-19 patients according to different SARS-CoV-2 variants. Sci Rep 13, 3612 (2023). https://doi.org/10.1038/s41598-023-30859-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-30859-7
This article is cited by
-
Pharmacological evaluation of vitamin D in COVID-19 and long COVID-19: recent studies confirm clinical validation and highlight metformin to improve VDR sensitivity and efficacy
Inflammopharmacology (2024)
-
The impact of ACE2 polymorphisms (rs1978124, rs2285666, and rs2074192) and ACE1 rs1799752 in the mortality rate of COVID-19 in different SARS-CoV-2 variants
Human Genomics (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.