Abstract
Deep learning has been used extensively in histopathological image classification, but people in this field are still exploring new neural network architectures for more effective and efficient cancer diagnosis. Here, we propose multi-scale, multi-view progressive feature encoding network (MSMV-PFENet) for effective classification. With respect to the density of cell nuclei, we selected the regions potentially related to carcinogenesis at multiple scales from each view. The progressive feature encoding network then extracted the global and local features from these regions. A bidirectional long short-term memory analyzed the encoding vectors to get a category score, and finally the majority voting method integrated different views to classify the histopathological images. We tested our method on the breast cancer histology dataset from the ICIAR 2018 grand challenge. The proposed MSMV-PFENet achieved 93.0\(\%\) and 94.8\(\%\) accuracies at the patch and image levels, respectively. This method can potentially benefit the clinical cancer diagnosis.
Similar content being viewed by others
Introduction
Cancer is one of the most lethal diseases for human beings. According to the report from WHO, the number of diagnosed patients with cancer reached 19.3 million, and its casualty had increased to 10 million1. The gold standard for the clinical diagnosis of cancer is histopathological examination in which pathologists look into the prepared tissue sections to determine whether the tumor is benign or malignant2. The examination done by pathologists has two shortcomings. First, pathologists sometimes need to carefully examine many places in histological image with high magnification and large field of view. This operation is labor-intensive and time-consuming. Second, for high sensitivity and specificity, the examination demands profound professional knowledge and experience - more than 100,000 examinations often required in practice3. Pathologists without many practices are prone to the wrong diagnosis, causing patients to miss a valuable period of early treatment.
Benefiting from the development of image recognition technology, the computer-aided histological examination has made a breakthrough in clinical application4,5,6,7. It is not only improving the efficiency of examination but also reducing the rate of misdiagnosis. The computer-aided cancer diagnosis decides by analyzing the features in the image. For example, the local binary patterns, gray level co-occurrence matrix, the opponent colour local binary pattern, and many other techniques can be used to extract the features and help achieve a classification accuracy of 85\(\%\)8,9. The features extracted by these methods usually are very useful in terms of classifying a specific lesion type but are not enough for classifying multiple lesion types in one classifier.
In recent years, deep learning has achieved tremendous success in image classification10,11 and image enhancement12,13. It can extract abstract features from images with little human intervention14,15. Image classification methods based on deep learning frameworks, such as the convolutional neural network (CNN) and recurrent neural network (RNN), have been widely used in the pathological classification of cancer16,17,18,19,20,21,22. CNN, as a feed-forward neural network relying on its filtering and pooling layers, is good at extracting the features at a local level23. In contrast, RNN as a feed-backward neural network regarding the previous output as a part of its input, is good at processing the information at a global level24. Both frameworks have attracted the attention from the healthcare industry. Before they can be formally deployed in clinical applications, some issues must first be addressed. For example, the useful information in each input image is quite limited, due to the small image size sent to the neural networks25,26; however, sending the whole-slide image (WSI) into the neural network leads to a significant computational cost27.
There are three common ways to solve this dilemma. The first one is using a low-latency discriminator which can find out the suspicious area in the original image28,29. Then, a neural network can classify each case with respect to the suspicious area. The second way is segmenting the original image into multiple patches30. The neural network then analyzes each patch independently. In the end, the program will summarize the result of each patch in the original image and draw a conclusion about the class of this case. The third way is extracting features at multiple scales and analyzing the local and global features simultaneously31,32. However, using the deep learning method to select suspicious areas from the high-resolution pathological image is time-consuming. Patch-based approach30 will lose feature context information if the relationship between patches cannot be well solved. In addition, the methods based on multi-scale31,32 fail to provide targeted models for multi-scale feature representation.
Furthermore, the breast cancer datasets such as BACH33 and BreakHis8 often have a limited volume. Using the limited data to train the network can easily lead to over-fitting and become useless in the application34. Transfer learning35 and data augmentation36 can somehow solve this problem. In the transfer learning, the network model is pre-trained on a generalized dataset and then trained on the specific dataset where your application is defined. In the image augmentation, the dataset is effectively expanded by various affine transformations37. This method is especially effective for the large-scale pathological images.
Here, we propose multi-scale, multi-view progressive feature encoding network (MSMV-PFENet). To extract more effective suspicious region, the key-region-extraction module (KREM) selects the image patches from the original pathological images, according to the density of cell nuclei. The progressive feature encoding network (PFENet), as our multi-scale feature representation model, progressively extracts features from the images and encodes the features into vectors. The feature discrimination network (FDNet) integrates encoding vectors of multiple-scales through bidirectional long short-term memory (BiLSTM)), mining potential feature relationships between local and global, to avoid loss of information between patches. We combine the advantages of CNN and RNN to preserve the short-term and long-term spatial correlation between the features at different scales. In addition, the majority-voting method summarizes the results of six views to determine the final category, avoiding one-sidedness under a single view. Testing the model on the breast cancer histology (BACH) dataset33 and Yan’s dataset30, MSMV-PFENet can achieve a good performance in terms of accuracy, precision, recall, and F1 score.
Methods
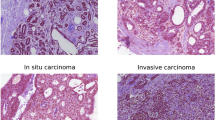
The proposed MVMS-PFENet took up the high-resolution HE staining pathological images (2048 \(\times \) 1536) and would classify the images into four categories—normal, benign, carcinoma in situ, and invasive carcinoma. As shown in Fig. 1, we performed six view transformations (raw, 90\(^\circ \), 180\(^\circ \), 270\(^\circ \), x-flip, y-flip) on an input image after preprocessing. In each view, the key regions at three scales were selected from the region with a high density of cell nuclei (Fig. 1a), we got six groups of multi-scale key image patches. The PFENet encoded each group of patches into vectors (Fig. 1b), then a two-layer BiLSTM fused the encoding vectors of the patches at different scales to get category scores in each view (Fig. 1c). Finally, the FDNet analyzed the scores obtained from the six views and determined the type of cancer according to majority voting (Fig. 1c). The detailed structure of MVMS-PFENet is shown in Table 1.
In KREM, inspired by the standard histological examination, we created a module to extract the suspicious areas in a large original image for further analysis to reduce computational cost. The designed module selected the image patches at different scales from the original image with respect to the density of cell nuclei due to its underlying relation with the lesion.
As shown in Fig. 2, the KREM selection process consists of four key steps: Eq. (1) We first converted the colorful input image to a grayscale image and set a threshold according to the mean of pixel values. As a result, we obtained a binary image in which the white pixels represented the area occupied by the nuclei. Equation (2) Via the statistical analysis of all the histological images at 200\(\times \) magnification in the dataset, we found that the majority of nuclei were in an oval shape with an area of more than 80 pixels and the ratio of area to circumference was less than 2.5. According to this criterion, irrelevant regions were removed to refine the region of the nucleus. Equation (3) We used a sliding window to search in the image and calculated the number of detected nuclei in the window. Equation (4) The image patches with the highest number of nuclei would be selected. Within a selected region, a smaller sliding window repeated the previous process to select the image patches at the next scale. In this paper, we used sliding windows at three scales. The sizes of sliding windows at the three scales were 897 \(\times \) 897, 400 \(\times \) 400, and 224 \(\times \) 224, respectively. Their corresponding step sizes respectively were 80, 40 and 20 pixels.
In the training phase, KREM first rotated each pathological image by 90\(^\circ \), 180\(^\circ \), 270\(^\circ \) , and inverted relative to the x- and y-axes to augment the limited dataset. Then we picked out 12 regions with the densest nuclei from candidate windows at the first selecting level to further expand the amount of data. Unlike other data augmentation methods, this step is based on the data augmentation achieved by our proposed sliding window nuclei density detection technique. This can reduce invalid patches and preserve the information of the lesions. After that, our pathological image dataset was amplified by 72 times. In the testing phase, KREM performed six view transformations on a testing image, and only picked the densest region of cell nuclei in each view. We observe from 6 views, thereby avoiding the sliding step length too long and could not cover the area with dense nuclei. Figure 3 shows the image patches selected by the KREM.
The structure of PFENet was composed of three parallel networks. The PFENet represented the image patches at multiple scales by vectors to facilitate the fusion of features of different scales (Fig. 4). Before inputting the network, we used bilinear interpolation to subsample the image patches at the ratios of 4\(\times \), 1.79\(\times \), and 1\(\times \). The three image patches produced in each view were used as a group, corresponding to the three inputs of the PFENet. The PFENet at all scales shares the same structure similar to ResNet5038. We did not adopt a network structure larger than ResNet50 to limit the computation cost in the feature extraction. The PFENet progressively extracted low-level color, texture, and deep semantic features layer by layer through convolution to learn pathological information at various scales. Finally, three 2048-dimensional encoding vectors were obtained via the adaptive average pooling layer. This would reduce the amount of calculation in the BiLSTM to get category scores.
The encoding vectors at different scales represent global and local features, and they have strong correlations at one view. We used two layers of BiLSTM to share and learn the feature representation. The two-layer BiLSTM is a part of FDNet, as shown in Fig. 1c, the input is three consecutive sequences corresponding to the output of PFENet. BiLSTM is composed of two LSTM (Fig. 5). A directed ring is formed between LSTM units, which creates the internal state of the network and makes information flow between units. The output of LSTM depends on the previous input and the current internal state. The encoding vectors were fed into the network, and the information flowed in three channels to establish a global and local relationship, thereby making up for the limitations of features at different scales. In this way, BiLSTM fused multi-scale pathological features and mined the potential feature relationships between input sequences, and then got pathological image category scores in each view. Finally, we got scores on six views. To realize the end-to-end classification, PFENet and BiLSTM were jointly trained.
Pathological images are special in that they have no directional specification. The diagnosis results should not be affected by the view and should be consistent. However, images clipped from the WSI may have blank areas, as shown in the last column of Fig. 6. The existence of the blank areas will cause inaccurate extraction of image patches in key areas. In addition, the window has a stride in sliding and cannot traverse the image completely, which will also affect the acquisition of key areas. Therefore we need to further discriminate categories from different views. The final category is determined by the results of six views using the majority voting method.
Results
Experimental setup
We trained and tested the MSMV-PFENet on a platform equipped with one NVIDIA Tesla V100 GPU (32G memory) and 24-core Intel Xeon Platinum 8168 Processor (33M Cache, 2.70 GHz). We trained the network with a batch size of 64 through 27,000 iterations, using stochastic gradient descent (SGD) in which we set the momentum to 0.9 and weight decay to 1e-4. To accelerate the training, we set the initial learning rate at 0.01 for the first 30 batches and then divided the rate by 10 after every 10 batches, until the rate was equal to 1e-4. The BACH dataset33 used in our experiment contained 400 training images and 100 testing images (Fig. 6, Table 2). The dimensions of images are 2048 \(\times \) 1536 pixels, and the pixel size is \(0.42\,\upmu \mathrm{m} \times 0.42\,\upmu \mathrm{m}\). We divided training images into two sets with a ratio equal to 4:1 for training and validation.
To avoid over-fitting due to the small size of the dataset, we used transfer learning35 and data augmentation36 techniques. PFENet originated from ResNet50 pre-trained from ImageNet. PFENet did not have the average pooling layers and fully connected layers that were essential for ResNet50 and replaced by an adaptive average pooling layer. We randomly initialized the parameters in FDNet and trained FDNet with PFENet together. PFENet encoded the features in the image into a vector, and FDNet took the vector in for classification. We fine-tuned the whole model according to the classification feedback so that FDNet could discriminate the various features and images correctly. The specific details of dataset augmentation are presented in the Methods section. We also normalized the color in the images to avoid the inconsistency caused by different staining protocols39.
To demonstrate the classification capabilities of MSMV-PFENet, we used the same image-level and patch-level as in30,31 to evaluate the network from multiple aspects. We defined the accuracy (Acc), precision (Pre), recall (Rec), and F1 score (F1) as
Here, TP, TN, FP, and FN are the true-positive, true-negative, false-positive, and false-negative, respectively. These criteria had also been adopted elsewhere40,41.
Comparison with other methods
To show the classification capability of MSMV-PFENet, we compared the accuracy given by our network with that in the other published30,39,42,43,44. Our MSMV-PFENet could achieve an accuracy of 93.0\(\%\) at patch-level (Table 3); the value was higher than those reported elsewhere. The comparison illustrated that a well-trained MSMV-PFENet could effectively extract the most important features in the original images at both the local and global levels. Moreover, the BiLSTM and majority voting mechanism in our network further boosted the accuracy through comprehensive consideration of local and global features. As a result, MSMV-PFENet achieved an accuracy of 94.8\(\%\) at image-level.
Results on the key regions extraction
To check the influence of various methods extracting the image patches from the original images, we compared our method guided by the density of cell nuclei with other methods of randomly selecting patches. When we chose the random selection, training MSMV-PFENet was difficult, because not all of the lesions in the original image would be sent to the classifier for analysis, which led to relatively low accuracy ( 85.4\(\%\)). In comparison, our method made the training process much easier than the random selection method and benefited the classification power of MSMV-PFENet, based on the fact that cancer cells were often associated with increased nuclear size, irregular nuclear contours, and disturbed chromatin distribution. At the patch-level, the accuracy could reach 93.0\(\%\).
Results on different network combinations
To find an optimal design for feature encoding and discriminating in MSMV-PFENet, we carried out a comprehensive study comparing various architectures (Table 4). For feature encoding, we chose a fully connected layer as the feature discriminator and tested FENet, PFENet, VGG1626, GoogLeNet45, and ResNet-10138 as the feature encoder (Table 4, rows 1–6). PFENet in this test provided the best accuracy at both the patch and image levels among the five architectures because the PFENet containing 3 CNNs in parallel could encode the features from the local and global levels simultaneously. The global features represented at low resolution also reduced the computational cost and made the classifier more efficient than analyzing a high-resolution image as a whole.
After we settled down the issue with feature encoding, we tested several candidates, including the FC, support vector machine (SVM) and BiLSTM, for the classifier (Table 4, rows 6–10). BiLSTM outperformed the other two candidates, because it could effectively associate the information at the local and global levels. Unfortunately, training BiLSTM was difficult. The deeper the BiLSTM network was, the harder the network converged to a qualified discriminator. Balancing the tradeoff between the accuracy and feasibility of training, we finally chose a network design with a double-layer BiLSTM for the following demonstrations.
Results of the whole network
Figure 7a shows the confusion matrix calculated in our test on the BACH test dataset. The results confirmed that our MSMV-PFENet could accurately associate the histological images with their clinical outcomes, including normal, benign, in situ carcinoma, and invasive carcinoma. The precision, recall, and F1 score as additional references were also calculated, and these scores had always been higher than 92.8\(\%\) (Fig. 7b). We noticed that the misclassification rate was higher for the normal tissue, which perhaps was induced by the subtle inconsistency in the tissue morphology among the populations with different genotypes.
In addition, we also verified our method on the Yan’s dataset30. This dataset contains 3771 diversified high-resolution pathological images. Figure 8a shows the confusion matrix calculated on the test dataset. The precision, recall, and F1 score were still maintained at higher than 92\(\%\) (Fig. 8b), and the accuracy reached 95.6\(\%\). The results demonstrated that MVMS-PFENet also can achieved good performance on a larger dataset.
Conclusion
In this paper, we proposed MSMV-PFENet to classify the high-resolution breast histopathological images. We first selected the image patches at multiple scales from the high-resolution images, using a sliding window and the statistics of cell nuclei, so as to avoid selecting invalid image patches and preserve the data information of lesions. The PFENet then extracted the color, texture and deep semantic features of pathological images and converted these image patches into the encoding vectors to improve the efficiency of multi-scale feature processing. The BiLSTM fused the encoding vectors and mined the potential feature between sequences to get category scores in each view. Transfer learning accelerated the convergence of PFENet as well as BiLSTM and improved the generality of the model. The image augmentation expanded the dataset through various rotations and flips and reduced the likelihood of over-fitting. The majority voting method combined the category scores from six views (original, 90\(^\circ \) rotated, 180\(^\circ \) rotated, 270\(^\circ \) rotated, x-flip, y-flip) and further improved the accuracy of classification. The results were verified on the BACH dataset, and the accuracy of patch-level and image-level reached 93\(\%\) and 94.8\(\%\) respectively. We also verified our method on the dataset released by Yan et al.30, and achieved the state-of-the-art results. Our method therefore could be a potentially useful tool for pathologists working on cancer.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Ferlay, J. et al. Cancer statistics for the year 2020: An overview. Int. J. Cancer 1, 1. https://doi.org/10.1002/ijc.33588 (2021).
Sari, C. T. & Gunduz-Demir, C. Unsupervised feature extraction via deep learning for histopathological classification of colon tissue images. IEEE Trans. Med. Imaging 38, 1139–1149. https://doi.org/10.1109/TMI.2018.2879369 (2018).
Jin, X. et al. Survey on the applications of deep learning to histopathology. J. Image Graph. 25, 1982–1993 (2020).
Zhang, Z. et al. A survey on computer aided diagnosis for ocular diseases. BMC Med. Inform. Decis. Mak. 14, 1–29. https://doi.org/10.1186/1472-6947-14-80 (2014).
El-Baz, A. et al. Computer-aided diagnosis systems for lung cancer: Challenges and methodologies. Int. J. Biomed. Imaging 1–46, 2013. https://doi.org/10.1155/2013/942353 (2013).
Lee, H. & Chen, Y. P. P. Image based computer aided diagnosis system for cancer detection. Expert Syst. Appl. 42, 5356–5365. https://doi.org/10.1016/j.eswa.2015.02.005 (2015).
Huang, P. et al. Added value of computer-aided ct image features for early lung cancer diagnosis with small pulmonary nodules: A matched case-control study. Expert Syst. Appl. 286, 286–295. https://doi.org/10.1148/radiol.2017162725 (2018).
Spanhol, F. A., Oliveira, L., Petitjean, C. & Laurent, H. A dataset for breast cancer histopathological image classificationl pulmonary nodules: A matched case–control study. IEEE Trans. Biomed. Eng. 63, 1455–1462. https://doi.org/10.1109/TBME.2015.2496264 (2015).
Gupta, V. & Bhavsar, A. Breast cancer histopathological image classification: Is magnification important?. In Computer Vision & Pattern Recognition Workshops, vol. 1 769–776. https://doi.org/10.1109/CVPRW.2017.107 (2017).
Affonsoa, C., Rossia, A. L. D., Vieiraa, F. H. A. & de Leon Ferreira de Carvalho, A. C. P. Deep learning for biological image classification. Expert Syst. Appl. 85, 114–122. https://doi.org/10.1016/j.eswa.2017.05.039 (2017).
Cengil, E., Çınar, A. & Özbay, E. Image classification with caffe deep learning framework. In 2017 International Conference on Computer Science and Engineering (UBMK) 440–444 https://doi.org/10.1109/UBMK.2017.8093433 (2017).
Wang, Y., Zhang, J. & Cao, Z. & Wang, Y. A deep cnn method for underwater image enhancement. In 2017 IEEE International Conference on Image Processing (ICIP) 1382–1386. https://doi.org/10.1109/ICIP.2017.8296508 (2017).
Rawat, S., Rana, K. P. S. & Kumar, V. A novel complex-valued convolutional neural network for medical image denoising. Biomed. Signal Process. Control 69, 102859. https://doi.org/10.1016/j.bspc.2021.102859 (2021).
Shan, K., Guo, J., You, W. & Bie, R. Automatic facial expression recognition based on a deep convolutional-neural-network structure. In 2017 IEEE 15th International Conference on Software Engineering Research, Management and Applications (SERA), vol. 13 123–128. https://doi.org/10.1109/SERA.2017.7965717 (2017).
Litjens, G. et al. A survey on deep learning in medical image analysis. Med. Image Anal. 42, 60–88. https://doi.org/10.1016/j.media.2017.07.005 (2017).
Alom, M. Z., Yakopcic, C., Nasrin, M. S., Taha, T. M. & Asari, V. K. Breast cancer classification from histopathological images with inception recurrent residual convolutional neural network. J. Digit. Imaging 32, 605–617. https://doi.org/10.1007/s10278-019-00182-7 (2019).
Budak, Ü., Cömert, Z., Rashid, Z. N., Şengür, A. & Çıbuk, M. Computer-aided diagnosis system combining fcn and bi-lstm model for efficient breast cancer detection from histopathological images. Appl. Soft Comput.https://doi.org/10.1016/j.asoc.2019.105765 (2019).
Yao, H., Zhang, X., Zhou, X. & Liu, S. Parallel structure deep neural network using cnn and rnn with an attention mechanism for breast cancer histology image classification. Cancershttps://doi.org/10.3390/cancers11121901 (2019).
Kaur, K. & Mittal, S. K. Classification of mammography image with cnn-rnn based semantic features and extra tree classifier approach using lstm. Mater. Today Proc.https://doi.org/10.1016/j.matpr.2020.09.619 (2020).
Yoon, H. et al. Tumor identification in colorectal histology images using a convolutional neural network. J. Digit. Imaging 32, 131–140. https://doi.org/10.1007/s10278-018-0112-9 (2019).
Iizuka, O. et al. Deep learning models for histopathological classification of gastric and colonic epithelial tumours. Sci. Rep. 10, 1–11. https://doi.org/10.1038/s41598-020-58467-9 (2020).
Gopan, N. K., Shajee, M. & Shanthini, K. S. Classification of nuclei in colorectal cancer histology images. In National Systems Conference-2018, Vikram Sarabhai Space Centre Thiruvananthapuram (2019).
Modarres, C., Astorga, N., Droguett, E. L. & Meruane, V. Convolutional neural networks for automated damage recognition and damage type identification. Struct. Control Health Monit. 25, 1–17. https://doi.org/10.1002/stc.2230 (2018).
Dhande, G. & Shaikh, Z. Recurrent neural networks for end-to-end speech recognition: A comparative analysis. Int. J. Recent Innov. Trends Comput. Commun. 6, 88–93. https://doi.org/10.17762/IJRITCC.V6I4.1523 (2018).
Krizhevsky, A., Sutskever, I. & Hinton, G. E. Imagenet classification with deep convolutional neural networks. Commun. ACM 60, 84–90. https://doi.org/10.1145/3065386 (2017).
Simonyan, K. & Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv:1409.1556 (2014).
Hou, L. et al. Patch-based convolutional neural network for whole slide tissue image classification. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition 2424–2433. https://doi.org/10.1109/CVPR.2016.266 (2016).
Xu, B. et al. Look, investigate, and classify: A deep hybrid attention method for breast cancer classification. In 2019 IEEE 16th International Symposium on Biomedical Imaging (ISBI 2019) 914–918. https://doi.org/10.1109/ISBI.2019.8759454 (2019).
Xu, B. et al. Attention by selection: A deep selective attention approach to breast cancer classification. IEEE Trans. Med. Imaging 39, 1930–1941. https://doi.org/10.1109/TMI.2019.2962013 (2019).
Yan, R. et al. Breast cancer histopathological image classification using a hybrid deep neural network. Methods 173, 52–60. https://doi.org/10.1016/j.ymeth.2019.06.014 (2020).
Sheikh, T. S., Lee, Y. & Cho, M. Histopathological classification of breast cancer images using a multi-scale input and multi-feature network. Cancers 12, 2031. https://doi.org/10.3390/cancers12082031 (2020).
Nazeri, K., Aminpour, A., Ebrahimi, M. Two-stage convolutional neural network for breast cancer histology image classification. In International Conference Image Analysis and Recognition 717–726. https://doi.org/10.1007/978-3-319-93000-8_81 (2018).
Aresta, G. et al. Bach: Grand challenge on breast cancer histology images. Med. Image Anal. 56, 122–139. https://doi.org/10.1016/j.media.2019.05.010 (2019).
Wang, L., Ouyang, W., Wang, X. & Lu, H. Stct: Sequentially training convolutional networks for visual tracking. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, vol. 56 1373–1381. https://doi.org/10.1109/CVPR.2016.153 (2016).
Yosinski, J., Clune, J., Bengio, Y. & Lipson, H. How transferable are features in deep neural networks?. Adv. Neural Inf. Process. Syst. 27, 3320–3328 (2014).
Perez, L. & Wang, J. The effectiveness of data augmentation in image classification using deep learning. arXiv:1712.04621 (2017).
Mutasa, S., Sun, S. & Ha, R. Understanding artificial intelligence based radiology studies: What is overfitting?. Clin. Imaging 65, 96–99. https://doi.org/10.1016/j.clinimag.2020.04.025 (2020).
He, K., Zhang, X., Ren, S. & Sun, J. Deep residual learning for image recognition. In 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 770–778. https://doi.org/10.1109/CVPR.2016.90 (2016).
Kohl, M., Walz, C., Ludwig, F., Braunewell, S. & Baust, M. Assessment of breast cancer histology using densely connected convolutional networks. In International Conference Image Analysis and Recognition 903–913. https://doi.org/10.1007/978-3-319-93000-8_103 (2018).
Motlagh, M. H. et al. Breast cancer histopathological image classification: A deep learning approach. In 2018 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) 2405–2412. https://doi.org/10.1109/BIBM.2018.8621307 (2018).
Mishra, S., Sharma, L., Majhi, B. & Sa, P. K. Microscopic image classification using dct for the detection of acute lymphoblastic leukemia (all). In Proceedings of International Conference on Computer Vision and Image Processing 171–180. https://doi.org/10.1007/978-981-10-2104-6_16 (2017).
Araújo, T. et al. Classification of breast cancer histology images using convolutional neural networks. PLoS ONE 12, e0177544. https://doi.org/10.1371/journal.pone.0177544 (2017).
Rakhlin, A., Shvets, A., Iglovikov, V. & Kalinin, A. A. Deep convolutional neural networks for breast cancer histology image analysis. In International Conference Image Analysis and Recognition 737–744. https://doi.org/10.1007/978-3-319-93000-8_83 (2018).
Golatkar, A., Anand, D. & Sethi, A. Classification of breast cancer histology using deep learning. In International Conference Image Analysis and Recognition 837–844. https://doi.org/10.1007/978-3-319-93000-8_95 (2018).
Szegedy, C. et al. Going deeper with convolutions. In 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 1–9. https://doi.org/10.1109/CVPR.2015.7298594 (2015).
Acknowledgements
The authors acknowledge financial supports from Foundation of National Facility for Translational Medicine (Shanghai) (No. TMSK-2020-129), Shanghai Pujiang Program (No. 20PJ1408700), and Natural Science Foundation of Shanxi Province (No. 202103021224015).
Author information
Authors and Affiliations
Contributions
J.Y. conceived the project. L.L. conceptualization, methodology, investigation, review and editing. W. F. conceptualization, methodology, investigation, formal analysis, writing. C.C. methodology, writing—review and editing. M.L. methodology, review and editing. Y.Q. supervision, writing—review and editing. J.Y. and L.L. project administration, funding acquisition.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, L., Feng, W., Chen, C. et al. Classification of breast cancer histology images using MSMV-PFENet. Sci Rep 12, 17447 (2022). https://doi.org/10.1038/s41598-022-22358-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-22358-y
This article is cited by
-
Deep learning infers clinically relevant protein levels and drug response in breast cancer from unannotated pathology images
npj Breast Cancer (2024)
-
Workflow for phenotyping sugar beet roots by automated evaluation of cell characteristics and tissue arrangement using digital image processing
Plant Methods (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.