Abstract
Herein, we examined emissions of CH4 and the community structures of methanogenic archaea and methanotrophic bacteria in paddy soils subjected to a novel irrigation system, namely continuous sub-irrigation with treated wastewater (TWW). This system has recently been developed by our group to effectively reuse TWW for the cultivation of protein-rich rice. The results showed that, despite not using mineral fertilisers, the wastewater reuse system produced a rice yield comparable to that of a conventional cultivation practice and reduced CH4 emissions from paddy fields by 80%. Continuous sub-irrigation with TWW significantly inhibited the growth of methanogens in the lower soil layer during the reproductive stage of rice plants, which was strongly consistent with the effective CH4 mitigation, resulting in a vast reduction in the abundance of methanotrophs in the upper soil layer. The compositions of the examined microbial communities were not particularly affected by the studied cultivation practices. Overall, this study demonstrated that continuous sub-irrigation with TWW was an effective method to produce high rice yield and simultaneously reduce CH4 emissions from paddy fields, and it also highlighted the potential underlying microbial mechanisms of the greenhouse gas mitigation.
Similar content being viewed by others
Introduction
Methane (CH4) is one of the major greenhouse gasses (GHGs) associated with human activity and has contributed significantly to global warming1. Rice cultivation is one of the largest anthropogenic sources of CH4, accounting for approximately 20% of its global emissions from agriculture2. The growing demand for rice to feed the ever-increasing global population has resulted in a continuous increase in rice production, which is most likely intensified CH4 emissions. In addition, increasing rice production inevitably requires higher inputs of mineral fertilisers into paddy soils, which is considered to be the key driver contributing to the overall C footprint of paddy rice farming3,4. Furthermore, the rapidly rising amount of fertilisers used in rice cultivation causes other problems, such as water pollution, soil acidification, soil mineral depletion, and rising cost of production5. Thus, there is a critical need for sustainable rice cultivation systems that effectively mitigate CH4 emissions and reduce the use of mineral fertilisers while maintaining or improving rice productivity.
We have recently introduced an innovative irrigation system for the cultivation of paddy rice6, namely continuous sub-irrigation with treated wastewater (hereinafter, referred to as CSI). The objective of this system is to promote the recycling of nutrients from municipal wastewater treatment plants (WWTPs) in a cost-effective manner and the production of high yielding protein-rich rice without the use of mineral fertilisers6,7,8. Importantly, although it was previously claimed that irrigation with wastewater would significantly increase CH4 emissions due to the high availability of organic C in irrigation wastewater9, our recent study found that CSI could markedly reduce CH4 emissions by up to 84% compared with conventional rice cultivation6. However, the underlying mechanisms for this mitigation have not yet been thoroughly investigated.
In general, CH4 emissions from rice paddy fields are strongly affected by microbial activity in soils. CH4 is produced by methanogenic archaea (methanogens) under anaerobic conditions in the rhizosphere; however, in these conditions, a portion of the produced gas is oxidised by methanotrophic bacteria (methanotrophs) before it is released into the atmosphere10. In addition, since the metabolic activity of soil microorganisms is particularly stimulated by irrigation with wastewater11, this wastewater reuse practice has been hypothesized to change soil microbial community structures and subsequently influence the emission patterns of GHGs from paddy soils9. The emission of CH4 in relation to changes in soil microbial communities caused by various agricultural management practices, particularly chemical and organic fertilisation, has been well documented12,13,14,15. However, to the best of our knowledge, no studies have been conducted on soil microbial communities associated with CH4 emissions from rice paddy fields irrigated with wastewater, especially those using CSI, in which TWW acts as the sole source of both irrigation and fertilisation for rice plants6.
Understanding how CH4 emissions are associated with the relevant microbial communities in the newly-introduced rice cultivation system remains a fundamental question that needs to be addressed. Therefore, the present study acts as a follow-up research of our previous work6 aiming (1) to verify the impacts of CSI on CH4 emissions in comparison with that of a conventional rice cultivation, and (2) to investigate the community structures of methanogens and methanotrophs in paddy soils under the two cultivation systems and the possible dominant microbial mechanisms involved in the CSI-induced influences on CH4 emissions.
Results
Seasonal dynamics of CH4 fluxes
Daily fluxes of CH4 estimated from CSI system and a conventional cultivation practice (Control) had similar patterns (Fig. 1a). CH4 emissions were not notably affected by the two treatments during the vegetative growth phase from 0 to 56 days after transplanting (DAT) and thereafter, substantial fluctuations were observed during the reproductive stage (64 DAT onwards).
Seasonal dynamics of CH4 fluxes (a) and cumulative emission of CH4 (b) from the paddy fields as influenced by the examined treatments. The fluxes are the means of the values measured within each treatment (n = 3). The grey belt indicates the time of the MSD period, while the error bars denote the standard deviation of the means. Different letters above the error bars identify a significant difference between the two treatments (p < 0.05).
In the early growth stage, CH4 emissions from the paddy fields were negligible (< 1.37 mg CH4 m−2 h−1), regardless of the treatments. However, the gas fluxes from both Control and CSI treatments markedly increased from the start of the booting time (63 DAT) and reached their peaks of 20.5 and 4.3 mg CH4 m−2 h−1 during the flowering stage (86 and 94 DAT, respectively). Thereafter, the fluxes from Control decreased sharply to their minimum (11.1 mg CH4 m−2 h−1) at the end of the crop season, whereas those from CSI gradually reduced and remained at considerably lower levels (2.1–3.3 mg CH4 m−2 h−1) during the late growing period. Overall, CSI exhibited a seasonal average flux of 1.1 ± 1.4 mg CH4 m−2 h−1, which was markedly lower than that of Control (5.5 ± 7.2 mg CH4 m−2 h−1). Consequently, CSI significantly reduced the cumulative emissions of CH4 compared with Control (212.8 kg ha−1) by 80% (p < 0.05; Fig. 1b).
Rice yield and yield-scaled emissions of CH4
Figure 2 presents the brown rice yield and yield-scaled emissions of CH4 from the examined treatments. CSI produced a rice yield of 10.7 t ha−1, which was comparable to that (11.4 t ha−1) recorded in Control (p > 0.05; Fig. 3a), while the trend in yield-scaled CH4 emissions between the two treatments was similar to that observed in the cumulative emissions of the gas. Particularly, CSI produced 3.9 kg CH4 t−1 of yield, which was 79% lower than that of Control (18.7 kg CH4 t−1 yield; p < 0.05; Fig. 3b).
Rice yield (a) and yield-scaled emission of CH4 (b) as influenced by the examined treatments. The data are the means of the values measured within each treatment (n = 3). Error bars denote the standard deviation of the means. Different letters above the error bars identify a significant difference between the two treatments (p < 0.05).
Abundance of methanogens and methanotrophs in paddy soils
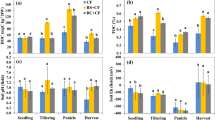
The abundance of methanogenic and methanotrophic communities varied considerably during the entire crop season (Fig. 3). Consistently in both treatments, the abundance of methanogenic archaea was markedly lower than that of methanotrophic bacteria, especially in the upper soil layer, where the loads of methanogens (3.3 × 106–1.3 × 107 gene copies g−1 soil) were vastly smaller than those of methanotrophs (1.5 × 107–8.7 × 107 gene copies g−1 soil). In addition, the target microbes generally had higher abundance in the lower soil layer than in the upper layer, regardless of the examined treatments.
Across six sampling time-points (0, 43, 52, 64, 107, and 126 DAT), CSI treatment tended to have a lower abundance of methanogens than Control. Particularly, the copy numbers of methanogenic archaeal 16S rRNA genes recorded in the upper and lower soil layers with CSI treatment, averaged 6.4 × 106 and 1.6 × 107 gene copies g−1 soil, respectively, which were 27 and 20% lower than those (8.8 × 106 and 2.0 × 107 gene copies g−1 soil) observed with Control (Fig. 3a,c). It was noteworthy that the gap in the numbers of methanogenic archaea between CSI and Control treatments were more prominent during the reproductive stage (from 64 DAT onwards, Fig. 3a,c). For methanotrophic bacteria, however, CSI immensely reduced the average abundance of methanotrophs (2.5 × 107 gene copies g−1 soil) by more than 43% relative to Control (4.4 × 107 gene copies g−1 soil) in the upper soil layer (Fig. 3b). In contrast, the methanotrophic community in the lower soil layer with CSI treatment (4.4 × 107 gene copies g−1 soil) was 18% more abundant than that (3.4 × 107 gene copies g−1 soil) with the Control (Fig. 3d). Furthermore, the pairwise correlation analyses (Table 1) showed that CH4 fluxes had a significantly positive relationship with the abundance of methanotrophic bacteria (r = 0.7, p < 0.05) while possessing no significant correlation with the methanogenic community in the upper soil layer. In the lower layer of paddy soils, however, the gas fluxes had no significant relationship with the methanotrophic community, but strongly and positively correlated (r = 0.82, p < 0.01) with the abundance of methanogenic community (Table 1). Overall, there were noticeable dynamic changes in the abundance of the soil microbial communities with CSI treatment in contrast to Control throughout the crop season.
Community composition of methanogens and methanotrophs in paddy soils
The methanogenic community consisted of 7 archaeal genera in all soil samples, as shown in Fig. 4, in which Methanolinea was irregularly found only in the lower soil layer. In the upper soil layer with CSI treatment, Methanocella and Methanobacterium were the predominant genera, accounting for 37% and 28% of the community, respectively, followed by Methanosarcina (16%), Methanosaeta (10%), and Ca. Methanoregula (8%). Methanospirillum had the lowest abundance, and it occasionally found at negligible loads (approximately 1%, Fig. 4a). This trend, with similar relative abundance of the genera, was also found with Control in the same soil layer, with the following genera (arranged in the order of dominance): Methanocella (38%), Methanobacterium (27%), Methanosarcina (16%), Methanosaeta (10%), Ca. Methanoregula (7%), and Methanospirillum (< 1%). Similarly, there was a consistency in the relative abundance of methanogenic genera between CSI and Control treatments in the lower soil layer (Fig. 4b). The three most abundant methanogens were Methanobacterium, Ca. Methanoregula, and Methanocella (approximately 21–26%), followed by Methanosaeta and Methanosarcina (14–16%), while the rest of the genera that were accounted for had negligible contributions (< 1%).
The methanotrophic community consisted of 6 bacterial genera as presented in Fig. 5, in which Ca. Methylomirabilis was detected only in the lower soil layer. Consistently in both the upper and lower soil layers, the most dominant methanotrophic bacteria were Methylosinus and Crenothrix, regardless of the treatments. In the upper soil layer, the copy numbers of Methylosinus averaged 55% and 53% for CSI and Control treatments, respectively, whereas Crenothrix had average copy numbers of 35% and 37% for the two treatments, respectively (Fig. 5a). In contrast to the upper soil layer, the growth of Crenothrix was stimulated in the lower layer, accounting for 54% and 56% of the methanotrophic communities treated with CSI and Control, respectively. The relative abundance of Methylosinus treated with CSI (31%) was comparable to that of the Control (33%; Fig. 5b). The rest of the methanotrophic genera had uniform contributions at minor proportions (< 10%) to the total methanotrophic bacterial communities, regardless of the examined cultivation practices. Thus, while no notable distinction in the microbial compositions was observed between CSI and Control, there was an apparent variation in the compositions of the identified genera between the upper and lower soil layers in both cultivation systems.
Discussion
High levels of fertilisation are expected to result in high yields of forage rice16. The rice plants used in this study were cv. Bekoaoba, which is a local forage rice that generally requires a high dose of mineral fertiliser supplementation. However, in spite of zero use of mineral fertilisers, CSI treatment still produced a comparable yield that were much higher than the target of 8 t ha−1 for the variety Bekoaoba17 and the average yield (7.3 t ha−1) obtained from conventional paddy fields in the same region7. The high yield obtained from CSI treatment was primarily attributed to the effective assimilation of the plant nutrients, especially the high concentrations of N (Table 2) derived from the irrigation TWW during the entire crop season6. This finding was in agreement with that recorded in our previous investigations on the paddy fields employing the same wastewater reuse system6,7,8. In addition, the effective mitigation of CH4 emissions under CSI was consistent with our preliminary findings6, in which CH4 emissions from a conventional cultivation practice were also reduced by 70–84%, with the use of CSI. Therefore, this follow-up study has verified the appealing advantages of CSI in rice paddy farming, i.e., producing high rice yields with reduced emissions of CH4 and simultaneously eliminating the use of mineral fertilisers.
Rice paddy fields are among the most common habitats suitable for the growth and development of methanogens that are obligate anaerobic microbes producing CH4 as an essential component of their energy metabolism10. The methanogenic genera identified in this study were those commonly found in rice paddy fields, sewage sludge, and freshwater sediments18. In contrast to methanogens, methanotrophs are classified as aerobic bacteria frequently found at the oxic-anoxic interfaces of specific environments like wetlands, aquatic sediments, and rice paddy fields, where they can oxidize 10–90% of the CH4 produced by methanogens in lower anoxic zones before the gas reaches the atmosphere10,19. The methanogenic and methanotrophic genera identified in the present study have been clearly described elsewhere10,18,19,20. The similarity on the relative abundance of these microbes under CSI and Control treatments was probably a result of the predominant flooding condition consistently in both cultivation systems, which thereby determined the soil microbial compositions in a similar fashion, despite their differences in fertilisation. This was likely due to the fact that flooding had a greater effect than nutrient loading on altering both the composition as well as the functional components of soil microbial communities21.
The explicit dissimilarity in the structures of the examined communities between the upper and lower soil layers was supported by the non-uniformed relationship between the CH4 fluxes and the abundance of methanogenic and methanotrophic communities in the two soil layers. In particular, the significantly positive correlation between the gas fluxes and the abundance of methanotrophic bacteria in the upper soil layer was most likely owing to the shallow oxic surface of the flooded soil in the upper layer that is suitable for the growth of aerobic methanotrophs but not for anaerobic methanogens10. This also explains the lower abundance of methanogens in the upper soil layer compared with the lower layer, regardless of the treatments. On the other hand, the strong and positive relationship between the CH4 fluxes and the abundance of methanogens in the lower soil layer was primarily attributed to the anoxic and reduced conditions in the deep soil layer, which is suitable for the growth and development of anaerobic methanogens, but inhibits the aerobic methanotrophic community10. Our results were in line with a previous report demonstrating that CH4 emissions were positively correlated with the transcripts of the mcrA and pmoA genes22, which were used as phylogenetic markers and a mean to calculate the abundance of methanogens and methanotrophs, respectively23. However, these results were in contrast with another study13, which claimed that CH4 fluxes were positively related to the mcrA gene copy numbers but negatively related to the pmoA gene copy numbers in paddy soils.
In the lower soil layer, the higher copy number of methanogenic 16S rRNA genes with CSI treatment during the vegetative growth period (0–52 DAT) was likely attributed to the higher input of total organic carbon (TOC) in TWW (Table 2), compared to the tap water used in Control. However, this difference did not translate into a notable variation in CH4 fluxes, which were comparable between CSI (0.09, 0.44, and − 0.004 mg CH4 m−2 h−1) and Control treatments (0.09, 1.04, and 0.37 mg CH4 m−2 h−1) across the first three times of sampling (0, 44, and 52 DAT, respectively). These CH4 fluxes were tremendously lower than those measured during the reproductive stage (around 64 DAT onwards, Fig. 1a) regardless of the treatments, which was in line with the lower and higher abundance of methanogens in the former and later growth periods, respectively. Regardless of the examined treatments, the lower CH4 emissions and the lower abundance of methanogens in the deep soil during the vegetative stage relative to those in the reproductive period were likely a result of lower availability of C substances in the soils. It was demonstrated that almost 100% of the CH4 produced in the early growth period were originated from rice straw and soil organic matter (SOM)24. In this microcosm experiment, the soils were homogenized and all plant residues were carefully removed before potting, thus leaving a relatively low content of organic matter in the soils. Later in the crop season, however, photosynthesis became a more important source for CH4 production. The high peaks recorded for CH4 fluxes from both CSI and Control treatments during the grain filling period (around 86 DAT onwards) were most likely attributed to high availability of C substances in the paddy soils. These C substances primarily originated from root exudates and decaying root debris25, which were at the highest rates during the flowering period (about 80 DAT) when compared to the other growth stages of the rice plants26. Since the photosynthetic products derived from root exudates were generally the main C substrates contributing to 65–70% of total CH4 emissions during this period24, higher root exudation in the flowering time could greatly stimulate CH4 fluxes during the following growth stage26. From 64 DAT onwards, the lower abundance of methanogens with CSI treatment relative to Control was likely a result of lower amounts of C substances available for the microbial metabolism. Consistently with our previous findings6, CSI was found to postpone the physiological senescence of rice plants as having higher levels of leaf greenness27, which was expressed as Soil Plant Analysis Development-SPAD value (Fig. 6), thereby maintaining effective photosynthetic activities of the rice plants when the maturity approached. As a result, CSI could likely reduce the rate of root exudation and decaying in the rhizosphere, which could inhibit CH4 production in the soils by limiting C substrates of the methanogenesis process28,29.
In the upper soil layer, on the other hand, the higher abundance of methanotrophs under Control treatment was probably owing to the larger amounts of CH4 produced in the deep soil compared to CSI treatment. Larger amounts of CH4 could be then diffused upwards, resulting in the stimulated methanotrophy process in the upper soil layer in Control. Overall, the CSI-induced reduction of CH4 was attributed to the inhibition of methanogenic community in the lower soil layer, while the reduced abundance of methanotrophic community under CSI was probably due to the lower CH4 production in the deep soil layer and subsequently the lower CH4 diffusion into the upper layer of the paddy soils.
In conclusion, we have demonstrated an effective method of wastewater reuse to produce high rice yields and reduce CH4 emissions while diminishing the dependence of paddy rice farming on mineral fertilisers and fresh water from conventional sources. The effective mitigation of CH4 emissions from rice paddy fields using CSI has been verified herein and the possible underlying mechanism of such the mitigation has been identified. Given that the patterns in CH4 emissions from rice paddy fields are associated with real-time variation in other soil chemical properties during the entire crop season, such as soil pH, dissolved oxygen (DO), porosity, N status, soil organic acids, and SOM15,23,30, follow-up studies are highly recommended to focus on GHG emissions linking with dynamic changes in soil microbial communities as well as other soil physicochemical properties in real-scale rice fields using CSI.
Methods
Experimental design and crop establishment
A microcosm experiment was conducted at Yamagata University, Tsuruoka City, Japan, from May to October 2019, with six growth containers (36 cm in height, 30 cm in width, 60 cm in length) to simulate paddy fields of 0.18 m2 in area (see Supplementary Fig. S1). The experiment was laid out in a completely randomized design with three replications of two treatments: (1) rice cropping under CSI and (2) conventional rice cultivation fertilized with mineral fertilisers and irrigated with tap water (Control).
Each container was filled with 32 kg of a paddy soil collected from an experimental field in the university farm and transplanted with four hills of 30-day-old seedlings (Oryza sativa L., cv. Bekoaoba) on 27th May 2019. The experiment was performed in accordance with relevant guidelines and regulations for research involving plants. The experimental soil was classified as loamy soil (air-dried, 20% moisture) with the following basic properties: pH (H2O) of 5.78, electrical conductivity (EC) of 0.09 dS m−1, SOM of 4.9%, and a total N, P, and K of 1.46, 0.88, and 3.17 g kg−1, respectively. The TWW used in the CSI system was collected from a local WWTP and monitored weekly for its basic properties (Table 2) following our previous studies6,7. In brief, pH, EC, and DO of water samples were measured on-site using pH/conductivity and DO portable meters (D-54 and OM-51, HORIBA, Ltd., Kyoto, Japan), whereas TOC and total N were analyzed using a TOC analyzer (TOC-VCSV, Shimadzu Corp., Kyoto, Japan) attached to a total N measuring unit (TNM-1, Shimadzu Corp., Kyoto, Japan). After a standard acid-digestion of water samples6, the concentration of P was measured using a portable colorimeter (DR/890, HATCH, USA), and the concentration of K was measured using an inductively coupled plasma mass spectrometry (ICP-MS ELAN DRCII, PerkinElmer Japan Co., Ltd.). The tap water used in this study was also tested on a regular basis and found to be stable throughout the crop season, with the following properties: pH of 7.8, EC of 0.095 dS m−1, DO, TOC, N, and P of 6.85, 0.49, 0.06, and 0.07 mg L−1, respectively, with K being below the ICP-MS detection limit (< 0.3 mg L−1).
The CSI system has been described thoroughly in our previous study6. Briefly, TWW was supplied to the experimental containers through underground perforated pipes from which TWW infiltrated the soil layers and eventually overflowed out of the simulated paddy fields (see Supplementary Fig. S1). The optimal irrigation rate suggested in our prior investigation was adopted as follows: a flow rate of 25 L m−2 day−1 was implemented during the active tillering and reproductive periods, and a lower rate of 8.3 L m−2 day−1 was implemented at the early tillering and late ripening stages6. Irrigation with TWW was initiated 3 DAT and maintained continuously throughout the crop season. No exogenous fertiliser was applied with CSI treatment. In the Control, rice plants were conventionally supplemented with high doses of mineral fertilisers for basal (160 kg N–P2O5–K2O ha−1) and topdressing fertilisation (100 kg N–K2O ha−1), while daily irrigation was conducted by manually adding tap water to maintain 5 cm of standing water above the soil surface. Consistent with both treatments, irrigation was withheld from 43 to 52 DAT for a mid-season drainage (MSD). The rice plants were harvested on 1st October 2019 and rice yields were assessed as the weight of brown rice at 15% moisture.
CH4 emission measurement
The static closed chamber method31 was used to sample the gas as described in our previous study6. Briefly, before the gas sampling, six transparent static chambers were mounted securely on the growth containers at the water-seal collars (see Supplementary Fig. S1). Gas samples were extracted from the chambers 0, 15, and 30 min after the chamber deployment and immediately transferred to the laboratory for CH4 measurement using a gas chromatograph (GC-2014, Shimadzu Corp., Kyoto, Japan). The gas was collected from around 10:00 a.m. to 11:00 a.m. once a week throughout the crop season, while the sampling frequency was increased to once every 2 days during the MSD period.
Daily fluxes and seasonal cumulative emissions of CH4 were calculated following the standard procedures as described previously31. In order to correlate the cumulative emission of CH4 to the rice production, the yield-scaled emission was calculated using the following equation:
where, YSE, CE and Y represents the yield-scaled emission (kg CH4 t−1 yield), the cumulative emission (kg CH4 ha−1) and the yield of brown rice (t ha−1), respectively.
Soil sampling
The paddy soils were sampled at six time-points, which were representative of different soil conditions and growth stages of the rice plants during the crop season: pre-transplantation, maximum tillering, panicle initiation, booting, grain filling, and ripening stages (0, 43, 52, 64, 107, and 126 DAT, respectively). Samples were collected in each container using a soil core (1.5 cm in diameter × 15 cm in depth) from which the upper and lower layers of the paddy soil (0–1 and 12–15 cm, respectively) were collected separately. The respective soil layers from three replicates were mixed to form one composite sample of each treatment, placed in a 50 mL commercial centrifuge tube (VWR SuperClear Ultra-High Performance Freestanding Centrifuge Tubes with Flat Caps, VWR international, USA) and then transferred to the laboratory and immediately stored at − 80 °C.
DNA extraction, PCR, and quantitative PCR assays
Soil DNA was extracted from 24 samples (2 treatments × 2 layers × 6 time points) using 0.25 g of the frozen soils as the input for the DNeasy PowerSoil Kit (QIAGEN, Hilden, Germany) following the manufacturer’s instructions. Next, DNA concentration was determined using Qubit 4 Fluorimeter (Thermo Fisher Scientific, Waltham, Massachusetts, USA) and the total DNA extracts were stored at − 80 °C until further analysis.
The 24 total DNA extracts were all used for Illumina MiSeq 16S rRNA gene sequencing. Amplicon library preparation and Illumina MiSeq sequencing were performed by Fasmac Co., Ltd. (Atsugi, Japan). The universal primers U515F (5′-ACACTCTTTCCCTACACGACGCTCTTCCGATCTGTGCCAGCMGCCGCGGTAA-3′) and U806R (5′-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTGGACTACHVGGGTWTCTAAT-3′) were used following a previous study32. They targeted the V4 hypervariable regions of the archaeal and bacterial 16S rRNA genes and were used for the first 16S rRNA gene amplification33. Operational taxonomic units (OTUs) were defined at the sequence similarity level of 97% and a representative sequence from each OTU was assigned to a taxonomic identity using the Quantitative Insights into Microbial Ecology (QIIME) software package34. Compositions of the archaeal and bacterial communities (at the genus level) in the soil samples were determined by classifying the taxa of each OTU using the Greengenes database at Fasmac Co., Ltd., Japan.
In order to evaluate the total load of archaeal and bacterial communities in the soil samples, two real-time PCR (qPCR) assays targeting the 16S rRNA genes of the archaeal and bacterial DNA were performed, respectively, using a CFX96 Touch Real-Time Detection System (Bio-Rad Laboratories, Inc. Hercules, CA, USA). The primers and probes used for the assays were previously developed35,36. The standards (102–106 gene copies) for the qPCR assays of archaea were prepared using the strain NBRC110930 Haloarchaeobius iranensis, while those for the qPCR assay of bacteria were prepared using the strain NBRC3301 Escherichia coli K12. The reaction conditions for the amplification of archaea were as follows: 95 °C for 10 min, followed by 40 cycles at 95 °C for 20 s and 60 °C for 1 min; in contrast, the conditions for bacterial amplification were as follows: 50 °C for 2 min and 95 °C for 10 min, followed by 50 cycles of 95 °C for 15 s and 60 °C for 1 min. The abundance of methanogenic and methanotrophic genera was calculated based on their proportions compared to the quantity of the total archaeal and bacterial communities in the soil samples.
Statistical analysis
Differences in CH4 emissions between the examined treatments were evaluated using Student’s t tests at a significance level of 0.05. The Pearson correlation analysis was carried out between CH4 fluxes and the abundance of methanogenic and methanotrophic communities, using the data derived from the 6 time-points across the treatments. All statistical analyses were performed using IBM SPSS Statistics 24.0.
References
Smith, K. R., Desai, M. A., Rogers, J. V. & Houghton, R. A. Joint CO2 and CH4 accountability for global warming. Proc. Natl. Acad. Sci. U. S. A. 110, E2865–E2874 (2013).
FAOSTAT. http://www.fao.org/faostat/en/#data/GT. Accessed 29 Jan 2021.
Hasler, K., Bröring, S., Omta, O. S. W. F. & Olfs, H. W. Eco-innovations in the German fertilizer supply Chain: Impact on the carbon footprint of fertilizers. Plant Soil Environ. 63, 531–544 (2017).
Xu, X., Zhang, B., Liu, Y., Xue, Y. & Di, B. Carbon footprints of rice production in five typical rice districts in China. Shengtai Xuebao Acta Ecol. Sin. 33, 227–232 (2013).
Savci, S. Investigation of effect of chemical fertilizers on environment. APCBEE Proc. 1, 287–292 (2012).
Phung, L. D. et al. Continuous sub-irrigation with treated municipal wastewater for protein-rich rice production with reduced emissions of CH4 and N2O. Sci. Rep. 10, 1–10 (2020).
Tran, L. D. et al. High yield and nutritional quality of rice for animal feed achieved by continuous irrigation with treated municipal wastewater. Paddy Water Environ. 17, 507–513 (2019).
Pham, D. D. et al. Rice cultivation without synthetic fertilizers and performance of microbial fuel cells (MFCs) under continuous irrigation with treated wastewater. Water (Switzerland) 11, 1–15 (2019).
Zou, J., Liu, S., Qin, Y., Pan, G. & Zhu, D. Sewage irrigation increased methane and nitrous oxide emissions from rice paddies in southeast China. Agric. Ecosyst. Environ. 129, 516–522 (2009).
Conrad, R. Microbial ecology of methanogens and methanotrophs. Adv. Agron. 96, 1–63 (2007).
Toze, S. Reuse of effluent water—Benefits and risks. Agric. Water Manag. 80, 147–159 (2006).
Liu, J. et al. Methane emission and soil microbial communities in early rice paddy as influenced by urea-N fertilization. Plant Soil 445, 85–100 (2019).
Kong, D. et al. Linking methane emissions to methanogenic and methanotrophic communities under different fertilization strategies in rice paddies. Geoderma 347, 233–243 (2019).
Yuan, J., Yuan, Y., Zhu, Y. & Cao, L. Effects of different fertilizers on methane emissions and methanogenic community structures in paddy rhizosphere soil. Sci. Total Environ. 627, 770–781 (2018).
Fan, X. et al. Effects of fertilization on microbial abundance and emissions of greenhouse gases (CH4 and N2O) in rice paddy fields. Ecol. Evol. 6, 1054–1063 (2016).
Riya, S. et al. Mitigation of CH4 and N2O emissions from a forage rice field fertilized with aerated liquid fraction of cattle slurry by optimizing water management and topdressing. Ecol. Eng. 75, 24–32 (2015).
Watanabe, T. et al. Nitrogen removal and power generation from treated municipal wastewater by its circulated irrigation for resource-saving rice cultivation. Water Sci. Technol. 75, 898–907 (2017).
Kim, W. & Whitman, W. B. Methanogens. Encycl. Food Microbiol. Second Ed. 2, 602–606 (2014).
Op den Camp, H. J. M. et al. Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ. Microbiol. Rep. 1, 293–306 (2009).
Stein, L. Y., Roy, R. & Dunfield, P. F. Aerobic methanotrophy and nitrification: Processes and connections. eLS https://doi.org/10.1002/9780470015902.a0022213 (2012).
Mentzer, J. L., Goodman, R. M. & Balser, T. C. Microbial response over time to hydrologic and fertilization treatments in a simulated wet prairie. Plant Soil 284, 85–100 (2006).
Lee, H. J., Kim, S. Y., Kim, P. J., Madsen, E. L. & Jeon, C. O. Methane emission and dynamics of methanotrophic and methanogenic communities in a flooded rice field ecosystem. FEMS Microbiol. Ecol. 88, 195–212 (2014).
Seo, J., Jang, I., Gebauer, G. & Kang, H. Abundance of methanogens, methanotrophic bacteria, and denitrifiers in rice paddy soils. Wetlands 34, 213–223 (2014).
Watanabe, A., Takeda, T. & Kimura, M. Evaluation of origins of CH4 carbon emitted from rice paddies. J. Geophys. Res. Atmos. 104, 23623–23629 (1999).
Lu, Y., Wassmann, R., Neue, H.-U. & Huang, C. Dynamics of dissolved organic carbon and methane emissions in a flooded rice soil. Soil Sci. Soc. Am. J. 64, 2011–2017 (2000).
Aulakh, M.S., Wassmann, R. & Rennenberg, H. Methane emissions from rice fields—quantification, role of management, and mitigation options. In Advances in Agronomy (ed. Sparks, D.L.) 70, 193–260 (Academic Press, 2001).
Ata-Ul-Karim, S. T. et al. Non-destructive assessment of plant nitrogen parameters using leaf chlorophyll measurements in rice. Front. Plant Sci. 7, 1–14 (2016).
Lu, W. F. et al. Methane emissions and mitigation options in irrigated rice fields in southeast China. Nutr. Cycl. Agroecosyst. 58, 65–73 (2000).
Lu, Y. & Conrad, R. In situ stable isotope probing of methanogenic Archaea in the rice rhizosphere. Science (80-). 309, 1088–1090 (2005).
Wang, P. X. et al. Manure amendment increased the abundance of methanogens and methanotrophs but suppressed the type I methanotrophs in rice paddies. Environ. Sci. Pollut. Res. 27, 8016–8027 (2020).
Minamikawa, K., Tokida, T., Sudo, S., Padre, A. & Yagi, K. Guidelines for Measuring CH4 and N2O Emissions from Rice Paddies by a Manually Operated Closed Chamber Method (National Institute of Agro-Environmental Sciences, 2015).
Caporaso, J. G. et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U. S. A. 108, 4516–4522 (2011).
Miya, M. et al. MiFish, a set of universal PCR primers for metabarcoding environmental DNA from fishes: Detection of more than 230 subtropical marine species. R. Soc. Open Sci. 2, 150088 (2015).
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010).
Nadkarni, M. A., Martin, F. E., Jacques, N. A. & Hunter, N. Determination of bacterial load by real-time PCR using a broad-range (universal) probe and primers set. Microbiology 148, 257–266 (2002).
Yu, Y., Lee, C., Kim, J. & Hwang, S. Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol. Bioeng. 89, 670–679 (2005).
Acknowledgements
This study was partially supported by Ministry of Land, Infrastructure, Transportation and Tourism, Japan. We also thank Bureau of Sewerage System, Tsuruoka City Government for providing treated municipal wastewater for our experiment.
Author information
Authors and Affiliations
Contributions
L.D.P. and T.W. conceived the experiment, L.D.P., M.M., M.N., and D.V.P. conducted the experiment, L.D.P. analysed the results, S.M. and F.T. provided resources, L.D.P. prepared a manuscript draft, T.W. and L.D.P. edited the manuscript. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Phung, L.D., Miyazawa, M., Pham, D.V. et al. Methane mitigation is associated with reduced abundance of methanogenic and methanotrophic communities in paddy soils continuously sub-irrigated with treated wastewater. Sci Rep 11, 7426 (2021). https://doi.org/10.1038/s41598-021-86925-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-86925-5
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.