Abstract
The correct identification of mosquito vectors is often hampered by the presence of morphologically indiscernible sibling species. The Maculipennis complex is one of these groups that include both malaria vectors of primary importance and species of low/negligible epidemiological relevance, of which distribution data in Italy are outdated. Our study was aimed at providing an updated distribution of Maculipennis complex in Northern Italy through the sampling and morphological/molecular identification of specimens from five regions. The most abundant species was Anopheles messeae (2032), followed by Anopheles maculipennis s.s. (418), Anopheles atroparvus (28) and Anopheles melanoon (13). Taking advantage of ITS2 barcoding, we were able to finely characterize tested mosquitoes, classifying all the Anopheles messeae specimens as Anopheles daciae, a taxon with debated rank to which we referred as species inquirenda (sp. inq.). The distribution of species was characterized by Ecological Niche Models (ENMs), fed by recorded points of presence. ENMs provided clues on the ecological preferences of the detected species, with An. daciae sp. inq. linked to stable breeding sites and An. maculipennis s.s. more associated to ephemeral breeding sites. We demonstrate that historical Anopheles malaria vectors are still present in Northern Italy.
Similar content being viewed by others
Introduction
In early 1900, after the incrimination of Anopheles mosquito as a malaria vector, malariologists made big attempts to solve the puzzling phenomenon of “Anophelism without malaria”, that is, the absence of malaria in areas with an abundant presence of mosquitoes that seemed to transmit the disease in other areas1. The enigma was solved after the description of Anopheles maculipennis sensu lato (s.l.) as a species complex (i.e., the Maculipennis complex) which includes sibling species indistinguishable as imago, but differing in distribution, behaviour, ecology and in vectorial capacity for malaria1. Falleroni2 was the first to observe differences in the shape and exochorion patterns of eggs oviposited by engorged females. While partially overlapping between different species, for the first time this character allowed differentiation within the species complex. Attempts to find other diagnostic characters to separate the species complex were undertaken with particular attention to chaetotaxy in larvae and male genitalia. However, these characters frequently partially overlap between different species3, making morphological identification inconclusive. The molecular barcoding technique overcomes these morphological inconsistencies by sequencing evolutionary conserved loci. The internal transcribed spacer 2 (ITS2) marker proved particularly useful for discerning species in mosquito complexes4.
Two of the main malaria vectors in historical times in Italy, Anopheles labranchiae Falleroni, 1926, and Anopheles sacharovi Favre, 1903, belonged to the Maculipennis complex4. In Italy, the complex also enumerated Anopheles atroparvus Vann. Thiel, 1927, which was considered a secondary vector in Italy, yet an important vector in northern Europe5. Other species present were Anopheles maculipennis sensu stricto (s.s.) Meigen, 1818, Anopheles melanoon Hakett, 1934, Anopheles messeae Falleroni, 1926, and the presumptive species Anopheles subalpinus Hackett & Lewis, 1935, now synonymised with An. melanoon6,7. Other species in the complex were described in the Palearctic region based on cytogenetic (e.g., polytene chromosome inversions)8,9,10 and barcoding approaches11,12,13: Anopheles beklemishevi, Stegnii & Kabanova, 1976, Anopheles martinius, Shingarev, 1926, Anopheles artemievi, Gordeyev, Zvantsov, Goryacheva, Shaikevich & Yezhov, 2005, Anopheles persiensis, Linton, Sedaghat & Harbach, 2003, Anopheles daciae, Linton, Nicolescu & Harbach, 2004, although the validity of some of these taxa is still debated. For instance, the species An. messeae was tentatively split into two species (An. messeae and An. daciae)11 but the hierarchic status of these taxa within the complex is controversial. We overcame this critical issue by referring to these taxa as An. messeae s.s. and An. daciae species inquirenda (sp. inq.) and using An. messeae to mean both taxa.
Studies, which started in the 1920s, characterized the distribution of the Italian species14: An. labranchiae was present in central and southern Italy, while An. atroparvus was reported in northern regions. Anopheles messeae and An. maculipennis s.s. were recorded along the Italian peninsula, as was An. melanoon that, however, was characterized by a fragmentary presence. Anopheles sacharovi was reported in Italy but it seems to have disappeared following malaria eradication campaigns. Its last recorded date in Northern Italy dates back to 195915,16.
Malaria was endemic in several areas of Italy until the 1950s, when the disease was eradicated following a strong campaign against mosquitoes and changes in people’s lifestyles17; the WHO finally declared the country free from malaria in the late 1970s18. Where mosquito vectors are present, locally-acquired malaria cases still occur after the arrival of affected persons, as reported in Greece since 200919. Moreover, areas with relevant residual malariogenic potential are still present in Italy20 and several cases of cryptic malaria were recorded in this country from 1997 until 201721,22,23. In this scenario, the characterization of species distribution is strongly recommended to support a correct risk assessment23,24.
After the eradication of malaria, the interest in anopheline fauna characterization decreased progressively and little data were available to corroborate the historical distribution of malaria vectors in northern Italy. The first aim of this study is to fill this knowledge gap by an extensive field sampling and identifying the Maculipennis complex species present by means of the barcoding technique. Furthermore, we used obtained presence points to model the environmental suitability of the surveyed territory for the different species detected, providing a useful tool for assessing the possible health risk associated with these vectors. The Ecological Niche Models (ENM) obtained provide interesting insights about characterizing different ecological traits of the species complex.
Results
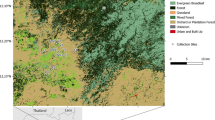
We collected more than 23,000 mosquitoes of the Maculipennis complex from the Po Valley, the widest plain in Italy. A large part of these mosquitoes was collected through attraction traps in fixed sites, which were sampled several times during the favourable season, i.e., between June and September. We used the geometric mean of Maculipennis complex mosquitoes collected per night of sampling in these sites in 2017–18 as a proxy for their abundance (Fig. 1). We molecularly characterized 2490 mosquitoes of the Maculipennis complex: 2031 An. messeae (81.6%), 418 An. maculipennis s.s. (16.8%), 28 An. atroparvus (1.1%) and 13 An. melanoon (0.5%) (Fig. 2, Table 1, Supplementary Table S1, Supplementary Figure S1). We were able to classify, through ITS2 haplotype screening, all An. messeae specimens as An. daciae sp. inq. Of the identified mosquitoes, 17 specimens were collected as larvae in five sites, 846 were collected in 101 sites with manual aspirations specifically performed to collect them, and 1,627 were sampled through attraction traps in 198 sites, mainly within the frame of West Nile Virus (WNV) surveillance plans. The majority of tested mosquitoes were collected in 2017 and 2018, 1,046 and 1,277 respectively, while 125 specimens were collected before 2017 and 42 in 2019 (details in Supplementary Table S2). While sampling effort was focused in plain area, two larvae of An. maculipennis s.s. were also collected in a site at 1225 m a.s.l.
Location of sampled sites on the Northern Italy map, with reference to the boundaries of the Pianura Padana, type of sampling and the geometric mean of mosquitoes of the Maculipennis complex collected per night in the 2017–18 seasons. Map created using QGIS 3.10 (www.qgis.org).
Proportion of different species identified in sampled sites. Abbreviations of Italian provinces (NUT3) with identified specimens are shown. The points of presence of every species are reported in Supplementary Figure S1. Map created using QGIS 3.10 (www.qgis.org); raster layers: Globe DEM (www.ngdc.noaa.gov), EU-Hydro (www.copernicus.eu).
Barcoding
We obtained 2330 ITS2 sequences (Table 2) which allowed for 93.6% of the achieved identification, confirming the discriminatory power of this marker for species complexes. The remaining 160 specimens were identified by Cytochrome C Oxidase-I (COI) (33) sequencing or by real-time PCR (127).
The ITS2 sequences obtained in this study were aligned with homologue sequences of Italian species of the complex available in GenBank and employed to obtain the phylogenetic tree in Fig. 3. The sequences were grouped into four well-supported clades with conspecific reference sequences, enabling the unambiguous classification of our specimens in the four taxa: An. maculipennis s.s., An. messeae, An. melanoon, An. atroparvus. All ITS2 sequences originated from An. messeae showed the same pattern in two diagnostic positions, with an A in position 362 and a C in position 382 (Table 2), allowing the classification of these specimens as An. daciae sp. inq.10,25.
All ITS2 sequences were referred to a specific haplotype comprising sequences with ambiguous bases (Table 2), due to the presence of unequal double peaks systematically recorded in specific positions of electropherograms (Supplementary Figure S2). We obtained a univocal haplotype, 292 base pair (bp) long (excluding the 5.8 S and 28 S motifs in the obtained amplicon sequences) for the species An. maculipennis s.s., identical to the available public sequences. We observed polymorphisms among sequences ascribable to An. daciae sp. inq. (305 bp long) An. atroparvus (307 bp) and An. melanoon (302 bp). We observed this variable pattern in three sites of An. daciae sp. inq. sequences, as previously reported10,25, in three sites of An. atroparvus and one site in An. melanoon (Table 2). The latter site was the same already reported as inter-individually polymorphic26. We did not observe other differences in obtained sequences falling into the same clade.
Ecological niche modelling
Following the screening of 148 available variables, a set of 34 were selected to be included in the Ecological Niche Models (ENM) (Table 3). As the number of capture sites for An. atroparvus and An. melanoon were scarce (eight and nine for the two species respectively, Fig. 4a), ENMs were only built for An. daciae sp. inq. and An. maculipennis s.s. Overall performance for the ENMs was good, with Area Under Curve (AUC) values of 0.80 for An. daciae sp. inq., and 0.74 for An. maculipennis s.s. The environmental suitability map suggested higher probabilities of the presence of An. daciae sp. inq. in the proximity of large breeding sites (Fig. 4): westward near the rice fields in the Piedmont and Lombardy Regions, eastward near the rice fields between the Emilia-Romagna and Veneto regions and close to wetlands. This is also confirmed by the relative contribution of the environmental variables to the ENM (Table 4), with distance to potential breeding sites (Proximity to rice fields, Proximity to water bodies < 1 km2, and Proximity to wetlands) providing a cumulative contribution of 41.4%. In addition, covariates related to elevation proved to be a relevant contribution to define the presence/absence of the species (Table 4, Altitude and Slope; cumulative contribution: 15.5%); more suitable areas had an average altitude of 49 m a.s.l. and ranged from 1 to 385 m a.s.l. Proximity to rice fields also resulted in being the covariate with the highest model gain when used alone; moreover, when the factor was omitted, the model had the lowest gain, according to jackknife test of variable importance. This indicates that the covariate provided the most important pieces of information for defining the presence of An. daciae sp. inq.
(a) Site of detection of An. atroparvus (red) and An. melanoon (green). Output of ENM obtained by Maxent for An. daciae sp. inq. (b) and An. maculipennis s.s. (c). Map created using QGIS 3.10 (www.qgis.org); raster layers: Globe DEM (www.ngdc.noaa.gov), EU-Hydro (www.copernicus.eu).
The most suitable area for An. maculipennis s.s. resulted located in the central and northern part of the Po Valley (Fig. 4). For this species, the contribution of proximity to apparent breeding sites (rice fields, wetlands and water bodies < 1 km2) was less important (cumulative contribution: 13.3%), while covariates related to altitude and slope resulted more relevant (27.2% contribution in total) (Table 3). However, soil type also showed a marked contribution in itself (9.5%), with a higher relevance of moisty soils characterized by the likely presence of surface water and a negative influence of arid soils on the model (Supplementary Figure S3). Also, factors describing the effect of vegetation (NDVI and middle-red reflectance) appeared to have an important cumulative contribution (12.9% in total) in the ENM for An. maculipennis s.s., in marked contrast to what was observed for An. daciae sp. inq. (cumulative contribution 4.9%), indicating different vegetation requirements for the two species (Table 4). In more detail, the response curve of the model to the minimum NDVI values in the study period indicates that habitats with denser vegetation (NDVI > 0.5) have a higher suitability for An. maculipennis s.s. than for An. daciae sp. inq. (Supplementary Figure S4). According to the jackknife test, the covariate with the highest model gain when used by itself for An. maculipennis s.s. was altitude, indicating that altitude alone could provide the most useful information when defining the presence of the species. In areas more suitable for An. maculipennis s.s., altitude had an average of 29 m a.s.l. (range 1–284 m a.s.l.) and was more predictive itself in foreseeing the presence of this species compared to An. daciae sp. inq. (Table 4). Furthermore, the two models showed a slightly different answer to the altitude, highlighting areas with a lower altitude as more suitable for An. maculipennis s.s. (Supplementary Figure S4). On the contrary, the greater loss of gain in a multi-variable model was observed if Proximity to water bodies < 1 km2 was omitted which, therefore, appeared to be the factor bearing the most important information that is not captured by the other covariates included in the ENM.
The contribution to the models of all employed covariates is reported in Supplementary Table S3.
Discussion
This study provides detailed data on the distribution of species of the Maculipennis complex in Northern Italy, filling the gap in the knowledge gathered over the last decades.
The use of the ITS2 marker allowed the identification of An. daciae sp. inq., An. maculipennis s.s. and the rarer An. melanoon and An. atroparvus, suggesting the disappearance of An. sacharovi in the surveyed area.
While ITS2 sequences ascribable to An. maculipennis s.s. were all identical, we recorded an unexpected variability in some positions of the ITS2 sequences in other species. The ITS2 marker is widely used for low intraspecific variability, but it could be present in hundreds of copies in a single genome4, and these copies can differ at individual level for single mutations, insertions or deletions, as extensively reported in mosquitoes29. The presence of a single mutation in the same specimen systematically caused a double peak of different intensity in a particular position of the electropherogram. We recorded these multiple peaks in three positions of An. daciae sp. inq., three positions of An. atroparvus and one position of An. melanoon. These variable bases, caused by an intra-individual polymorphism, are not reliable as diagnostic markers for definition at species level. While these particular polymorphisms were already reported in An. messeae, we reported the polymorphic sites in another two species of the Maculipennis complex, An. melanoon and An. atroparvus.
The ITS2 polymorphic sites were used to define new species. The taxon An. daciae was earlier defined through the presence of five polymorphisms in An. messeae from Romania11. As observed in our data and demonstrated by cloning25, three of these polymorphisms were intra-individual, making only the last two sites reliable for biomolecular identification. In the last two polymorphic sites, our specimens show the same haplotype, strongly suggesting that we faced a single taxonomic entity (species or form) in the study area, namely An. daciae sp. inq.
The taxonomy of An. messeae is puzzling: the existence of two taxa, named A and B chromosomal forms, was already suggested30. The identity of form A with An. daciae sp. inq. was then postulated31, but the rank of the two taxa, An. daciae/An. messeae or A/B forms, is still debated. The described intragenomic heterogeneity raised doubts on the use of ITS2 as a univocal species marker for the definition of An. daciae sp. inq.32. Besides, sequences bearing both variants in the two above-mentioned diagnostic sites were reported10,31,32, strongly suggesting some extent of interbreeding between An. daciae sp. inq. and An. messeae s.s.10, and unequivocal diagnostic morphological traits were not identified between the two taxa11. On the other hand, significantly different relative proportions of chromosomal inversions were observed between A and B forms33. In addition, a fixed chromosomal inversion (X1) was observed in An. messeae s.s., in addition to a relevant genetic distance between An. messeae s.s. and An. daciae sp. inq., mainly on the X chromosome10. We suggest that a critical review of the An. messeae status, with more robust evidences based on strong genetic and ecological data, is needed before unambiguously splitting An. messeae into two species.
Moreover, the definition of the taxa of the Maculipennis complex is a difficult task, as highlighted by the absence of clear morphological differences and the incomplete reproductive barriers among members of the complex34. This is probably due to the recent, or incipient, process of speciation, which pose several taxa of the complex in the “grey zone”35 that are difficult to characterize. Nevertheless, the definition of taxonomic relationship inside the complex is a stimulating task, because Anopheles complexes are an ideal context to define the boundaries of sibling species36,37,38,39.
The elucidation of relations inside this complex is not only a taxonomic curiosity, since the identification of a new species can have strong epidemiological implications. Ecological (as suitable breeding sites) and behavioural (as host preferences) characteristics of different species may affect the capacity to transmit pathogens, such as Plasmodium parasites5,40. The “anophelism without malaria” phenomenon demonstrated the epidemiological importance of finely characterizing species in the complex.
Part of the specimens used were obtained by exploiting existing surveillance plans, mainly targeting the WNV, also demonstrating the usefulness of these plans for tasks that are outside of the main scope. Although traps used in WNV surveillance targeting Culex mosquitoes were not expressly studied for trapping Anopheles mosquitoes, they collected a relevant number of the latter. Moreover, we regularly monitored WNV sites for two years, fortnightly over the summer season, thus obtaining comparable data on the abundance of Anopheles from monitored sites. This data demonstrated that mosquitoes of the Maculipennis complex are more abundant in areas rich in large breeding sites (such as rice fields and wetlands).
Even if a comparison with historical data is not easy, due to the different sampling strategies and identification protocols, our data suggest that An. melanoon and An. atroparvus are less widespread than in the past. Only few specimens of An. atroparvus, a historical malaria vector in northern Europe, were collected in the eastern part of the surveyed area, supporting evidence of the scarce presence of the species that seems rarer than in the past, particularly in the Po Delta area. The same scenario is described for An. melanoon, recorded in the western part of the monitored area, which seems to have reduced its diffusion when compared to the past14.
The presence of the more abundant species An. daciae sp. inq. and An. maculipennis s.s. appeared to be more likely in plain areas, as demonstrated by the importance of altitude and slope in defining environmental suitability. In particular An. maculipennis s.s. seems to favour lower altitudes, although this does not prevent its finding even at higher altitudes41. Nevertheless, the two species showed a different geographical distribution. While An. daciae sp. inq. resulted in having a higher chance of occurrence in western and eastern parts of the monitored area where large breeding sites were also reported, An. maculipennis s.s. appeared more diffused in the central section of the study area where larger breeding sites are rare and many of these are transient (Fig. 2). In fact, the two species seem to show different preferences in breeding site typologies. Anopheles daciae sp. inq. was more related to consistent breeding sites such as rice fields, wetlands and sparse water bodies. Anopheles maculipennis s.s. seems to exploit ephemeral breeding sites, as indicated by the relevance in the ENM of slope and soil types characterized by the propensity to host surface waters, then linked to the presence of transient breeding habitats. Furthermore, the results of our ecological niche models suggests that An. maculipennis s.s. might have a higher propensity to occupy areas with denser vegetation, compared to An. daciae sp. inq. These results, obtained by modelling, are consistent with the two species’ known preferences, with An. messeae preferring large water bodies with irregular water levels rich in vegetation (as ponds, ditches, natural and artificial pools), and An. maculipennis s.s. occupying small water bodies with scarce vegetation and a widely-fluctuating daily temperature41,42.
After the strong campaign of malaria eradication in the post-World War II, the anophelines not represented the target of insecticidal treatments in northern Italy. Anopheline larvae are often killed by mosquito control plans targeting annoying species exploiting the same habitats (e.g., larvicidal treatment with Bacillus thuringiensis var. israelensis in rice fields to limit Aedes caspius)43. Nevertheless, the acquired knowledge on ecological preferences could be useful for planning specific treatments. In particular characterization of breeding sites can permit to treat preferentially one species, thus offering the opportunity to select the correct active ingredient and formulation, or to use alternative approaches, e.g. monomolecular film in transient environment, water level regulation or biological control.
The presence of these potential malaria vectors highlights epidemiological importance for an understanding of their distribution, a fundamental prerequisite to evaluate the risk of malaria introduction via Plasmodium carriers. The distribution of potential vectors differed from the past and is constantly changing according to ecological mutations, such as land use modifications and climatic change. The presence of competent vectors, together with favourable conditions, can lead to the occurrence of locally-acquired malaria, as demonstrated in Italy in 199721, and more recently and impressively in Greece with 109 cases classified as locally-acquired from 2009 to 201919. A deadly malaria case that involved a child in Italy in 2017, and which successively proved to be nosocomial, aroused a great deal of public opinion23,24. Previous knowledge of the Anopheles fauna would have provided the basic data for a first rapid assessment of the case’s origin and a prompt and clear risk communication to the people.
Species of the Maculipennis complex exhibit a different vector competence for Plasmodium species. Furthermore, Plasmodium strains that can be potentially imported are completely different to those that historically circulated in the country and are no longer present. Despite further information on this aspect are needed, experimental infections and xenodiagnoses demonstrated the ability of European An. messeae, An. atroparvus, and An. sacharovi to transmit Asian Plasmodium vivax44. While European An. atroparvus, and An. labranchiae seem refractory to African strains of Plasmodium falciparum22,45,46,47,48,49.
In order to assess the risk of locally-acquired malaria cases, these fragmentary data must be corroborated by comprehensive studies to characterize the vectorial competence of local mosquitoes, particularly for new taxa, when they have been described with exotic and potentially importable strains of Plasmodium. All these data, although difficult to obtain, are necessary to improve our preparedness to face potential emergencies.
Methods
Study area
The surveyed area included the widest plain in Italy, the Po Valley (Pianura Padana or Pianura Padano-Veneta), which includes parts of the Piedmont, Lombardy, Emilia-Romagna, Veneto and Friuli Venezia Giulia regions. This area, of about 46,000 km2, enumerates more than 20 million inhabitants and is the most densely populated in Italy, with some of the largest Italian cities (Milan, Turin, Bologna, and Venice). This territory is geared to agriculture, characterized by intensive farming and animal husbandry, with few hedges, rare scattered trees and a dense irrigation network. Industrial settlements and residential areas frequently interweave this agricultural environment. Natural areas are rare, mainly represented by river borders, characterized by riparian vegetation, or protected and re-naturalized areas.
The same section of the surveyed area presents widespread presence of breeding sites suitable for Anopheles mosquitoes, such as the rice field areas (2250 km2 between eastern Piedmont and western Lombardy, 110 km2 between the Ferrara and Rovigo provinces and 40 km2 between the Mantua and Verona provinces, besides other smaller areas). These areas were endemic for malaria in the past, particularly in the eastern part bordering the Adriatic Sea, at the Po River Delta. Potential mosquito breeding sites such as restored natural areas, quarries, irrigation channels and natural and artificial wetlands are abundant and sparsely present in the Po Valley.
Entomological collection
Mosquito samples belonging to the Maculipennis complex were collected mainly in 2017 and 2018 by active sampling or attraction traps, in addition to stored samples from previous years (Table S2). A large part of tested mosquitoes was retrieved from entomological monitoring in the frame of the WNV surveillance, in which adult mosquitoes were sampled by traps (CDC-like) baited with dry ice as a source of carbon dioxide in fixed stations of the Po Valley50. Mosquitoes derived from entomological samplings performed for other purposes were identified to a lesser extent (e.g., Leishmania infantum surveillance, nuisance monitoring), sometimes sampled by CDC-light traps (15 specimens). Sites surveyed for WNV were sampled fortnightly from June to September for two seasons. The abundance of specimens belonging to the Maculipennis complex was expressed as the geometric mean of mosquitoes in the complex per sampling (Fig. 1). To complete the picture, we actively collected larvae and adult Anopheles mosquitoes in areas where WNV traps were unsuccessful. The survey was carried out by actively collecting larvae by dipping and adult mosquitoes by direct aspiration in resting sites, from May to October. These sites included animal shelters (bovines, equines, goats and poultry) in farms to collect engorged and host-seeking mosquitoes, and uninhabited buildings to collect mosquitoes ready for overwinter. We separated Maculipennis complex specimens from other mosquitoes according to morphological keys3,42. Sites and relative methods of sampling are depicted in Fig. 1.
Barcoding and sequence analysis
We selected part of the collected specimens and analysed a maximum of 13 specimens belonging to the Maculipennis complex per sample (namely a collection made in one site in one day). We used the entire bodies or just one leg of these mosquitoes for the biomolecular analysis; in this case, the rest of the body was individually stored in a cryotube at − 80 °C.
We extracted the DNA which was submitted to traditional PCR protocol for amplification of the internal transcribed sequences 2 (ITS2) according to Marinucci et al.51; this PCR amplified the DNA encoding for ITS2 and part of ribosomal 5.8 S and 28 S rRNA subunits. The obtained amplicons were then sequenced and obtained electropherograms were screened with the Lasergene 10.0 software (DNAStar) with default setting to obtain sequences. Part of the samples were identified by sequencing of the Cytochrome C Oxidase-I (COI) marker52 or real-time PCR using An. maculipennis s.s. MP and An. messeae DMP probes53.
We defined all obtained haplotypes, also comprising ambiguous bases, and ascribed to them all the sequences obtained in a Region (Table 2). In order to prevent the incorporation of possible sequencing errors, we set the arbitrary threshold of 1% of conspecific sequences to insert ambiguous bases in one haplotype. These sequences were used in a BLAST search to obtain a homologue sequence of vouchered specimens from different countries. Only the ITS2 part of obtained sequences was aligned with the MAFFT algorithm54. We used the alignment to infer a phylogenetic tree by means of the maximum likelihood method implemented in PhyML 3.1 software55 with 1000 bootstrap replicates. The model TPM2 + G was selected according to the jModelTest2 software56. The tree was visualized with ITol software57. These representative sequences were deposited in the European Nucleotide Archive (EBI) database under accession numbers from LR898482 to LR898499.
Ecological niche modelling
We used the presence points of the detected species to model the environmental suitability of the different species in the surveyed area according to the maximum entropy approach, using MaxEnt software v3.4.158. This software estimates environmental suitability for a species departing from a set of occurrence locations and gridded covariates, maximising the entropy in geographic space or, in other terms, minimizing relative entropy between covariates59. A series of meteorological (such as temperatures and precipitations) and ecological variables (such as soil type and indexes) or covariates, were obtained from different sources (Table 3). Proximity maps were obtained by utilizing vector files for different categories of breeding sites (rice fields, wetlands, sparse water bodies under 1 km2 of surface and riverbeds).
All covariates were acquired as raster, then rescaled and aligned at 1 km2 spatial resolution in ASCII format on the surveyed area extent (WGS84 projection) by using QGIS 3.10. We defined the reference raster area by enlarging the boundaries of the plain area by 15 km, represented as the vector file already described50.
An explorative analysis was run with all covariates. Those with no contribution in the models, and thus ecologically irrelevant, were excluded from the final analysis. We screened covariates for correlation, selecting those having a Pearson’s correlation coefficient of less than 0.7. In order to avoid potential collinearity problems within the set of covariates used in the model, we assessed collinearity with the variance inflation factor (VIF) which is a measure of correlation between pairs of covariates. Correlation and collinearity analysis were performed in R version 3.5 and ENM Tools60. The set of covariates used in the models is reported in the supplemental material (Supplementary Table S3).
Ten replicates were done with the cross-validation run and the cloglog model output grid format. In order to overcome the different sampling efforts in different areas, a bias file was utilized categorizing provinces into three groups (100%, 60% and 40%) according to densities of observations.
Model performance was evaluated using the AUC value, a measure of the model’s sensitivity. AUC was used to test the model’s performance with real observations in the training area. An AUC value of 0.5 shows that the model predicts randomly, while a value close to 1 indicates optimal model performance. AUC > 0.75 was considered as a good value58.
The better covariates’ relative contributions to the MaxEnt model were estimated using percent contribution. Covariate contribution was tested by jackknife analysis in the MaxEnt model to get alternate estimates of the most important covariates in the model61. Altitude averages and ranges of areas with more suitable conditions were obtained overlapping areas with a host suitability index over 0.75 to the DEM raster. We performed a GIS analysis with QGIS 3.10 software.
References
Kettle, D.S. Medical and Veterinary Entomology 2nd edn (CAB International, 1995).
Falleroni, D. Fauna anofelica italiana e suo “habitat” (paludi, risaie, canali). Metodi di lotta contro la malaria. Riv. Malariol. 5, 553–559 (1926).
Severini, F., Toma, L., Di Luca, M. & Le, R. R. Zanzare Italiane: generalità e identificazione degli adulti (Diptera, Culicidae). Fragmenta Entomologica. 41(2), 213–372. https://doi.org/10.4081/FE.2009.92 (2009).
Beebe, N. W. DNA barcoding mosquitoes: advice for potential prospectors. Parasitology 145(5), 622–633. https://doi.org/10.1017/S0031182018000343 (2018).
Manguin, S. et al. Biodiversity of Malaria in the World (John Libbey Eurotext, 2008).
Linton, Y. M., Smith, L. & Harbach, E. Observations on the taxonomic status of Anopheles subalpinus Hackett & Lewis and An. melanoon Hacket. Eur. Mosq. Bull. 13, 1–7 (2002).
Boccolini, D., Di Luca, M., Marinucci, M. & Romi, R. Further molecular and morphological support for the formal synonymy of Anopheles subalpinus Hackett & Lewis with An. melanoon Hackett. Eur. Mosq. Bull. 16, 1–5 (2003).
Andreeva, I. V., Sibataev, A. K., Rusakova, A. M. & Stegniĭ, V. N. Morpho-cytogenetic characteristic of the mosquito Anopheles artemievi (Diptera: Culicidae), a malaria vector from the complex maculipennis. Parazitologiia. 41(5), 348–363 (2007).
Artemov, G. N. et al. A standard photomap of ovarian nurse cell chromosomes and inversion polymorphism in Anopheles beklemishevi. Parasites Vectors 11(1), 211. https://doi.org/10.1186/s13071-018-2657-3 (2018).
Naumenko, A. N. et al. Chromosome and genome divergence between the cryptic Eurasian malaria vector-species Anopheles messeae and Anopheles daciae. Genes (Basel) 11(2), 165. https://doi.org/10.3390/genes11020165 (2020).
Nicolescu, G., Linton, Y. M., Vladimirescu, A., Howard, T. M. & Harbach, R. E. Mosquitoes of the Anopheles maculipennis group (Diptera: Culicidae) in Romania, with the discovery and formal recognition of a new species based on molecular and morphological evidence. Bull. Entomol. Res. 94(6), 525–535. https://doi.org/10.1079/ber2004330 (2004).
Gordeev, M. I., Zvantsov, A. B., Goriacheva, I. I., Shaĭkevich, E. V. & Ezhov, M. N. Description of the new species Anopheles artemievi sp.n. (Diptera, Culicidae). Med. Parazitol. (Mosk). 2, 4–5 (2005).
Djadid, N. D. et al. Molecular identification of Palearctic members of Anopheles maculipennis in northern Iran. Malar. J. 6, 6. https://doi.org/10.1186/1475-2875-6-6 (2007).
Bietolini, S., Candura, F. & Coluzzi, M. Spatial and long term temporal distribution of the Anopheles maculipennis complex species in Italy. Parassitologia 48(4), 581–608 (2006).
Romi, R. et al. Status of malaria vectors in Italy. J. Med. Entomol. 34(3), 263–271. https://doi.org/10.1093/jmedent/34.3.263 (1997).
Zamburlini, R. & Cargnus, E. Residual mosquitoes in the northern Adriatic seacoast 50 years after the disappearance of malaria. Parassitologia 40, 431–437 (1998).
Gratz, N. G. Vector- and Rodent-Borne Diseases in Europe and North America: Distribution, Public Health Burden, and Control (Cambridge University Press, 2006).
Zahar, A. R. The WHO European region and the two Eastern Mediterranean Region. Applied field studies. In Vector Bionomics in the Epidemiology and Control of Malaria. Part II. WHO/VBC/90.1 (World Health Organization, 1990).
NPHO Annual Epidemiological Surveillance Report Malaria in Greece, 2019. https://eody.gov.gr/wp-content/uploads/2019/01/MALARIA_ANNUAL_REPORT_2019_ENG.pdf (2019).
Romi, R. et al. Assessment of the risk of malaria re-introduction in the Maremma plain (Central Italy) using a multi-factorial approach. Malar. J. 11, 98. https://doi.org/10.1186/1475-2875-11-98 (2012).
Baldari, M. et al. Malaria in Maremma, Italy. Lancet 351(9111), 1246–1247. https://doi.org/10.1016/S0140-6736(97)10312-9 (1998).
Romi, R., Boccolini, D., Menegon, M. & Rezza, G. Probable autochthonous introduced malaria cases in Italy in 2009–2011 and the risk of local vector-borne transmission. Euro Surveill. 17(48), 20325 (2012).
Boccolini, D. et al. Non-imported malaria in Italy: paradigmatic approaches and public health implications following an unusual cluster of cases in 2017. BMC Public Health 20(1), 857. https://doi.org/10.1186/s12889-020-08748-9 (2020).
European Centre for Disease Prevention and Control. Multiple reports of locally-acquired malaria infections in the EU—20 September 2017. (ECDC, 2017).
Lilja, T., Eklöf, D., Jaenson, T. G. T., Lindström, A. & Terenius, O. Single nucleotide polymorphism analysis of the ITS2 region of two sympatric malaria mosquito species in Sweden: Anopheles daciae and Anopheles messeae. Med. Vet. Entomol. 34(3), 364–368. https://doi.org/10.1111/mve.12436 (2020).
Di Luca, M., Boccolini, D., Marinucci, M. & Romi, R. Intrapopulation polymorphism in Anopheles messeae (An. maculipennis complex) inferred by molecular analysis. J. Med. Entomol. 41(4), 582–586. https://doi.org/10.1603/0022-2585-41.4.582 (2004).
Scharlemann, J. P. et al. Global data for ecology and epidemiology: a novel algorithm for temporal Fourier processing MODIS data. PLoS ONE 3(1), e1408. https://doi.org/10.1371/journal.pone.0001408 (2008).
Fick, S. E. & Hijmans, R. J. WorldClim 2: new 1km spatial resolution climate surfaces for global land areas. Int. J. Climatol. 37(12), 4302–4315 (2017).
Batovska, J., Cogan, N. O., Lynch, S. E. & Blacket, M. J. Using Next-Generation Sequencing for DNA Barcoding: Capturing Allelic Variation in ITS2. G3 (Bethesda) 7(1), 19–29. https://doi.org/10.1534/g3.116.036145 (2017).
Novikov, Iu. M. & Kabanova, V. M. Adaptive association of inversions in a natural population of the malaria mosquito Anopheles messeae Fall. Genetika 15(6), 1033–1045 (1979).
Vaulin, O. V. & Novikov, Y. M. Polymorphism and interspecific variability of cytochrome oxidase subunit I (COI) gene nucleotide sequence in sibling species of A and B Anopheles messeae and An. beklemishevi (Diptera: Culicidae). Russ. J. Genet. Appl. Res. 2(6), 421–429. https://doi.org/10.1134/S2079059712060159 (2012).
Bezzhonova, O. V. & Goryacheva, I. I. Intragenomic heterogeneity of rDNA internal transcribed spacer 2 in Anopheles messeae (Diptera: Culicidae). J. Med. Entomol. 45(3), 337–341. https://doi.org/10.1603/0022-2585(2008)45[337:ihorit]2.0.co;2 (2008).
Novikov, Y. M. & Shevchenko, A. I. Inversion polymorphism and the divergence of two cryptic forms of Anopheles messeae (Diptera, Culicidae) at the level of genomic DNA repeats. Russ. J. Genet. 37, 754–763 (2001).
Kitzmiller, J. B., Frizzi, G. & Baker, R. Evolution and speciation within the Maculipennis complex of the genus Anopheles. In Genetics of Insect Vectors of Disease ed. (ed. Wright, J.W. & Pal, R.) (Elsevier Publishing, 1967).
De Queiroz, K. Species concepts and species delimitation. Syst. Biol. 56(6), 879–886. https://doi.org/10.1080/10635150701701083 (2007).
Alquezar, D. E., Hemmerter, S., Cooper, R. D. & Beebe, N. W. Incomplete concerted evolution and reproductive isolation at the rDNA locus uncovers nine cryptic species within Anopheles longirostris from Papua New Guinea. BMC Evol. Biol. 10, 392. https://doi.org/10.1186/1471-2148-10-392 (2010).
Mallet, J., Besansky, N. & Hahn, M. W. How reticulated are species?. BioEssays 38(2), 140–149. https://doi.org/10.1002/bies.201500149 (2016).
Fouet, C., Kamdem, C., Gamez, S. & White, B. J. Genomic insights into adaptive divergence and speciation among malaria vectors of the Anopheles nili group. Evol Appl. 10(9), 897–906. https://doi.org/10.1111/eva.12492 (2017).
Pombi, M. et al. Dissecting functional components of reproductive isolation among closely related sympatric species of the Anopheles gambiae complex. Evol. Appl. 10(10), 1102–1120. https://doi.org/10.1111/eva.12517 (2017).
Cohuet, A., Harris, C., Robert, V. & Fontenille, D. Evolutionary forces on Anopheles: What makes a malaria vector?. Trends Parasitol. 26(3), 130–136. https://doi.org/10.1016/j.pt.2009.12.001 (2010).
Jetten, T. H., Takken, W. Anophelism Without Malaria in Europe: A Review of the Ecology and Distribution of the Genus Anopheles in Europe. (Wageningen Agricultural University, 1994).
Becker, N. et al. Mosquitoes and Their Control 2nd edn (Springer Science & Business Media, 2010).
Mosca, A., Balbo L., Grieco C. & Roberto P. Rice-field mosquito control in Northern Italy. In Proc. of 14th E-SOVE Int. Conf. 98 (2010).
Daskova, N. G. & Rasnicyn, S. P. Review of data on susceptibility of mosquitoes in the USSR to imported strains of malaria parasites. Bull. World Health Organ. 60(6), 893–897 (1982).
de Zulueta, J., Ramsdale, C. D. & Coluzzi, M. Receptivity to malaria in Europe. Bull. World Health Organ. 52(1), 109–111 (1975).
Ramsdale, C. D. & Coluzzi, M. Studies on the infectivity of tropical African strains of Plasmodium falciparum to some southern European vectors of malaria. Parassitologia 17(1–3), 39–48 (1975).
Teodorescu, C., Ungureanu, E., Mihai, M. & Tudose, M. Contributions to the study of the receptivity of the vector A. labranchiae atroparvus to two strains of P. vivax. Revista Medico-Chirurgicala din Iasi 52(1), 73–75 (1978).
Sousa, C. A. G. Malaria Vectorial Capacity and Competence of Anopheles atroparvus Van Thiel, 1927 (Diptera, Culicidae): Implications for the Potential Re-emergence of Malaria in Portugal (Thesis, Universidade Nova de Lisboa, Instituto de Higiene e Medicina Tropical, 2008).
Toty, C. et al. Malaria risk in Corsica, former hot spot of malaria in France. Malar. J. 9, 231. https://doi.org/10.1186/1475-2875-9-231 (2010).
Calzolari, M. et al. West Nile virus surveillance in 2013 via mosquito screening in Northern Italy and the influence of weather on virus circulation. PLoS ONE 10(10), e0140915. https://doi.org/10.1371/journal.pone.0140915 (2015).
Marinucci, M., Romi, R., Mancini, P., Di Luca, M. & Severini, C. Phylogenetic relationships of seven palearctic members of the maculipennis complex inferred from ITS2 sequence analysis. Insect. Mol. Biol. 8(4), 469–480. https://doi.org/10.1046/j.1365-2583.1999.00140.x (1999).
Jalali, S., Ojha, R. & Venkatesan, T. DNA barcoding for identification of agriculturally important insects. In New Horizons in Insect Science: Towards Sustainable Pest Management (ed. Chakravarthy, A. K.) (Springer, 2015).
Lühken, R. et al. Distribution of individual members of the mosquito Anopheles maculipennis complex in Germany identified by newly developed real-time PCR assays. Med. Vet. Entomol. 30, 144–154. https://doi.org/10.1111/mve.12161 (2016).
Katoh, K., Rozewicki, J. & Yamada, K. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4), 1160–1166. https://doi.org/10.1093/bib/bbx108 (2019).
Guindon, S. & Gascuel, O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52(5), 696–704. https://doi.org/10.1080/10635150390235520 (2003).
Darriba, D., Taboada, G. L., Doallo, R. & Posada, D. jModelTest 2: more models, new heuristics and parallel computing. Nat. Methods 9(8), 772. https://doi.org/10.1038/nmeth.2109 (2012).
Letunic, I. & Bork, P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucl. Acids Res. 47(W1), W256–W259. https://doi.org/10.1093/nar/gkz239 (2019).
Phillips, S. J., Anderson, R. P., Dudík, M., Schapire, R. E. & Blair, M. E. Opening the black box: an open-source release of Maxent. Ecography 40, 887–893. https://doi.org/10.1111/ecog.03049 (2017).
Elith, S. J. et al. A statistical explanation of MaxEnt for ecologists. Divers. Distrib. 17, 43–57. https://doi.org/10.1111/j.1472-4642.2010.00725.x (2011).
Warren, D. L., Glor, R. E. & Turelli, M. ENMTools: a toolbox for comparative studies of environmental niche models. Ecography 33, 607–611. https://doi.org/10.1111/j.1600-0587.2009.06142.x (2010).
Phillips, S., Anderson, R. P. & Schapire, R. E. Maximum entropy modeling of species geographic distributions. Ecol. Model. 190(3–4), 231–259. https://doi.org/10.1016/j.ecolmodel.2005.03.026 (2006).
Acknowledgements
Authors would thank all field and laboratory technicians who collaborated to this study. This work was supported by the PRC2015017 (E88C16000190001) the “Arbovirus surveillance plans” implemented by Lombardy, Emilia-Romagna, Veneto, Friuli Venezia Giulia Regions, the Piedmont Region—LR 75/95.
Author information
Authors and Affiliations
Contributions
M.C., R.D, D.L. and F.M. contributed to the conception of the study; E.C., S.C., M.B., P.R. and A.M. contributed to the design the work and carried out the entomological part; V.B., P.B., S.P., S.R. and D.T. contributed to the biomolecular analysis and results interpretation; M.C and S.C. contributed to phylogenetic analysis; A.A., M.C., and P.M. contributed to GIS analysis and interpretation of the data; M.C. drafted the manuscript, M.P. and M.D.L. have substantially revised the work. All authors contributed to final revision of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Calzolari, M., Desiato, R., Albieri, A. et al. Mosquitoes of the Maculipennis complex in Northern Italy. Sci Rep 11, 6421 (2021). https://doi.org/10.1038/s41598-021-85442-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-85442-9
This article is cited by
-
Characterization of environmental drivers influencing the abundance of Anopheles maculipennis complex in Northern Italy
Parasites & Vectors (2024)
-
Trends in mosquito species distribution modeling: insights for vector surveillance and disease control
Parasites & Vectors (2023)
-
Updated occurrence and bionomics of potential malaria vectors in Europe: a systematic review (2000–2021)
Parasites & Vectors (2022)
-
Comparative efficacy of BG-Sentinel 2 and CDC-like mosquito traps for monitoring potential malaria vectors in Europe
Parasites & Vectors (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.