Abstract
ARID1A loss-of-function mutation accompanied by a loss of ARID1A protein expression is considered one of the most important driver events in endometriosis-associated ovarian cancer. Although our recent genomic study clarified that ARID1A loss-of-function mutations were detected in 13% of ovarian endometriosis, an association between the ARID1A mutation status and ARID1A protein expression in ovarian endometriosis remains unclear. We performed immunohistochemical staining for ARID1A in 78 ovarian endometriosis samples and 99 clear cell carcinoma samples. We revealed that not only 70 endometriosis samples without ARID1A mutations but also eight endometriosis samples with ARID1A loss-of-function mutations retained ARID1A protein expression. On the other hand, most of clear cell carcinomas with ARID1A loss-of-function mutations showed a loss of ARID1A protein expression. In particular, clear cell carcinoma samples which harbor multiple ARID1A loss-of-function mutations or both a single ARID1A loss-of-function mutation and ARID1A allelic imbalance lost ARID1A protein expression. However, ARID1A protein expression was retained in seven clear cell carcinomas with ARID1A loss-of-function mutations. These results suggest that a single ARID1A loss-of-function mutation is insufficient for ARID1A loss in ovarian endometriosis and some clear cell carcinoma. Further driver events may be needed for the malignant transformation of ovarian endometriosis with ARID1A loss-of-function mutations.
Similar content being viewed by others
Introduction
The AT-rich interaction domain 1A (ARID1A) gene is located on chromosome 1p36.11 and encodes ARID1A, a key component of the SWI/SNF complex1. The SWI/SNF complex plays an important role in chromatin remodeling and is associated with numerous biological functions, such as differentiation and proliferation2. Therefore, aberrations in the SWI/SNF complex subunits have the potential to cause cancer. In particular, ARID1A, which is well known as a tumor suppressor gene, is frequently mutated in a wide variety of cancers3,4. COSMIC data demonstrated that more than half of ARID1A mutations are loss-of-function mutations, including frameshift indels mutations, and nonsense mutations, that lead to a loss of ARID1A protein expression in cancer cells5.
ARID1A mutation is considered one of the most important driver events in endometriosis-associated ovarian cancer6,7,8. According to previous studies, including ours6,7,9,10,11,12,13, 46–70% of clear cell carcinomas and 30–46% of endometrioid carcinomas harbor ARID1A mutations, and immunohistochemical analysis has demonstrated that ARID1A loss-of-function mutations are strongly correlated with the loss of ARID1A protein expression in endometriosis-associated ovarian cancer6,7. On the other hand, our recent genomic study clarified that ARID1A loss-of-function mutations are detected in 13% of ovarian endometriosis cases14. Some deep infiltrating endometriosis cases also harbor ARID1A mutations15,16. Although several previous studies demonstrated that ARID1A was expressed in endometriosis by immunohistochemical analysis17,18,19,20,21,22, the mutation status of ARID1A in endometriotic epithelial cells was not investigated. The association of ARID1A protein expression with ARID1A mutations in benign endometriosis remains unclear.
In this study, we performed immunohistochemical staining for ARID1A in ovarian endometriosis samples whose ARID1A mutation status was determined by whole-exome sequencing or target gene sequencing to clarify the correlation between ARID1A protein expression and the ARID1A mutation status in ovarian endometriosis. Additionally, we evaluated an association between ARID1A protein expression and the ARID1A mutation status in ovarian clear cell carcinomas by immunohistochemical analysis. We demonstrated that ARID1A protein expression was retained in all ovarian endometriosis samples and a small portion of ovarian clear cell carcinoma samples harboring ARID1A loss-of-function mutations.
Results
ARID1A protein expression in ovarian endometriosis
We assessed ARID1A immunoreactivity in 15 frozen section samples derived from six ovarian endometriosis patients (Fig. 1). We performed multiregional sampling from ovarian endometriosis tissues in three patients (Table 1). A representative image of ARID1A immunostaining for a multisampling case (ENDO_3) is shown in Fig. 2. Both the ARID1A wild-type region and the ARID1A mutated regions showed positive immunoreactivity for ARID1A. Immunohistochemical analysis demonstrated positive immunoreactivity for ARID1A in all eight frozen section samples harboring ARID1A loss-of-function mutations and seven frozen section samples without ARID1A mutations. Table 1 shows the mutation status of other cancer-associated genes in samples with ARID1A mutations. While mutations in oncogenes such as PIK3CA and KRAS were detected in two samples (ENDO1 and ENDO3), mutations in tumor suppressor genes such as PTEN, ATM, and TP53 were not detected in any of the 15 ovarian endometriosis samples.
Final analysis set of ovarian endometriosis in this study. We recruited 54 ovarian endometriosis patients for which whole-exome sequencing or target gene sequencing was conducted in our previous study14. Then, we collected 16 frozen tissue section samples from 7 patients with ARID1A mutations and 69 formalin-fixed paraffin-embedded (FFPE) samples from 47 patients without ARID1A mutations.
Next, we assessed ARID1A protein expression in 63 FFPE samples derived from 41 ovarian endometriosis patients without ARID1A mutations (Fig. 1). As expected, ARID1A protein expression was detected in all 63 FFPE tissue samples. In summary, all 78 endometriosis samples retained ARID1A protein expression regardless of the ARID1A mutation status.
To clarify the significance of ARID1A loss-of-function mutations in endometriosis, we compared the clinicopathological features of ovarian endometriosis patients with ARID1A loss-of-function mutations to those without ARID1A mutations (Supplementary Table 1). Interestingly, ovarian endometriosis patients with ARID1A loss-of-function mutations had a higher frequency of endometriosis lesions in bilateral ovaries (P = 0.006). There were no differences in other characteristics according to the ARID1A mutation status (Supplementary Table 1).
Correlation between ARID1A protein expression and the ARID1A mutation status in ovarian clear cell carcinoma
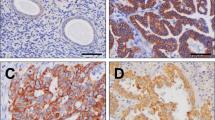
To evaluate an association between ARID1A protein expression and the ARID1A mutation status, we performed immunohistochemical analysis for 99 ovarian clear cell carcinoma samples whose ARID1A mutation status was already investigated in our previous study (Supplementary Table 2) 12. Nine clear cell carcinomas with ARID1A mutations and four clear cell carcinomas without ARID1A mutations were excluded from this analysis because of the low antigenicity or low quality of FFPE section samples (Fig. 3). Of 60 clear cell carcinoma samples with ARID1A mutations, 49 (81.7%) showed a loss of ARID1A protein expression (Table 2). Specifically, 47 of 56 samples (83.9%) with ARID1A loss-of-function mutations showed a loss of ARID1A protein expression. On the other hand, 6 of 26 samples (23.1%) without ARID1A mutations also demonstrated a loss of ARID1A protein expression. The presence of ARID1A loss-of-function mutations was significantly associated with the loss of ARID1A protein expression in clear cell carcinomas (P < 0.001) (Fig. 4A) and the representative ARID1A staining images correspond to four patterns on the basis of ARID1A mutations and ARID1A protein expression (Fig. 4B).
Final analysis set of ovarian clear cell carcinoma in this study. We enrolled 99 patients with ovarian clear cell carcinoma which were already sequenced in our previous study12. Additionally, we prepared FFPE tissue sections from 99 clear cell carcinoma cases for immunohistochemical analysis.
The association between ARID1A loss-of-function mutations and ARID1A protein expression in ovarian clear cell carcinomas. (A) The number of clear cell carcinomas with or without ARID1A protein expression and/or ARID1A mutations is shown. (B) Representative ARID1A staining images correspond to four patterns on the basis of ARID1A mutations and ARID1A protein expression. The scale bars represent 100 µm.
We examined the correlation of ARID1A loss-of-function mutations with the loss of ARID1A protein expression. Figure 5 depicts the correlation of ARID1A allelic imbalance or the number of ARID1A loss-of-function mutations with ARID1A protein expression. All samples that harbored ARID1A allelic imbalance showed a loss of ARID1A protein expression. In addition, 14 of 15 samples that harbored multiple loss-of-function mutations showed a loss of ARID1A protein expression.
Strong correlation between ARID1A allelic imbalance or the number of ARID1A loss-of-function mutations and ARID1A protein expression in clear cell carcinoma. The heatmap shows the landscape of the ARID1A mutation status, allelic imbalance, ARID1A protein expression and other cancer-associated gene mutations in each clear cell carcinoma sample.
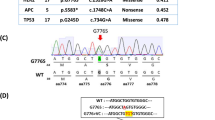
ARID1A protein expression was observed in 9 of 56 clear cell carcinoma samples (16.1%) with ARID1A loss-of-function mutations. Because we could not use serial sections for both immunohistochemical analysis in this study and target gene sequencing in the previous study, we validated the ARID1A mutation status of FFPE tissue samples in nine clear cell carcinoma samples with ARID1A truncating mutations. We macrodissected cancer cells, extracted DNA, and performed Sanger sequencing for ARID1A (Table 3). Although PCR was not successful in one sample due to poor DNA quality, we validated that seven FFPE samples harbored ARID1A loss-of-function mutations. In only one sample, the targeted ARID1A mutation was not detected by Sanger sequencing, probably because the mutation was in a subclonal state (MAF = 0.27).
To confirm concordance of the ARID1A staining level between frozen sections and FFPE samples, we prepared frozen sections and FFPE samples from the same patient. ARID1A immunohistochemical staining of frozen sections was similar to that of FFPE samples in two clear cell carcinomas and ovarian endometriosis case (Supplementary Fig. 2).
Finally, we compared the clinicopathological features of ovarian clear cell carcinoma with ARID1A loss-of-function mutations to those of ovarian clear cell carcinoma without ARID1A mutations (Supplementary Table 3). Although the optimal rate of primary debulking surgery was marginally lower in the ARID1A loss-of-function mutation group than in the ARID1A wild-type group (P = 0.052), no significant differences in any clinicopathological characteristics, including prognosis, were observed (Supplementary Fig. 3).
Discussion
Strong evidence for an association between ovarian endometriosis and ovarian clear cell and endometrioid carcinomas has been established in many studies23,24,25,26. In particular, there is epidemiological evidence that a personal history of endometriosis increases the risk of clear cell and endometrioid carcinomas27,28,29. Pathological studies have also demonstrated that atypical endometriosis merging between endometriosis and carcinoma exists in ovarian clear cell and endometrioid carcinoma cases30,31. In addition, there is accumulating molecular evidence linking endometriosis with clear cell carcinoma7,14,26. Wiegand et al. demonstrated that the loss of ARID1A caused by ARID1A loss-of-function mutations is observed in clear cell carcinoma and contiguous atypical endometriosis but not in distant endometriosis7. It is well known that ARID1A mutations are frequently detected in ovarian clear cell and endometrioid carcinomas but not in high-grade serous ovarian carcinomas7,12,28,32. These results suggest that ARID1A loss-of-function mutations are a driver event in endometriosis-associated ovarian cancer6,7. On the other hand, our recent studies clarified that cancer-associated genes such as ARID1A, PIK3CA and KRAS are frequently mutated not only in ovarian clear cell carcinoma but also in ovarian endometriosis12,14. Specifically, ARID1A loss-of-function mutations were detected in 7 of 54 ovarian endometriosis patients in our previous study14. Additionally, ovarian endometriosis samples harboring a single ARID1A loss-of-function mutation had mutations in oncogenes such as PIK3CA and KRAS and maintained benign conditions pathologically (Table 1)14. On the other hand, mutations in tumor suppressor genes, such as PTEN, ATM, and TP53, were not detected in these ovarian endometriosis samples. The significance of ARID1A mutations in the malignant transformation of ovarian endometriosis remains unclear.
Several studies have focused on ARID1A protein expression in endometriosis or ovarian cancer. Immunohistochemical analyses of ARID1A in ovarian cancer demonstrated that 0–40% of endometriosis lesions adjacent to ovarian cancer showed a loss of ARID1A protein expression, whereas all distant endometriosis lesions in ovarian cancer expressed ARID1A7,17,19,21,33. Similarly, several studies showed that ARID1A was expressed in almost all benign endometriosis lesions if ARID1A protein expression in stromal cells was correctly assessed as an internal positive control17,18,19,20,21,22. Although immunohistochemical staining and assessment protocols were not unified between studies, there was an obvious difference in ARID1A protein expression between benign endometriosis and endometriosis-associated ovarian cancer. These findings suggest that the loss of ARID1A protein expression might be an early driver event in the malignant transformation of ovarian endometriosis. However, the mutation status of ARID1A in endometriotic epithelial cells was not examined in these studies. Furthermore, the mechanism by which ARID1A protein expression is lost has not been sufficiently discussed. Wiegand et al. showed that 25% of clear cell carcinomas with loss-of-function mutations in one ARID1A allele retained ARID1A protein expression7. They also found that both mutant and wild-type alleles of ARID1A were expressed by using RNA sequencing data derived from nine clear cell carcinomas with ARID1A loss-of-function mutations. Based on these results, Wiegand et al. concluded that ARID1A could function as a haploinsufficient tumor suppressor. On the other hand, our study demonstrated that ARID1A protein expression was retained in 16% of clear cell carcinomas harboring ARID1A loss-of-function mutations (Fig. 4A). Moreover, ARID1A protein expression was retained in all benign endometriosis samples with ARID1A loss-of-function mutations. These findings are inconsistent with the concept of haploinsufficiency proposed by Wiegand et al.7. Our study also demonstrated that 14 of 15 samples that harbored multiple loss-of-function mutations showed a loss of ARID1A protein expression. In particular, all clear cell carcinoma samples harboring both ARID1A loss-of-function mutations and ARID1A allelic imbalance showed a loss of ARID1A protein expression. These findings suggest that the “two-hit” hypothesis can explain the cause of ARID1A loss in cancer cells34,35,36. In Knudson’s two-hit hypothesis34, germline mutation in tumor suppressor gene lead to a hereditary susceptibility to cancer and the inactivation of both alleles of tumor suppressor genes is essential to cause a phenotypic chance, leading to carcinogenesis. In other words, the “two-hit” hypothesis can explain why ARID1A protein expression was retained in all benign endometriosis samples with ARID1A loss-of-function mutations and a portion of clear cell carcinoma with ARID1A loss-of-function mutations. Taken together, these results suggest that the two-hit would be necessary for benign endometriosis with ARID1A heterozygous mutation to transform into malignant tumor.
Consistent with Wiegand et al.7, we also observed a portion of clear cell carcinomas without ARID1A mutations showed a loss of ARID1A protein expression, suggesting that epigenetic silencing, posttranscriptional and posttranslational regulation as well as genomic alterations might be important for the loss of ARID1A protein expression in clear cell carcinoma.
In this study, the sample size of ovarian endometriosis patients with ARID1A mutations was limited. It is necessary to assess ARID1A protein expression in ovarian endometriosis samples with ARID1A mutations in independent data sets. Although we used serial sections to assess ARID1A mutations and ARID1A protein expression in endometriosis, we could not extract DNA, RNA and protein from the same tissue simultaneously. There may be room for improvement not only in the number of samples but also in the extraction of DNA/RNA/protein for further study.
In conclusion, we clarified that ARID1A protein expression was retained in ovarian endometriosis samples harboring ARID1A loss-of-function mutations. The mechanism of ARID1A loss, which occurs specifically in endometriosis-associated ovarian cancer but not in ovarian endometriosis, is an important key for elucidating the pathogenesis of the malignant transformation of ovarian endometriosis.
Material and methods
Tissue samples
This study was performed in conformity with the Declaration of Helsinki and approved by the institutional ethics review boards of Niigata University, Niigata Chuo General Hospital, and the National Institute of Genetics. All patients provided written informed consent for the collection of samples and subsequent analyses.
We recruited 54 ovarian endometriosis patients for which whole-exome sequencing or target gene sequencing was conducted in our previous study14. We defined frameshift indels mutations, and nonsense mutations as ARID1A loss-of-function mutations. Then, we collected 16 frozen tissue section samples from seven patients with ARID1A mutations and 69 formalin-fixed paraffin-embedded (FFPE) samples from 47 patients without ARID1A mutations (Fig. 1). Frozen tissue samples were obtained from the same tissue blocks used for sequencing in our previous study14. Of these samples, one frozen tissue sample and six FFPE tissue samples were excluded from this study because there were no endometriotic epithelial cells in either the frozen tissue or FFPE sample.
We also enrolled 99 patients with ovarian clear cell carcinoma in this study to compare the association between ARID1A protein expression and the ARID1A mutation status with that in ovarian endometriosis. These clear cell carcinoma samples were already sequenced in our previous study12. Sixty-nine of 99 (69.7%) ovarian clear cell carcinoma samples harbored ARID1A mutations (Supplementary Table 2). Additionally, we prepared FFPE tissue sections from 99 clear cell carcinoma cases for immunohistochemical analysis (Fig. 3). We also prepared frozen tissue sections from two clear cell carcinomas to assess the concordance of ARID1A immunoreactivity between frozen tissue and FFPE samples in the same patient.
Hematoxylin and eosin-stained sections of all tissues used in this study were histologically reviewed by an experienced gynecologic pathologist (T.M.). All frozen tissue samples were cut from surgical specimens, embedded in Tissue-Tek O.C.T. compound (Sakura Finetek, Torrance, CA, USA) in a Tissue-Tek Cryomold (Sakura Finetek) and quickly frozen in liquid nitrogen as described in our previous study14.
Immunohistochemical staining for ARID1A protein expression
Immunohistochemical analysis of ARID1A protein expression was performed for frozen tissue section and FFPE tissue section samples. A polyclonal rabbit anti-ARID1A antibody (HPA005456, Sigma-Aldrich, St. Louis, MO, USA) was used for immunostaining as a primary antibody. Frozen tissue sections (6 µm) and FFPE tissue sections (5 µm) were cut with a cryostat and a microtome, respectively. FFPE tissue sections were stained as previously described37,38. Briefly, after deparaffinization, antigen retrieval was carried out with Target Retrieval Solution (10 mM citrate buffer, pH 6.0; Dako, Tokyo, Japan) in a microwave for 20 min at 98 °C. Subsequently, the sections were incubated with the primary antibody (1:500 dilution) overnight and biotinylated secondary antibodies (Vector Laboratories, Burlingame, CA, USA) for 1 h, followed by incubation with ABC reagent (Dako) and 3,3′-diaminobenzidine (Sigma-Aldrich) for 3 min. Slides were counterstained with hematoxylin.
We fixed frozen tissue sections with 4% paraformaldehyde at 4 °C for 20 min followed by methanol at − 20 °C for 10 min. The immunohistochemical staining protocol after fixation was the same as the protocol for FFPE tissue sections.
We assessed normal nonepithelial cells, including endothelial cells, fibroblasts, and lymphocytes, as positive internal controls. The immunostaining was decided as positive if epithelial cells showed definite nuclear staining by two investigators (Y.N. and R.T.) The distribution of the percentage of positive cells showed bimodality as a previous study39. We evaluated samples with more than 80% positive cells as ARID1A positive and samples with under 20% positive cells as ARID1A loss (Supplementary Fig. 1). Samples in which normal cells in the stroma had no immunoreactivity were defined as having low antigenicity or low quality and excluded from the subsequent analysis.
Validation of mutations by Sanger sequencing
To validate the mutation status of ovarian clear cell carcinoma FFPE samples, we prepared FFPE serial section following the one used for immunohistochemistry assay to perform Sanger sequencing per FFPE sample. We isolated tumor cells by needle macrodissection and extracted DNA using a QIAamp DNA FFPE Tissue Kit (QIAGEN Ltd., Manchester, UK) according to the manufacturer’s instructions.
We performed polymerase chain reaction (PCR) using a KAPA Taq EXtra HotStart ReadyMix PCR Kit, and the primers used are listed in Supplementary Table 4. We designed PCR primers using Primer3 software (https://bioinfo.ut.ee/primer3-0.4.0/). PCR products were purified and sequenced by GENEWIZ (Saitama, Japan).
Statistical analysis
We conducted all standard statistical tests with the R program (https://www.r-project.org). We compared categorical variables between two groups by Fisher’s exact test and continuous variables between two groups by the Wilcoxon rank-sum test. Progression-free survival (PFS) and overall survival (OS) were estimated using the Kaplan–Meier method. Deviation in the mutant allele frequency (MAF) of the somatic mutation from 0.5 was assessed by a one-sided binomial test. A P value < 0.05 was considered allelic imbalance40.
References
Chunder, N. et al. Deletion mapping of chromosome 1 in early onset and late onset breast tumors—A comparative study in eastern India. Pathol. Res. Pract. 199, 313–321. https://doi.org/10.1078/0344-0338-00423 (2003).
Wilson, B. G. & Roberts, C. W. M. SWI/SNF nucleosome remodellers and cancer. Nat. Rev. Cancer 11, 481–492. https://doi.org/10.1038/nrc3068 (2011).
Kadoch, C. et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat. Genet. 45, 592–601. https://doi.org/10.1038/ng.2628 (2013).
Lawrence, M. S. et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501. https://doi.org/10.1038/nature12912 (2014).
Tate, J. G. et al. COSMIC: The catalogue of somatic mutations in cancer. Nucleic Acids Res. 47, D941–D947. https://doi.org/10.1093/nar/gky1015 (2019).
Jones, S. et al. Frequent mutations of chromatin remodeling gene ARID1A in ovarian clear cell carcinoma. Science 330, 228–231. https://doi.org/10.1126/science.1196333 (2010).
Wiegand, K. C. et al. ARID1A mutations in endometriosis-associated ovarian carcinomas. N. Engl. J. Med. 363, 1532–1543. https://doi.org/10.1056/NEJMoa1008433 (2010).
Yamamoto, S., Tsuda, H., Takano, M., Tamai, S. & Matsubara, O. PIK3CA mutations and loss of ARID1A protein expression are early events in the development of cystic ovarian clear cell adenocarcinoma. Virchows Arch. 460, 77–87. https://doi.org/10.1007/s00428-011-1169-8 (2012).
Murakami, R. et al. Exome sequencing landscape analysis in ovarian clear cell carcinoma shed light on key chromosomal regions and mutation gene networks. Am. J. Pathol. 187, 2246–2258. https://doi.org/10.1016/j.ajpath.2017.06.012 (2017).
Shibuya, Y. et al. Identification of somatic genetic alterations in ovarian clear cell carcinoma with next generation sequencing. Genes Chromosomes Cancer 57, 51–60. https://doi.org/10.1002/gcc.22507 (2018).
Su, Y. F., Tsai, E. M., Chen, C. C., Wu, C. C. & Er, T. K. Targeted sequencing of a specific gene panel detects a high frequency of ARID1A and PIK3CA mutations in ovarian clear cell carcinoma. Clin. Chim. Acta 494, 1–7. https://doi.org/10.1016/j.cca.2019.03.003 (2019).
Sugino, K. et al. Germline and somatic mutations of homologous recombination-associated genes in Japanese ovarian cancer patients. Sci. Rep. 9, 17808. https://doi.org/10.1038/s41598-019-54116-y (2019).
Kim, S. I. et al. Genomic landscape of ovarian clear cell carcinoma via whole exome sequencing. Gynecol. Oncol. 148, 375–382. https://doi.org/10.1016/j.ygyno.2017.12.005 (2018).
Suda, K. et al. Clonal expansion and diversification of cancer-associated mutations in endometriosis and normal endometrium. Cell Rep. 24, 1777–1789. https://doi.org/10.1016/j.celrep.2018.07.037 (2018).
Anglesio, M. S. et al. Cancer-associated mutations in endometriosis without cancer. N. Engl. J. Med. 376, 1835–1848. https://doi.org/10.1056/NEJMoa1614814 (2017).
Lac, V. et al. Iatrogenic endometriosis harbors somatic cancer-driver mutations. Hum. Reprod. 34, 69–78. https://doi.org/10.1093/humrep/dey332 (2019).
Yamamoto, S., Tsuda, H., Takano, M., Tamai, S. & Matsubara, O. Loss of ARID1A protein expression occurs as an early event in ovarian clear-cell carcinoma development and frequently coexists with PIK3CA mutations. Mod. Pathol. 25, 615–624. https://doi.org/10.1038/modpathol.2011.189 (2011).
Samartzis, E. P. et al. Loss of ARID1A/BAF250a-expression in endometriosis: A biomarker for risk of carcinogenic transformation?. Mod. Pathol. 25, 885–892. https://doi.org/10.1038/modpathol.2011.217 (2012).
Ayhan, A. et al. Loss of ARID1A expression is an early molecular event in tumor progression from ovarian endometriotic cyst to clear cell and endometrioid carcinoma. Int. J. Gynecol. Cancer 22, 1310–1315. https://doi.org/10.1097/IGC.0b013e31826b5dcc (2012).
Xaio, W., Awadallah, A. & Xin, W. Loss of ARID1A/BAF250a expression in ovarian endometriosis and clear cell carcinoma. Int. J. Clin. Exp. Pathol. 5, 642–650 (2012).
Chene, G. et al. The ARID1A pathway in ovarian clear cell and endometrioid carcinoma, contiguous endometriosis, and benign endometriosis. Int. J. Gynaecol. Obstet. 130, 27–30. https://doi.org/10.1016/j.ijgo.2015.02.021 (2015).
Borrelli, G. M. et al. (Partial) Loss of BAF250a (ARID1A) in rectovaginal deep-infiltrating endometriosis, endometriomas and involved pelvic sentinel lymph nodes. Mol. Hum. Reprod. 22, 329–337. https://doi.org/10.1093/molehr/gaw009 (2016).
Van Gorp, T., Amant, F., Neven, P., Vergote, I. & Moerman, P. Endometriosis and the development of malignant tumours of the pelvis. A review of literature. Best. Pract. Res. Clin. Obstet. Gynaecol. 18, 349–371, https://doi.org/10.1016/j.bpobgyn.2003.03.001 (2004).
Prowse, A. H. et al. Molecular genetic evidence that endometriosis is a precursor of ovarian cancer. Int. J. Cancer 119, 556–562. https://doi.org/10.1002/ijc.21845 (2006).
Yamamoto, S. et al. PIK3CA mutation is an early event in the development of endometriosis-associated ovarian clear cell adenocarcinoma. J. Pathol. 225, 189–194. https://doi.org/10.1002/path.2940 (2011).
Anglesio, M. S. et al. Multifocal endometriotic lesions associated with cancer are clonal and carry a high mutation burden. J. Pathol. 236, 201–209. https://doi.org/10.1002/path.4516 (2015).
Munksgaard, P. S. & Blaakaer, J. The association between endometriosis and gynecological cancers and breast cancer: A review of epidemiological data. Gynecol. Oncol. 123, 157–163. https://doi.org/10.1016/j.ygyno.2011.06.017 (2011).
Pearce, C. L. et al. Association between endometriosis and risk of histological subtypes of ovarian cancer: A pooled analysis of case–control studies. Lancet Oncol. 13, 385–394. https://doi.org/10.1016/s1470-2045(11)70404-1 (2012).
Heidemann, L. N., Hartwell, D., Heidemann, C. H. & Jochumsen, K. M. The relation between endometriosis and ovarian cancer—A review. Acta Obstet. Gynecol. Scand. 93, 20–31. https://doi.org/10.1111/aogs.12255 (2014).
LaGrenade, A. & Silverberg, S. G. Ovarian tumors associated with atypical endometriosis. Hum. Pathol. 19, 1080–1084. https://doi.org/10.1016/s0046-8177(88)80090-x (1988).
Fukunaga, M., Nomura, K., Ishikawa, E. & Ushigome, S. Ovarian atypical endometriosis: Its close association with malignant epithelial tumours. Histopathology 30, 249–255. https://doi.org/10.1046/j.1365-2559.1997.d01-592.x (1997).
Lal, N. et al. KRAS mutation and consensus molecular subtypes 2 and 3 are independently associated with reduced immune infiltration and reactivity in colorectal cancer. Clin. Cancer Res. 24, 224–233. https://doi.org/10.1158/1078-0432.CCR-17-1090 (2018).
Stamp, J. P. et al. BAF250a expression in atypical endometriosis and endometriosis-associated ovarian cancer. Int. J. Gynecol. Cancer 26, 825–832 (2016).
Knudson, A. G. Jr. Mutation and cancer: Statistical study of retinoblastoma. Proc. Natl. Acad. Sci. U S A 68, 820–823. https://doi.org/10.1073/pnas.68.4.820 (1971).
Sato, N. et al. Loss of heterozygosity on 10q23.3 and mutation of the tumor suppressor gene PTEN in benign endometrial cyst of the ovary: possible sequence progression from benign endometrial cyst to endometrioid carcinoma and clear cell carcinoma of the ovary. Cancer Res. 60, 7052–7056 (2000).
Manderson, E. N., Presneau, N., Provencher, D., Mes-Masson, A. M. & Tonin, P. N. Comparative analysis of loss of heterozygosity of specific chromosome 3, 13, 17, and X loci and TP53 mutations in human epithelial ovarian cancer. Mol. Carcinog. 34, 78–90. https://doi.org/10.1002/mc.10051 (2002).
Yamawaki, K. et al. Sox2-dependent inhibition of p21 is associated with poor prognosis of endometrial cancer. Cancer Sci. 108, 632–640. https://doi.org/10.1111/cas.13196 (2017).
Tamura, R. et al. XCL1 expression correlates with CD8-positive T cells infiltration and PD-L1 expression in squamous cell carcinoma arising from mature cystic teratoma of the ovary. Oncogene https://doi.org/10.1038/s41388-020-1237-0 (2020).
Khalique, S. et al. Optimised ARID1A immunohistochemistry is an accurate predictor of ARID1A mutational status in gynaecological cancers. J. Pathol. Clin. Res. 4, 154–166. https://doi.org/10.1002/cjp2.103 (2018).
Buil, A. et al. Gene-gene and gene-environment interactions detected by transcriptome sequence analysis in twins. Nat. Genet. 47, 88–91. https://doi.org/10.1038/ng.3162 (2015).
Acknowledgements
We are grateful to Anna Ishida and Kenji Ohyachi for technical assistance. This work was supported in part by JSPS KAKENHI grant number JP16H06267 (Grant-in-Aid for Young Scientists A for KY), JP16H06279 (Grant-in-Aid for Scientific Research on Innovative Areas—Platforms for Advanced Technologies and Research Resources for HN and KY), JP18K16760 (Grant-in-Aid for Young Scientists for RT), JP19K18633 (Grant-in-Aid for Young Scientists for KS), and JP19K09822 (Grant-in-Aid for Scientific Research C for KY) and by "Challenging Exploratory Research Projects for the Future" grant from ROIS (Research Organization of Information and Systems) for HN.
Author information
Authors and Affiliations
Contributions
N.Y. and K.Y. designed and performed experiments, analyzed data and co-wrote the paper. K.S. performed experiments and collected samples and the data. H.N. performed bioinformatics analysis. H.U., K.S., M.Y., Y.M., K.Y. and M.I. collected samples. R.T., T.I. served as scientific advisors. M.T. reviewed stained sections histologically. I.I. and T.E. critically reviewed the study proposal.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yachida, N., Yoshihara, K., Suda, K. et al. ARID1A protein expression is retained in ovarian endometriosis with ARID1A loss-of-function mutations: implication for the two-hit hypothesis. Sci Rep 10, 14260 (2020). https://doi.org/10.1038/s41598-020-71273-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-71273-7
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.