Abstract
The common grove snail Cepaea nemoralis displays a stable pigmentation polymorphism in its shell that has held the attention of scientists for decades. While the details of the molecular mechanisms that generate and maintain this diversity remain elusive, it has long been employed as a model system to address questions related to ecology, population genetics and evolution. In order to contribute to the ongoing efforts to identify the genes that generate this polymorphism we have tested the long-standing assumption that melanin is the pigment that comprises the dark-brown bands. Surprisingly, using a newly established analytical chemical method, we find no evidence that eumelanin is differentially distributed within the shells of C. nemoralis. Furthermore, genes known to be responsible for melanin deposition in other metazoans are not differentially expressed within the shell-forming mantle tissue of C. nemoralis. These results have implications for the continuing search for the supergene that generates the various pigmentation morphotypes.
Similar content being viewed by others
Introduction
Many molluscs adorn their shells with complex and colourful geometric patterns that fulfil a variety of ecological roles. Despite considerable interest in the aesthetic beauty of these pigments (which can also be of significant commercial relevance), little is known about their chemical nature or the molecular mechanisms that generate them. With highly variable and colourful patterns of shell pigmentation the common grove snail, Cepaea nemoralis, has long been a textbook example of phenotypic variability and a model system for evolution, ecology, population genetics and even global change1,2,3,4,5. Early studies established that the shell background colour, the number of dark pigmented bands on the shell and several other shell-pigment characteristics are inherited in a Mendelian fashion6,7, suggesting that the loci for these characters are tightly clustered into what can be referred to as a “supergene”8,9,10. In addition, it has long been assumed that melanin (specifically eumelanin) is the pigment that comprises the dark pigmented bands on shells of C. nemoralis11,12,13, and that melanins are common in mollusc shells11. If melanin is indeed the pigment that comprises the dark brown bands, this could greatly inform the ongoing search for the supergene that controls this polymorphism. Here we test this assumption using two techniques: a newly developed sensitive analytical chemistry method that we recently reported for melanins14; and RT-qPCR against melanin pathway genes with reference genes specifically validated for C. nemoralis15.
Results and Discussion
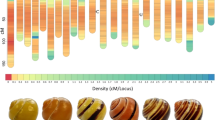
Our initial analytical chemistry experiments on whole shells of the common yellow banded morph of C. nemoralis detected all four common melanin oxidation products: pyrrole-2,3-dicarboxylic acid (PDCA) and pyrrole-2,3,5-tricarboxylic acid (PTCA) for eumelanin, and thiazole-4,5-dicarboxylic acid (TDCA) and thiazole-2,4,5-tricarboxylic acid (TTCA) for pheomelanin, albeit for eumelanin at levels significantly lower than the darkly pigmented shells of Mytillus edulis16. To further investigate the spatial distribution of eu- and pheomelanin in C. nemoralis shells we have now conducted quantitative measurements of colour-sorted shell fragments (brown-bands derived from pink background, pink background, brown-bands derived from yellow background, and yellow background; Fig. 1A). All four colour-sorted shell fractions contained the markers PDCA and PTCA (Fig. 1B), indicating a broad distribution of eumelanin, however we could not detect significant differences in the amounts of these eumelanin markers between brown-bands and background shell fractions for either yellow or pink shells (Fig. 1C,D). We also detected the pheomelanin markers TDCA and TTCA in both brown-bands derived from yellow background and yellow background shell fractions in relatively high abundance, but again with no significant difference between the two (Fig. 1D). In pink shells we detected far less TDCA in both fractions while TTCA was below the limit of quantitation but could still be detected (Table 1).
Quantification of eu- and pheomelanin markers from colour-sorted shell fragments of C. nemoralis. (A) Shell fragments used for LC-UV-MS were sorted by colour. (B) Mass spectra for all four melanin oxidation markers as measured in a representative yellow background sample (III from panel A). (C) Representative UV chromatograms used for quantitation of melanin oxidation markers. (D) Eumelanin and pheomelanin oxidation products were quantitated by HPLC-UV with external calibration. Eumelanin (PDCA and PTCA) and pheomelanin (TDCA and TTCA) oxidation markers were quantified for each shell colour fraction and normalised to initial sample weight. n = 3 for each shell colour fraction with each replicate comprised of 8 shells.
To further investigate the presence of melanin in the brown bands of C. nemoralis shells we studied the expression of four known melanin synthesis pathway genes in regions of the mantle responsible for depositing the brown bands in to the shell (Fig. 2A). We quantified gene expression via qPCR using reference genes we recently validated for C. nemoralis15. We selected the genes Yellow and Laccase 2 (known to be involved in melanin synthesis in insects17,18,19,20,21). Furthermore Miyashita and Takagi22 recently showed that a tyrosinase is involved in the subtle pigmentation of oyster pearls, while Vicario et al.13 previously suggested that either a Tyrosinase or a Tyrosinase related protein is involved in the melanic pigmentation of C. nemoralis shells. Phylogenetic analyses of the four genes that we studied are provided in the Supplementary Material (Supplementary Files 1–3).
qPCR analysis of normalised relative expression values (ddCt) of four melanin pathway genes. (A) RNA was extracted from pigmented and immediately adjacent non-pigmented mantle tissue (white boxed regions). (B) Expression levels were compared between dark-brown pigmented mantle tissue vs. non-pigmented mantle tissue, and between whole mantle vs. foot tissue. For each tissue n = 6. * indicates p ≤ 0.05, ** indicates p ≤ 0.01 as per a Mann-Whitney test.
Despite considerable variation Cnem-Yellow-like was significantly differentially expressed in pigmented vs. non-pigmented regions of the mantle (p < 0.05 but not <0.01; Fig. 2B). However Cnem-Yellow-like transcripts were significantly more abundant (approximately three fold) in foot tissue relative to mantle tissue overall. This suggests that either Cnem-Yellow-like has pleiotropic roles in the foot and mantle, or that there is another rate-limiting enzyme or process that masks the differential activity of Cnem-Yellow-like between mantle and foot tissues, or that Cnem-Yellow-like is not involved in melanogenesis in C. nemoralis. Supporting this latter scenario the Cnem-Yellow-like homolog we identified here has relatively low sequence similarity (25% identity) with insect isoforms known to be involved in melanogenesis, and does not phylogenetically group with canonical insect Yellow proteins (Supplementary File S2). Cnem-Tyrosinase related protein was significantly upregulated in mantle vs. foot, however comparisons between pigmented and non-pigmented mantle tissue revealed no significant differences in expression (Fig. 2B). In addition, Cnem-Tyrosinase and Cnem-Laccase 2 were equally expressed between foot and mantle tissues and also between pigmented and non-pigmented mantle tissue (Fig. 2). Although these qPCR results in themselves offer no support for the evidence of melanin synthesising activity in the mantle of C. nemoralis we must acknowledge that it remains possible that an inhibitory mechanism may differentially operate in pigmented and non-pigmented tissues.
While we do detect the presence of eu- and pheomelanin oxidation markers in shells of C. nemoralis, the spatially quantified LC-UV-MS and RT-qPCR data we present here suggests that eumelanin is not the dark banded pigment used by C. nemoralis. As far as we are aware the biochemical evidence that suggested melanin to be the responsible pigment in C. nemoralis was published more than 80 years ago12. Interestingly Comfort later states in regard to the banded pigment in C. nemoralis that “Attempts to attack the chemistry of the pigments with melanin inhibitors such as phenyl thiourea have not so far succeeded….” and that “Very little can be said of these colour patterns in terms of chemistry. The black pigment (‘melanocochlin’) is alkali-soluble12,23”. Nonetheless, subsequent works refer to pigmented cells in the mantle as melanocytes24 and have worked from the assumption that melanin is the brown-banded pigment. For example the upregulation of either tyrosinase or tyrosinase-related genes in C. nemoralis mantle tissue has been interpreted as evidence that melanin synthesis is occurring there13. However it is known that these enzymes also function to sclerotize the periostracum and are not necessarily involved in melanin synthesis25,26. We previously demonstrated that the brown-banded pigment in C. nemoralis is apparently not a chromoprotein27, and the results we present here indicate that a non-melanogenic mechanism is operating in the mantle tissue of C. nemoralis to deposit a dark-brown pigment in a banded fashion. A recent review on molluscan shell colour by Williams28 highlighted the lack of data that unequivocally demonstrates the presence of melanin in molluscan shells, and our own recent survey of molluscan shells with brown/black pigments suggests that in many cases these pigments are not melanin16. What then are the brown-banded and background pigments in C. nemoralis shells? Currently our LC-UV-MS analyses do not reveal any hints as to the nature of these molecules, however a macromolecular composition may be supposed.
A chemical understanding of what the dark-banded pigment and the various background pigments are, together with all of their respective synthesis pathways and associated genes, would assist efforts in identifying the supergene that is thought to regulate the famous Cepaea colour polymorphism. Such a holistic understanding of this polymorphism would provide deep insight into the evolutionary biology of this system, and would allow C. nemoralis to join the short list of species for which a polymorphic trait is understood on molecular, phenotypic, population and ecological scales.
Methods
Cepaea nemoralis
Six living animals and approximately 100 empty shells of C. nemoralis were collected at University of Göttingen, Germany (51°33′24.0″N 9°57′27.3″E). Empty shells were cleaned and dried, then crushed. Shell pieces were sorted according to replicate group and colour fraction. Tissue samples were taken from fresh material by careful dissection of mantle and foot tissue.
Sample preparation, melanin oxidation and LC-UV-MS analyses
Two major morphs of C. nemoralis (pink banded and yellow banded) were investigated. Their corresponding colour-sorted shell fragments were as follows: pink background; brown band on pink background; yellow background; brown band on yellow background (see Fig. 1A). Three technical replicates of each colour group were performed, with each replicate comprised of up to 8 shells.
Analysis of eumelanin and pheomelanin oxidation products was carried out as previously described14 (see main text): In brief, shells were cleaned in deionized water and weighted. Cleaned shell pieces were dissolved in 6 M HCl and centrifuged at 13,000 rpm for 15 min. Residues were washed with HPLC grade water. Samples were treated with proteinase K in 1 M Tris-HCl buffer at 37 °C for 2 h. Treatment was stopped by acidification with 6 M HCl, and samples were centrifuged and washed as described above.
Oxidation reactions and solid-phase extractions were performed as previously described14. For each sample, oxidation was performed for 20 h at 25 °C under vigorous shaking using 100 μL H2O, 375 μL 1 M K2CO3 and 25 μL 30% H2O2. Remaining H2O2 was decomposed by the addition of 50 μL 10% Na2SO3 and the mixture was acidified with 140 μL 6 M HCl. Solutions were then centrifuged at 13,000 rpm for 40 min and the supernatants transferred to fresh tubes. Samples were then treated by solid-phase extraction on Phenomenex Strata-X 33 μm polymeric reversed phase columns under vacuum. Columns were first conditioned with 5 mL methanol (MeOH) followed by 5 mL H2O. Shell extracts were loaded onto the columns which were then washed with 5 mL 0.3% formic acid. The columns were then dried for 30 min and elution was carried out with 3 mL MeOH followed by 3 mL ethyl acetate. Solvents were removed under constant nitrogen stream at 40 °C and samples were dissolved in 200 μL H2O.
Measurements were also performed as previously described14. Briefly, a Thermo Fisher Scientific LC-MS system consisting of an Accela HPLC with a Finnigan Surveyor PDA Detector coupled to an LTQ Orbitrap XL mass spectrometer equipped with an electrospray ionisation (ESI) source was employed. Separation was performed on a Phenomenex Gemini C18 column (250 × 2 mm, 5 μm) at 45 °C using a flow rate of 0.2 mL/min. The mobile phase was 0.3% formic acid in H2O:MeOH (80:20). UV data were recorded between 200 and 400 nm. Mass spectra were acquired in negative ion mode over an m/z range of 120–220.
Reverse transcription quantitative PCR (qPCR) of melanin pathway genes in C. nemoralis
Four genes known to be involved in melanin synthesis and dark pigmentation were chosen for qPCR testing: Cnem-Tyrosinase (Tyr), Cnem-Tyrosinase related protein (TyrRP), Cnem-Yellow-like (Yellow) and Cnem-Laccase 2 (Lacc2). These sequences were identified from within a C. nemoralis mantle tissue transcriptome data set using tBLASTx. Primers for qPCR were designed with Primer3 (https://primer3plus.com). Primer sequences and Genbank accession codes are listed in Table 2.
The experimental design for RT-qPCRs followed the protocol described in Affenzeller et al.15: Six sub-adult individuals of C. nemoralis were collected at the University of Göttingen. Total RNA from pigmented mantle (producing the band in the shell), unpigmented mantle (producing background coloured shell) and foot tissue was extracted from each individual using Qiazol (Qiagen) according to the manufacturer’s instructions resulting in a total of twelve RNA extractions. These underwent a DNase treatment (RQ1 RNase-free DNase, Promega) according to the manufacturer’s instructions. Nanodrop and agarose gel electrophoresis were employed to verify quality and integrity of RNA. Synthesis of cDNA was carried out with 1 μg of total extracted RNA per sample using Promega M-MLV reverse transcriptase and oligo dTs. Reaction was run at 42 °C for 75 min, followed by 15 min at 70 °C to inactivate reverse transcriptase. The cDNA was stored at −20 °C until further use.
All qPCR runs followed a maximum sample layout, comply with the MIQE guidelines29 and included no template controls (NTC) for each primer pair and three inter run calibrators (IRC): elongation factor 1 alpha (EF1α); RNA-directed DNA polymerase (RNAP); and Ubiquitin (UBI). Samples were run in triplicate, NTC and IRCs were run in duplicate. Amplification reactions contained 5 μL 2× Rotor-Gene SYBR Green PCR Master Mix, 0.4 μL cDNA, 1 μM final Primer concentration and 4.4 μL ddH2O to a final volume of 10 μL. Reactions were run on a Rotor-Gene Q (Qiagen) using Rotor-Gene Q software (version 2.0.2) with the following temperature profile: 5 min initial activation and denaturation at 95 °C; 45 cycles of 5 sec denaturation at 95 °C, 10 sec annealing and extension at 60 °C (data collection at this step); a final melt curve analysis from 60 °C to 95 °C at a rate of 5 sec/1 °C.
Quantification and statistical analyses
LC-UV-MS of melanin oxidation products
Quantitation of melanin oxidation products was carried out by external calibration with standard mixtures (obtained from S. Ito). External calibration was set with 9-point calibration curves. Limit of quantitation (LOQ) was set at 10:1 signal-to-noise ratio. All manual peak integrations of chromatograms and analyses of mass spectra were done in Xcalibur 2.2 Qual Browser (Thermo Scientific, Waltham, USA). Quantitation was based on areas gained from peak integrations of UV chromatograms in a range of 250–290 nm. Each colour fraction was run for three replicates (comprised of up to eight shells each). Statistical analyses (mean and standard deviation calculations) were carried out in Microsoft Excel for Office 365 MSO (16.0.11629.20192).
qPCR
Raw fluorescence data were baseline and amplification efficiency corrected in LinRegPCR30. Inter run correction was performed using Factor-qPCR31. So gained corrected cycle threshold (Cq) values were used to calculate the geometric means of technical replicates. Normalisation and relative expression were calculated based on the Pfaffl method32 with beta-actin (BACT) and EF1α serving as reference genes as previously tested for mantle tissue in C. nemoralis15.
Descriptive statistical analyses (mean and standard deviation calculations) of six biological replicates for each sample set (pigmented mantle, unpigmented mantle, foot) were carried out in Microsoft Excel for Office 365 MSO (16.0.11629.20192). Statistical comparisons between pigmented mantle and unpigmented mantle, as well as between all mantle samples and foot tissue, were run in PAST 3.15 as t-tests using Mann-Whitney as a significance measure (*p ≤ 0.05, **p ≤ 0.01).
Data availability
All data generated or analysed during this study are included in this published article or are available from the Dryad repository (https://doi.org/10.5061/dryad.gf1vhhmjs).
References
Fisher, R. A. & Diver, C. Crossing-over in the Land Snail Cepaea nemoralis, L. Nature 133, 834 (1934).
Cain, A. J. & Sheppard, P. M. Natural Selection in Cepaea. Genetics 39, 89 (1954).
Clarke, B., Diver, C. & Murray, J. Studies on Cepaea VI. The spatial and temporal distribution of pheno-types in a colony of Cepaea nemoralis (L.). Philos. Trans. R. Soc. Lond. B. Biol. Sci. 253, 519 (1968).
Cook, L. M. A two-stage model for Cepaea polymorphism. Proc. R. Soc. Lond. B. Biol. Sci. 353, 1577 (1997).
Ożgo, M. & Schilthuizen, M. Evolutionary change in Cepaea nemoralis shell colour over 43 years. Glob. Change Biol. 18, 74 (2012).
Cain, A. J., King, J. M. & Sheppard, P. M. New Data on the Genetics of Polymorphism in the Snail Cepaea Nemoralis L. Genetics 45, 393 (1960).
Cook, L. M. The genetics of Cepaea nemoralis. Heredity 22, 397 (1967).
Davison, A., Jackson, H. J., Murphy, E. W. & Reader, T. Discrete or indiscrete? Redefining the colour polymorphism of the land snail Cepaea nemoralis. Heredity 123, 162 (2019).
Gonzalez, D. R., Aramendia, A. C. & Davison, A. Recombination within the Cepaea nemoralis supergene is confounded by incomplete penetrance and epistasis. Heredity 123, 153 (2019).
Richards, P. M. et al. RAD-Seq derived markers flank the shell colour and banding loci of the Cepaea nemoralis supergene. Mol. Ecol. 22, 3077 (2013).
Comfort, A. The pigmentation of molluscan shells. Biol. Rev. 26, 285 (1951).
Helmcke, J. G. Untersuchungen über die Schalen und Köperfärbung der Landschnecken. Zoologische Jahrbücher 60, 1 (1935).
Vicario, A., Mazon, L. I., Aguirre, A., Estomba, A. & Lostao, C. A tyrosinase-like enzyme in Cepaea nemoralis and its possible relation with shell banding polymorphism. J. Moll. Stud. 55, 441 (1989).
Affenzeller, S., Frauendorf, H., Licha, T., Jackson, D. J. & Wolkenstein, K. Quantitation of eumelanin and pheomelanin markers in diverse biological samples by HPLC-UV-MS following solid-phase extraction. PLoS One 14, e0223552 (2019).
Affenzeller, S., Cerveau, N. & Jackson, D. J. Identification and validation of reference genes for qPCR in the terrestrial gastropod Cepaea nemoralis. PLoS One 13, e0201396 (2018).
Affenzeller, S., Wolkenstein, K., Frauendorf, H. & Jackson, D. J. Eumelanin and pheomelanin pigmentation in mollusc shells may be less common than expected: insights from mass spectrometry. Front. Zool. 16, 47 (2019).
Arakane, Y., Muthukrishnan, S., Beeman, R. W., Kanost, M. R. & Kramer, K. J. Laccase 2 is the phenoloxidase gene required for beetle cuticle tanning. Proc. Natl. Acad. Sci. USA 102, 11337 (2005).
Futahashi, R. & Fujiwara, H. Regulation of 20-hydroxyecdysone on the larval pigmentation and the expression of melanin synthesis enzymes and yellow gene of the swallowtail butterfly, Papilio xuthus. Insec. Biochem. Mol. Biol. 37, 855 (2007).
Wittkopp, P. J., Vaccaro, K. & Carroll, S. B. Evolution of yellow gene regulation and pigmentation in Drosophila. Curr. Biol. 12, 1547 (2002).
Jimenez-Cervantes, C. et al. A new enzymatic function in the melanogenic pathway. The 5, 6-dihydroxyindole-2-carboxylic acid oxidase activity of tyrosinase-related protein-1 (TRP1). J. Biol. Chem. 269, 17993 (1994).
Kobayashi, T. et al. Tyrosinase related protein 1 (TRP1) functions as a DHICA oxidase in melanin biosynthesis. EMBO J. 13, 5818 (1994).
Miyashita, T. & Takagi, R. Tyrosinase causes the blue shade of an abnormal pearl. J. Moll. Stud. 77, 312 (2011).
Comfort, A. Observations on the shell pigments of land pulmonates. J. Moll. Stud. 29, 35 (1951).
Emberton, L. R. B. Relationships between pigmentation of shell and of mantle in the snails Cepaea nemoralis (L.) and Cepaea hortensis (Müll). Proc. Zool. Soc. Lon. 140, 273 (1963).
Miglioli, A., Dumollard, R., Balbi, T., Besnardeau, L., Canesi, L. Characterization of the main steps in first shell formation in Mytilus galloprovincialis: possible role of tyrosinase. Proc. R. Soc. Lond. B. Biol. Sci. 286, 20192043 (2019).
Zhang, C., Xie, L., Huang, J., Chen, L. & Zhang, R. A novel putative tyrosinase involved in periostracum formation from the pearl oyster (Pinctada fucata). Biochem. Biophys. Res. Commun. 342, 632 (2006).
Mann, K. & Jackson, D. J. Characterization of the pigmented shell-forming proteome of the common grove snail Cepaea nemoralis. BMC Genomics 15, 249 (2014).
Williams, S. T. Molluscan shell colour. Biol. Rev. 92, 1039 (2017).
Bustin, S. A. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611 (2009).
Ruijter, J. M. et al. Moorman, Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Research 37(6), e45–e45 (2009).
Ruijter, J. M., Villalba, A. R., Hellemans, J., Untergasser, A. & van den Hoff, M. J. B. Removal of between-run variation in a multi-plate qPCR experiment. Biomol. Detect. Quant. 5, 10 (2015).
Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29, e45 (2001).
Acknowledgements
We are grateful to Professors Shosuke Ito and Kazumasa Wakamatsu (Department of Chemistry, Fujita Health University School of Health Sciences) for donating standard solutions for melanin oxidation products. Furthermore thanks are due to Tobias Licha for valuable input during method development and for providing equipment and technical assistance. We thank Leyla Seyfullah and Volker Thiel for productive suggestions and discussions. This research was funded by DFG grant WO 1491/4-2 to KW, and JA 2108/2-1 and JA 2108/6-1 to DJJ.
Author information
Authors and Affiliations
Contributions
S.A. conceived the study design, prepared samples and contributed to data acquisition, analysed data and prepared figures and revised the manuscript. K.W. conceived the study design, provided equipment and funding, supervised experiments and contributed to data acquisition and revised the manuscript. H.F. carried out mass spectrometric measurements and revised the manuscript. D.J.J. conceived the study design, provided equipment and funding, supervised experiments, prepared figures, drafted and revised the manuscript. All authors gave final approval for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Affenzeller, S., Wolkenstein, K., Frauendorf, H. et al. Challenging the concept that eumelanin is the polymorphic brown banded pigment in Cepaea nemoralis. Sci Rep 10, 2442 (2020). https://doi.org/10.1038/s41598-020-59185-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-59185-y
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.