Abstract
The free-living amoebae Naegleria spp. and Acanthamoeba spp. exist in the natural environment and are sometimes causal agents of lethal primary amoebic meningoencephalitis (PAM), amoebic keratitis (AK) and granulomatous amebic encephalitis (GAE) in humans, respectively. To ascertain the existence of free-living amoebae in Korea, water samples were collected from the Korean hydrosphere, Namhangang (southern Han River), an active location for water skiing and recreation. Samples underwent two-step filtration and were cultured on non-nutrient agar medium with inactivated E. coli. The remaining samples were subjected to PCR for primarily the 18S small ribosomal RNA gene and gene sequencing. Similarities in 18S rDNA sequences, in comparison with various reference amoebae in GenBank, showed 86~99% homology with N. gruberi, N. philippinensis, N. clarki, A. polyphaga, A. castellannii, and Hartmannella (Vermamoeba) vermiformis. Therefore, this study will be useful for seasonal detection of free-living amoebae from various Korean hydrospheres in future studies.
Similar content being viewed by others
Introduction
The free-living amoebae (FLA) Naegleria spp. and Acanthamoeba spp. are mainly distributed in ponds, rivers, and fresh waters worldwide. Their existing stages are trophozoite and cyst and additional biflagellate form in in case of Naegleria. Trophozoites shows moving, feeding, and proliferation activity. However, cysts are formed in poor environments, such as under abrupt temperature changes, drying, and food depletion, and can survive for long periods1,2. N. fowleri is a pathogenic agent that causes primary amoebic meningoencephalitis (PAM), which is fatal to humans and laboratory animals2,3. Acanthamoeba spp. and Balamuthia mandrillaris cause chronic granulomatous amebic encephalitis (GAE)4,5,6. Further, A. castellanii and A. polyphaga can infect the eye, resulting in acanthamoeba keratitis (AK)2,7,8.
PAM is mainly associated with activities in amoeba-contaminated water (swimming or water leisure activity), use of Neti-pots in rhinitis treatment, and religious ceremonies in some Asian countries5,9,10. The amoeba enters through the nasal cavity to invade the mucosal membrane. Subsequently, it moves into the olfactory bulb and meninges via the nasal nerve system, leading to development of meningoencephalitis11,12,13. Symptoms of PAM include acute headache, anorexia, nausea, vomiting, high fever (38–40 °C), and limb dysfunction symptoms. It also progresses acutely, with a mortality rate of over 95%. Amphotericin B is mainly used as a therapeutic agent, as a combination treatment by mixing with micronazole, rifampin, and doxycycline; however, only limited therapeutic effects have been demonstrated1,2,14,15,16.
AK usually occurs after wearing contaminated contact lenses, improper ophthalmic surgery, or corneal injury in various cases. With the popularization of contact lenses and careless lens management, the number of AK patients continues to increase2,7,17,18.
PAM due to N. fowleri occurs annually worldwide. In the United States, 143 cases of PAM occurred from 1962 to 2017, of which 80% were in males. Further, infection was most reported in adolescents19,20,21,22. In Pakistan, 22 patients in 2012 were found to be infected with N. fowleri after washing their nostrils with tap water as a religious ceremony10,23. Additional PAM patients are expected worldwide. In Korea, there has been no case of PAM caused by N. fowleri; yet, one case of GAE due to Acanthamoeba sp. has been reported24. Conversely, multiple AK patients have been identified and reported25,26,27,28.
The occurrence of PAM cases continues to increase in the tropics and subtropics, with increasing risk worldwide due to global warming. In addition, as many people enjoy water-related sports, PAM occurrence has been a social issue through the broadcast media. While it has been reported in many countries, there has not yet been reported in South Korea. There, we wanted to find out what kinds of free living amoeba, especially N. fowleri, exist in South Korea. In addition, various FLAs are expected to reside in the Korean hydrosphere. Therefore, a distribution survey was conducted to identify Naegleria and Acanthamoeba species in southern Han River, an active location for water skiing and recreation.
Materials and Methods
Water sample collection
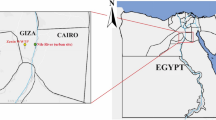
To investigate the distribution of free living amoeba in Yeoju city and Yangpyeong-gun, near southern Han River (Namhangang), which are geographically close to Seoul, we collected 1-L surface water samples with a 20-cm diameter scoop from 10 sites periodically from August 2015 to August 2016 (over 120 water samples) (Fig. 1). Sample collection sites where carries out water recreational activities were shown in Fig. 2. The atmospheric temperature at the water sampling sites was between −11 °C and 35 °C, and the water temperature ranged from 0.1 °C to 29 °C, especially 26–29 °C in August (Table 1).
Location of Namhangang River (nearby Yeoju city and Yangpyeong) in South Korea. Water sampling from Namhangang, location where water skiing and recreation are actively performed. Maps made by H-J Shin in Adobe photoshop (version 7.0.1, https://www.adobe.com/).
Water collection sites in Yeoju city (①~⑧) and Yangpyeong (⑨,⑩). The collection was repeated at the same sites for one year. Maps made by H-J Shin in Adobe photoshop (version 7.0.1, https://www.adobe.com/). Photograph by H-J Sohn.
Harvesting of FLA
To concentrate FLA from water samples, we used a two-step filtration system. The collected surface water was first filtered using a Whatman filter, followed by filtration (with a 0.4 μm pore-size bottle top filter (Thermo Fisher Scientific Inc., USA) (Fig. 3). After leaving about 30 mL of solution and thorough washing of the filter, the mixture was centrifuged at 1,500 rpm for 5 min. A portion of the pellet and filter paper were placed on a non-nutrient agar (NN-agar) plate for the amoeba culture, and the remaining pellet was subjected to PCR for amplification of the 18S RNA gene.
Free-living amoebae collection methods using two-filtration system. After the second filtration, a filter and some suspended water were cultured on NN-agar medium (B), and the remaining water pellet was subjected to PCR for 18s-rRNA gene amplification. The figure was prepared by H-J Shin using Adobe photoshop (version 7.0.1, https://www.adobe.com/).
Culturing of FLA
To culture FLA, the NN-agar plate was prepared with NN-agar medium. Briefly, 15 g of nutrient agar medium and 0.1 g of yeast extract were dissolved in 1,000 mL of distilled water. After sterilization and solidification, the agar medium was poured into 10 petri dishes. Escherichia coli was killed at 60 °C for 30 min, and then evenly coated on the surface of the prepared agar plate. The prepared specimen was dropped onto the medium, following which the culture dish was covered and incubated at 27 °C for 2–4 days. The amoebae were sub-cultured on fresh NN-agar plates two or three times. Further, N. fowleri, A. castellanii, and A. polyphaga used as positive control amoebae were cultured on Nelson’s and PYG medium according to previous reports29,30. The morphological identification of cultured amoebae was observed with an inverted microscope (Olympus CKX 31, Japan).
PCR identification of FLA
DNA was extracted from environmental sample according to the manufacturer’s protocol (Qiagen, USA). Next, molecular identification of FLA has been performed as described below.
Briefly, 2 uL of the extracted DNA was used as a template, and mixed with 10 μL of Noblezyme ™ PCR Plus Premix (Noble Bioscience Inc., Korea) in a PCR tube. Primer pairs to amplify the 18S rRNA gene of various FLAs, called pan primers (P-FLA) (2pi, Bioneer Inc., Korea), were according to previous studies31,32. The recommended amplicon sizes were as follows: Acanthamoeba spp., 1080–1500 bp; Vahlkampfia spp., 950 bp; N. fowleri, 900 bp; and Vermamoeba (Hartmannella) vermiformis, 800 bp. Amplification was performed with an initial polymerase activation step (5 min at 95 °C), followed by 35 cycles of denaturation (1 min s at 94 °C), primers hybridisation (1 min at 60 °C), and extension (3.5 min at 74 °C) in a G-Storm thermocycler (Genetechnologie, UK). All PCR experiments were performed with the inclusion of positive controls (DNA extracted from N. fowleri, A. polyphaga, and A. castellanii) to ensure correct functionality of the reaction.
After completion of the reaction, the PCR products were electrophoresed at 120 V for 20 min using a 1% agarose gel stained with ethidium bromide (0.005%) and then analyzed by Gel-doc (Bio-rad, USA).
18S rDNA sequencing and phylogenetic analysis
The FLA nucleotide sequences of the PCR-amplified 18S rRNA gene were obtained from the direct sequencing (Genotech, Daejeon, Korea), and homology against registered FLAs in GenBank was analyzed. Based on this, we conducted FLA phylogenetic analysis by estimating the neighbor-joining distance using the MEGA6 program33.
Results
Morphology of cultured FLA
FLA cultured from water samples showed a resemblance to the presumed genus Naegleria or Acanthamoeba, as trophozoites showing round pseudopodia or acanthopodia; cysts were also observed in the colony (Fig. 4).
18S rRNA gene sequence and alignment
Based on the PCR results using P-FLA primers, the main PCR reaction bands were 700–900 bp for Yeoju water samples collected throughout the year (Fig. 5). Further, a homology search of DNA sequences from 18S rRNA genes of FLA isolates indicated matches with N. gruberi (99%), N. philippinensis (99%), N. clarki (97%), A. polyphaga (98%). A. castellannii (99%), and Hartmannella (Vermamoeba) vermiformis (97%), as which one Yangpyeong sample showed the lowest homology with N. gruberi (86%), and one Yeoju sample showed the highest homology with N. clarki (99%) by sequence alignment (Figs. 6 and 7). And then, summary of results was showed in Table 1.
Phylogenetics of FLA 18S rDNA sequences
A neighbor-joining distance tree was constructed based on phylogenetic analysis of the DNA sequences from the amplified FLA 18S rRNA gene. Many Yeoju specimens clustered mainly with N. clarki and N. gruberi, whereas Yangpyeong samples were closely related to N. gruberi and N. australiensis (Fig. 8). However, one of the Yeoju samples clustered with A. castellanii and A. polyphaga (Fig. 8).
Discussion
Epidemiological studies of PAM have mainly occurred in the southeastern part of the United States, through swimming and watering activities in N. fowleri-contaminated freshwater, such as lakes and ponds, during the summer months20,34,35. Subsequently, in other countries, various survey have been conducted on ponds, lakes, rivers, swimming pools, hot springs, and contaminated sewage, which are known as FLA habitats.
In Thailand and Japan, Naegleria and Acanthamoeba species were detected in hot springs frequented by tourists and local residents36,37. In Italy, New Zealand, and California of the United States, N. fowleri was detected in a river area used for swimming and in a swimming pool38,39,40. In Taiwan, the nearest Asian country, N. fowleri was isolated from hot springs, and Acanthamoeba and Naegleria spp. from recreational water41. Acanthamoeba spp. was detected in water in neighboring China42. In Korea, Acanthamoeba spp. has been detected in amoeba-contaminated tap water in damaged water pipes and in several AK patients43,44.
We investigated only water samples where water sports or leisure activities were active, because we have the interest in public health which is caused by pathogenic free-living amoebae, especially Naegleria spp. and Acanthamoeba spp. Based on this survey on FLA distribution in the Korean hydrosphere, especially Namhangang, various species of Naegleria sand Acanthamoeba were found in Yeoju and Yangpyeong samples, especially in August. Notably, the highly virulent N. fowleri (which favors high temperatures) was not found in this survey, possibly because the temperature of the water system was inadequate (measured as 26–29 °C). A future detailed and extensive survey will be required to determine whether virulent N. fowleri inhabits this area. We conducted the survey over the course of a year to observe seasonal variations as well. However, the temperature in Korea sharply decreased in September, and the river surface water was frozen in December. As such, we could not observe FLA from September to June, although some bacteria and fungi were detected (data not shown).
Because many studies have not described the detailed process of FLA collection in an environmental water system, especially in rivers containing many floats, the culture protocol using NN-agar medium and the PCR protocol were difficult to conduct. By implementing the two-filtration system, we were able to overcome this limitation. After two filtrations, we were able to readily perform the amoeba culture and obtain PCR results. In addition, we attempted aseptic culture with Nelson’s and PYG medium, which are commonly used in Naegleria and Acanthamoeba culture protocols. However, much time and effort are required for successful aseptic culture. Several Acanthamoeba species were successfully cultured; however, no Naegleria spp. could be cultured aseptically. These results will be published in a future report.
On the results of PCR-based DNA amplification, amoebic 18S rRNA sizes amplified with PAN primer, especially Acanthamoeba species, from water samples and reference amoebae were smaller than that suggested in previous paper31,32. Although there were differences in size, sequencing results were consistent and complete in this study. This issue is worth further study. Another primer, Nfa1 and ITS primer for the amplification of Naegleria spp.29, and 18s-rDNA primer for Acanthamoeba spp.27, was applied in the preliminary experiment, but various amoebae were not amplified (data not shown). Otherwise, PAN primer amplified various species.
Considering that FLA was isolated in many countries worldwide, especially around the Korean peninsula that PAM cases or N. fowleri were detected in the natural environment in China and Japan, the survey of FLA distribution in the Korean hydrosphere was required. As suggested in this study, the discovery of pathogenic Acanthamoeba sp. in southern Han River poses a potential risk to Korea. Therefore, future studies are necessary to investigate its wide distribution in various water environments, such as rivers with frequent water activities, hot springs, and swimming pools, in Korea.
References
Ma, P. et al. Naegleria and Acanthamoeba infections: review. Rev. Infect. Dis. 12, 490–513 (1990).
Visvesvara, G. S., Moura, H. & Schuster, F. L. Pathogenic and opportunistic free-living amoebae: Acanthamoeba spp., Balamuthia mandrillaris, Naegleria fowleri, and Sappinia diploidea. FEMS Immunol. Med. Microbiol. 50, 1–26 (2007).
Carter, R. F. Description of a Naegleria sp. isolated from two cases of primary amoebic meningo-encephalitis, and of the experimental pathological changes induced by it. J. Pathol. 100, 217–244 (1970).
Schafer, K. R. et al. Disseminated Balamuthia mandrillaris Infection. J. Clin. Microbiol. 53, 3072–3076 (2015).
Schuster, F. L. & Visvesvara, G. S. Free-living amoebae as opportunistic and non-opportunistic pathogens of humans and animals. Int. J. Parasitol. 34(1001–1027), 2004.06.004 (2004).
Sutcu, M. et al. Granulomatous amebic encephalitis caused by Acanthamoeba in an immuncompetent child. Turk. J. Pediatr. 60, 340–343 (2018).
Alfonso-Munoz, E. A. et al. A report of 10 patients with Acanthamoeba keratitis. Arch. Soc. Esp. Oftalmol. 93(497-502), 2018.04.009 (2018).
Auran, J. D., Starr, M. B. & Jakobiec, F. A. Acanthamoeba keratitis. A review of the literature. Cornea 6, 2–26 (1987).
Ghanchi, N. K. et al. Case Series of Naegleria fowleri Primary Ameobic Meningoencephalitis from Karachi, Pakistan. Am. J. Trop. Med. Hyg. 97(1600–1602), 17–0110 (2017).
Siddiqui, R. & Khan, N. A. Primary amoebic meningoencephalitis caused by Naegleria fowleri: an old enemy presenting new challenges. PLoS Negl Trop Dis 8, e3017, 0003017 (2014).
Carter, R. F. Primary amoebic meningo-encephalitis. An appraisal of present knowledge. Trans. R. Soc. Trop. Med. Hyg. 66, 193–213 (1972).
Jarolim, K. L., McCosh, J. K., Howard, M. J. & John, D. T. A light microscopy study of the migration of Naegleria fowleri from the nasal submucosa to the central nervous system during the early stage of primary amebic meningoencephalitis in mice. J. Parasitol. 86, 50–55 (2000).
John, D. T. Primary amebic meningoencephalitis and the biology of Naegleria fowleri. Annu. Rev. Microbiol. 36, 101–123 (1982).
Jain, R., Prabhakar, S., Modi, M., Bhatia, R. & Sehgal, R. Naegleria meningitis: a rare survival. Neurol. India 50, 470–472 (2002).
Seidel, J. S. et al. Successful treatment of primary amebic meningoencephalitis. N. Engl. J. Med. 306, 346–348 (1982).
Wang, Q. et al. A case of Naegleria fowleri related primary amoebic meningoencephalitis in China diagnosed by next-generation sequencing. BMC Infect. Dis. 18, 349 (2018).
Fears, A. C., Metzinger, R. C., Killeen, S. Z., Reimers, R. S. & Roy, C. J. Comparative in vitro effectiveness of a novel contact lens multipurpose solution on Acanthamoeba castellanii. J. Ophthalmic Inflamm. Infect. 8, 19 (2018).
Lorenzo-Morales, J., Khan, N. A. & Walochnik, J. An update on Acanthamoeba keratitis: diagnosis, pathogenesis and treatment. Parasite 22, 10 (2015).
Matanock, A., Mehal, J. M., Liu, L., Blau, D. M. & Cope, J. R. Estimation of Undiagnosed Naegleria fowleri Primary Amebic Meningoencephalitis, United States (1). Emerg. Infect. Dis. 24, 162–164 (2018).
Centers for Disease Control and Prevention-USA Naegleria fowleri–case report data & graphs. [insert website/webpage name here], https://www.cdc.gov/parasites/naegleria/graphs.html#case-reports, Accessed [insert date here] (2019)
Visvesvara, G. S. & Stehr-Green, J. K. Epidemiology of free-living ameba infections. J. Protozool. 37, 25S–33S (1990).
Yoder, J. S., Eddy, B. A., Visvesvara, G. S., Capewell, L. & Beach, M. J. The epidemiology of primary amoebic meningoencephalitis in the USA, 1962–2008. Epidemiol. Infect. 138, 968–975 (2010).
Shakoor, S. et al. Primary amebic meningoencephalitis caused by Naegleria fowleri, Karachi, Pakistan. Emerg. Infect. Dis. 17, 258–261 (2011).
Ringsted, J., Jager, B. V., Suk, D. & Visvesvara, G. S. Probable acanthamoeba meningoencephalitis in a Korean child. Am. J. Clin. Pathol. 66, 723–730 (1976).
Kim, E. C. & Kim, M. S. Bilateral acanthamoeba keratitis after orthokeratology. Cornea 29, 680–682 (2010).
Lee, J. E. et al. Acanthamoeba keratitis related to orthokeratology. Int. Ophthalmol. 27, 45–49 (2007).
Xuan, Y. H. et al. Keratitis by Acanthamoeba triangularis: report of cases and characterization of isolates. Korean J. Parasitol. 46, 157–164 (2008).
Lee, J. K. & Lee, J. S. Two Cases of Corneal Toxicity in Acanthamoeba Keratitis by Combined Topical Anti-Acanthamoeba Keratitis Eye Solution. J. Korean Ophthalmol. Soc. 56, 280–284 (2015).
Kang, H. et al. Effective PCR-based detection of Naegleria fowleri from cultured sample and PAM-developed mouse. Eur. J. Protistol. 51, 401–408 (2015).
Sohn, H. J. et al. Efficient Liquid Media for Encystation of Pathogenic Free-Living Amoebae. Korean J. Parasitol. 55, 233–238 (2017).
Schroeder, J. M. et al. Use of subgenic 18S ribosomal DNA PCR and sequencing for genus and genotype identification of acanthamoebae from humans with keratitis and from sewage sludge. J. Clin. Microbiol. 39, 1903–1911 (2001).
Tsvetkova, N. et al. The identification of free-living environmental isolates of amoebae from Bulgaria. Parasitol. Res. 92, 405–413 (2004).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013).
De Jonckheere, J. F. Origin and evolution of the worldwide distributed pathogenic amoeboflagellate Naegleria fowleri. Infect. Genet. Evol. 11, 1520–1528 (2011).
Heggie, T. W. Swimming with death: Naegleria fowleri infections in recreational waters. Travel. Med. Infect. Dis. 8, 201–206 (2010).
Izumiyama, S. et al. Occurrence and distribution of Naegleria species in thermal waters in Japan. J. Eukaryot. Microbiol. 50(Suppl), 514–515 (2003).
Lekkla, A., Sutthikornchai, C., Bovornkitti, S. & Sukthana, Y. Free-living ameba contamination in natural hot springs in Thailand. Southeast. Asian J. Trop. Med. Public. Health 36(Suppl 4), 5–9 (2005).
Cursons, R., Sleigh, J., Hood, D. & Pullon, D. A case of primary amoebic meningoencephalitis: North Island, New Zealand. N. Z. Med. J. 116, U712 (2003).
Johnson, R. O. et al. Notes from the Field: Primary Amebic Meningoencephalitis Associated with Exposure to Swimming Pool Water Supplied by an Overland Pipe - Inyo County, California, 2015. MMWR Morb. Mortal. Wkly. Rep. 65, 424 (2016).
Montalbano Di Filippo, M., Novelletto, A., Di Cave, D. & Berrilli, F. Identification and phylogenetic position of Naegleria spp. from geothermal springs in Italy. Exp. Parasitol. 183, 143–149 (2017).
Tung, M. C. et al. Identification and significance of Naegleria fowleri isolated from the hot spring which related to the first primary amebic meningoencephalitis (PAM) patient in Taiwan. Int. J. Parasitol. 43, 691–696 (2013).
Lass, A. et al. Detection of Acanthamoeba spp. in water samples collected from natural water reservoirs, sewages, and pharmaceutical factory drains using LAMP and PCR in China. Sci. Total. Env. 15, 584–585 (2017).
Jeong, H. J. & Yu, H. S. The role of domestic tap water in Acanthamoeba contamination in contact lens storage cases in Korea. Korean J. Parasitol. 43, 47–50 (2005).
Kong, H. H. Molecular phylogeny of acanthamoeba. Korean J. Parasitol. 47(Suppl), S21–28 (2009).
Acknowledgements
This study was supported by the Research Program funded by the Korea Centers for Disease Control and Prevention (2015-E5400600). Operation of the MEGA6 program was partially supported by a grant (2018R1D1A1B07047302) from the Basic Science Research Program through the National Research Foundation (NRF) funded by the Ministry of Science and ICT, Republic of Korea.
Author information
Authors and Affiliations
Contributions
H.K.K., H.-J.S. and H.-J.S. conceived and designed the experiments. H.K.K., A.-Y.P. and G.-E.S. performed the experiments. H.K.K., H.-J.S., G.-S.S., S.-Y.J. and A.-J.H. Collection of water sample and analyzed the data. H.-J.S., S.-E.L. and S.-H.C. helping to many discussions. H.-J.S. contributed reagents/materials/analysis tools and illustration preparation. H.K.K. and H.-J.S. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kang, H., Sohn, HJ., Seo, GE. et al. Molecular detection of free-living amoebae from Namhangang (southern Han River) in Korea. Sci Rep 10, 335 (2020). https://doi.org/10.1038/s41598-019-57347-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-57347-1
This article is cited by
-
Prevalence of free-living amoebae in swimming pools and recreational waters, a systematic review and meta-analysis
Parasitology Research (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.