Abstract
Basidioradulum was morphologically considered to be a synonym of Xylodon. Here, its independence within Hymenochaetales is confirmed from a phylogenetic perspective. Basidioradulum radula, the generic type, is widely distributed in Northern Hemisphere. Two Southern Hemisphere species close to B. radula are newly described as B. mayi and B. tasmanicum, respectively, from Victoria and Tasmania, Australia. Basidioradulum mayi differs from B. radula by lack of cystidia. Moreover, the hymenial surface of B. radula is normally much more strongly hydnoid than that of B. mayi. Basidioradulum tasmanicum is distinct from B. radula and B. mayi by having capitate cystidia, ellipsoid to subglobose basidiospores, and crystal-covered hyphae. Although morphologically distinct, the two new species isolated by Bass Strait have an almost identical ITS region, and could not be differentiated by nLSU- and ITS-based phylogenetic analyses. This case reminds us that basing phylogeny simply on the ITS as a barcode region may underestimate fungal species diversity.
Similar content being viewed by others
Introduction
Basidioradulum Nobles was introduced for B. radula (Fr.) Nobles1. The generic type of the genus, B. radula, was combined as Xylodon radula (Fr.) Ţura, Zmitr., Wasser & Spirin based on morphological characters2. However, phylogenetic studies including this species all failed to group X. radula with other species of Xylodon (Pers.) Gray3,4,5. Meanwhile, morphologically fungal taxonomists have not widely accepted this species as a member of Xylodon or even of Schizoporaceae6,7. A further 10 species have been placed at some time in Basidioradulum. According to Index Fungorum (http://www.indexfungorum.org/), nine of these belong elsewhere. The remaining species, B. crustosum (Pers.) Zmitr., Malysheva & Spirin is an invalid combination in Basidioradulum according to the Arts. 41.5, 41.8(a), and 41.8(b) of the International Code of Nomenclature8.
During an examination of wood-inhabiting fungi from Australia, our attention was drawn to the relationships of five specimens with similarities to Basidioradulum radula. After taking Chinese specimens of B. radula into consideration together, we described the Australian specimens with almost identical ITS sequences as two new species based on extremely distinct morphological characters and localities separated by Bass Strait.
Results
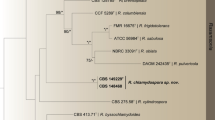
Nine specimens from China and Australia were sequenced for this study, which resulted in seven new nLSU sequences and six new ITS sequences (Table 1). For the nLSU alignment of 101 taxa with 929 characters, the ML search stopped after 400 BS replicates, and all chains converged indicated by the effective sample sizes (ESSs) of all parameters above 2500 and the potential scale reduction factors (PSRFs) equal to 1000. With regard to the ITS alignment of 13 taxa with 573 characters, after 300 BS replicates the ML search stopped, while the ESSs of all parameters above 6000 and the PSRFs equal to 1000 suggested the convergence of all chains. The two phylogenetic methods generated nearly congruent topologies for both nLSU and ITS alignments. Therefore, the topologies generated from the ML method are presented along with the statistical values at the nodes of BS and BPPs above 50% and 0.8, respectively (Figs. 1 and 2).
Phylogenetic relationship among species of Basidioradulum inferred from the ITS region. The topology generated from the maximum likelihood method is presented. Values at nodes are bootstrap and Bayesian posterior probability, respectively, above 50% and 0.8. Newly sequenced specimens are in bold. The voucher locations of sequences are labeled in parentheses if available. The photos of basidiospores of B. mayi and B. tasmanicum taken from the holotypes are presented after their species names.
The phylogeny inferred from the nLSU region did not classify families well within Hymenochaetales (Fig. 1), but did separate the fully supported Basidioradulum lineage from other species belonging to Schizoporaceae, including Xylodon quercinus (Pers.) Gray, the generic type of Xylodon. The Basidioradulum lineage was divided into two clades. The clade of B. radula with five Northern Hemisphere specimens received a moderate BS value of 68% and a high BPP value of 0.97. The Southern Hemisphere clade with three Australian specimens was strongly supported (99% BS, 1 BPP).
The midpoint-rooted tree inferred from the ITS region indicates that all 13 collections of Basidioradulum formed two fully supported clades (Fig. 2). One clade including eight collections originating from the Northern Hemisphere represented B. radula, while the other clade had five Southern Hemisphere collections.
The pairwise distance matrix for ITS sequences of Basidioradulum (Table 2) shows that the 13 collections were separated into two groups that are congruent with the two clades in the ITS-based phylogeny (Fig. 2). The distances among Group 1 including five Southern Hemisphere collections and among Group 2 including eight Northern Hemisphere collections were, respectively, 0.2% and 0.9%, while the distance between the two groups was 3.4%.
All the above molecular evidence supported the Southern Hemisphere specimens as distinct from Basidioradulum radula. Moreover, further morphological examinations indicated that among the Southern Hemisphere specimens the two Victorian specimens were extremely different to the three Tasmanian specimens. Therefore, two new species of Basidioradulum are described below.
Taxonomy
Basidioradulum mayi Xue W. Wang & L.W. Zhou, sp. nov. Figs. 3 and 4
Basidiocarps of Basidioradulum mayi and B. tasmanicum. (A) B. mayi (LWZ 20180510-23, holotype). (B) B. mayi (LWZ 20180510-18). (C) B. radula (Zhao 1043). (D) B. radula (Dai 15769). (E) B. tasmanicum (MEL 2403476). (F) B tasmanicum (MEL 2386000). (G) B. tasmanicum (MEL 2385925, holotype). Scale bars = 1 cm.
MycoBank: MB 833704.
Etymology: mayi (Latin), in honor of Australian mycologist, Dr. Tom May, who kindly arranged the author Li-Wei Zhou’s field trip in Victoria, Australia.
Type: AUSTRALIA: Victoria, Yarra Ranges National Park, Cora Lynn Falls, on the base of dead standing angiosperm, 10 May 2018, LWZ 20180510-23 (holotype in MEL, isotype in IFP).
Basidiocarps annual, effused, adnate, hard and cracked when dry. Hymenial surface smooth, odontoid to hydnoid, the teeth conical or irregular, up to 1–2 mm long and 0.5–1 mm wide, becoming gradually shorter towards margin, whitish-cream to light-ochraceous. Margin slightly fibrillose, paler or concolorous with fertile area. Subiculum homogeneous, whitish-cream, up to 0.8 mm thick.
Hyphal system monomitic; generative hyphae with clamp connections. Subicular hyphae hyaline, thin- to slightly thick-walled, occasionally branched, more or less parallel to substrate, 2.5–4.5 μm wide. Subhymenial hyphae somewhat horizontal to vertical along substrate, hyaline, agglutinated, 2.5–4.5 μm wide. Cystidia absent. Basidia clavate to subclavate, 20–25 × 5.5–7.5 μm, with 4 sterigmata each 2–3 μm long and a clamp connection at the base. Basidioles similar in shape to basidia, but smaller. Basidiospores cylindrical to slightly allantoid, hyaline, smooth, thin-walled, CB−, IKI–, (7.2−)7.3–9.5(−9.7) × (2.1−)2.2–3.5(−3.7) μm, L = 8.20 μm, W = 2.92 μm, Q = 2.80–2.82 (60/2).
Additional specimen (paratype) examined: AUSTRALIA: Victoria, Yarra Ranges National Park, Cora Lynn Falls, on fallen angiosperm branch, 10 May 2018, LWZ 20180510-18 (IFP, MEL).
Notes: The smooth, odontoid to hydnoid hymenial surface, clavate to subclavate basidia, and cylindrical to slightly allantoid basidiospores indicate that the new species is close to Basidioradulum radula. However, B. mayi differs by lack of cystidia and being found in Australia in the Southern Hemisphere. Moreover, the hymenial surface of B. radula as exemplified by Chinese specimens (Fig. 3) is normally much more strongly hydnoid than that of B. mayi.
Basidioradulum tasmanicum Xue W. Wang & L.W. Zhou, sp. nov. Figs. 3 and 5.
MycoBank: MB 833705.
Etymology: tasmanicum (Latin), refers to the island of Tasmania, Australia.
Type: AUSTRALIA: Tasmania, Sandspit Forest, Wielangta Rainforest Reserve, on fallen angiosperm branch, 8 August 2006, N. Hallenberg 15784 (holotype MEL 2385925).
Basidiocarps annual, effused, adnate, cracked when dry. Hymenial surface smooth or with various kinds of projections, cream to buff. Margin slightly pilose, paler or concolorous with fertile area. Subiculum homogeneous, whitish-cream.
Hyphal system monomitic; generative hyphae with clamp connections. Subicular hyphae hyaline, thin- to slightly thick-walled, occasionally branched, more or less parallel to substrate, 2.5–4.5 μm wide. Subhymenial hyphae somewhat horizontal to vertical along substrate, hyaline, agglutinated, usually covered with granular crystals, capitate terminal branches numerous, 2.5–4.5 μm wide. Capitate cystidia thin-walled, projecting for approximately half their lengths, 20–35 × 3.5–4.5 μm. Basidia clavate to subclavate, 20–25 × 5.5–7.5 μm, with 4 sterigmata each 2–3 μm long and a clamp connection at the base. Basidioles similar in shape to basidia, but smaller. Basidiospores ellipsoid or subglobose, with a large oil drop, hyaline, smooth, thin-walled, CB–, IKI−, (4.6–)4.7–5.8(−6.5) × (2.3−)2.4–3.7(−3.9) μm, L = 5.20 μm, W = 3.07 μm, Q = 1.60–1.80 (90/3).
Additional specimens (paratypes) examined: AUSTRALIA: Tasmania, Franklin-Gordon Wild Rivers National Park, Franklin River Nature Trail, on fallen angiosperm branch, 13 August 2006, N. Hallenberg 15892 (MEL 2386000); Tasmania, Franklin-Gordon Wild Rivers National Park, Franklin River Nature Trail, on dead wood, 15 April 2008, G.M. Gates & D.A. Ratkowsky FF328 (MEL 2403476).
Notes: Basidioradulum tasmanicum is distinct from B. radula and B. mayi by having capitate cystidia, ellipsoid to subglobose basidiospores, and crystal-covered hyphae.
Discussion
In this study, the phylogenetic position of Basidioradulum was reevaluated by sampling more collections of this genus in the nLSU-based phylogeny (Fig. 1). Although the resolution at the family level was poor, Basidioradulum was clearly separated from Xylodon quercinus, the generic type of Xylodon, as in previous studies3,4,5. Therefore, we reject the transfer of B. radula, the generic type of Basidioradulum to Xylodon proposed by Ţura et al.2, and treat Basidioradulum as a distinct genus from Xylodon. Because comprehensive phylogenetic sampling of taxa of the order Hymenochaetales is lacking, we consider Basidioradulum to be a genus incertae sedis at the family rank within Hymenochaetales.
Basidioradulum radula as defined morphologically is considered to be widely distributed in Northern Hemisphere2. The sequences of B. radula from Chinese, Korean and American collections analyzed in this study also confirm its wide distribution. Meanwhile, the two Southern Hemisphere relatives of B. radula are distinct, and newly described as B. mayi and B. tasmanicum. These two species cannot be differentiated by molecular evidence on the basis of ITS region, which is used as the fungal barcode (Fig. 2, Table 2). The situation where B. mayi and B. tasmanicum have distinct morphological characters but cannot be delimited by DNA data indicates that these two species isolated by Bass Strait may be undergoing an ongoing allopatric speciation event9. However, more evidence, such as mating test between these two species and multi locus-based phylogeny, is needed to further confirm this event. Nevertheless, the current case of Basidioradulum does stimulate consideration of the multitude of species concepts and recognition criteria as applied to fungi10,11. Although the molecular phylogenetic method normally has been considered to be a powerful means to discover new fungal lineages12,13, it is concluded from the current case that if we only consider differences in the ITS region, fungal diversity may be underestimated. The speciation of fungi needs time, and during the process of speciation the divergence of phenotype and genotype could happen at differentiated paces. Therefore, neither the genotype nor phenotype alone is suitable to be used as evidence to describe species. Instead, polyphasic evidence from morphology, phylogeny, ecology and so on, if available, should be used together to delimit fungal species.
Conclusion
Two wood-inhabiting basidiomycetous species Basidioradulum mayi and B. tasmanicum are newly described from both sides of Bass Strait, Australia. These two species have almost identical fungal barcoding ITS sequences but distinct morphological characters.
Materials and Methods
The studied specimens are deposited at the herbaria of the Institute of Applied Ecology, Chinese Academy of Sciences (IFP), Shenyang, China, the Institute of Microbiology, Beijing Forestry University (BJFC), Beijing, China, Southwest Forestry University (SWFC), Kunming, China, and the National Herbarium of Victoria (MEL), Melbourne, Australia.
The hymenial surfaces of basidiocarps were observed under a stereomicroscope. Special color terms followed Petersen14. Microscopic characters were examined using a Nikon Eclipse 80i microscope at magnification up to 1000×. Specimen sections were stained in Cotton Blue (CB), Melzer’s reagent (IKI) and 5% potassium hydroxide. All measurements were taken from CB-stained sections. The basidiospore size variation was presented by putting 5% of measurements from each end of the range in parentheses. Drawings were made with the aid of a drawing tube. Photos of basidiospores were taken using a Nikon Digital Sight DS-U3 camera. The following abbreviations are used in the text: L = mean basidiospore length (arithmetic average of all basidiospores), W = mean basidiospore width (arithmetic average of all basidiospores), Q = variation in the L/W ratios between the specimens studied, and n = number of basidiospores measured from given number of specimens.
Crude DNA was extracted from basidiocarps of dry specimens using FH Plant DNA Kit (Beijing Demeter Biotech Co., Ltd., Beijing, China), and then directly used as template for subsequent PCR amplifications. The nLSU and ITS regions were amplified and sequenced using primer pairs LR0R and LR715, and ITS1F16 and ITS417, respectively. The PCR procedure was as follows: for nLSU region initial denaturation at 94 °C for 1 min, followed by 34 cycles at 94 °C for 30 s, 47.2 °C for 1 min and 72 °C for 1.5 min, and a final extension at 72 °C for 10 min, while for ITS region initial denaturation at 95 °C for 3 min, followed by 34 cycles at 94 °C for 40 s, 57.2 °C for 45 s and 72 °C for 1 min, and a final extension at 72 °C for 10 min. The PCR products were sequenced at the Beijing Genomics Institute, Beijing, China. The newly generated sequences were deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/; Table 1).
The nLSU dataset was used to explore the phylogenetic position of Basidioradulum, and included sequences from all main lineages in Hymenochaetales and two species in Polyporales as ingroup taxa and Amaurodon viridis (Alb. & Schwein.) J. Schröt. in Thelephorales as an outgroup taxon. The ITS dataset was used to differentiate the phylogenetic relationships among specimens of Basidioradulum. The datasets were aligned using MAFFT 7.11018 under the g-ini-i option19, and the resulting alignments, after manual inspection, were deposited in TreeBASE (http://www.treebase.org; accession number 24505). jModelTest20,21 was used to estimate the best-fit evolutionary model of the two alignments. Maximum likelihood (ML) and Bayesian inference (BI) methods were utilized for phylogenetic analyses of the two alignments following the evolutionary models. The ML method was performed using raxmlGUI 1.222,23 with the calculation of bootstrap (BS) replicates under the auto FC option24. The BI method was carried out using MrBayes 3.225 with two independent runs, each including four chains of 10 million generations and starting from random trees. The first 25% of the sampled trees every 1000th generation was removed, and the other 75% trees were remained for constructing a 50% majority consensus tree and calculating Bayesian posterior probabilities (BPPs). Tracer 1.5 (http://tree.bio.ed.ac.uk/software/tracer/) was used to judge whether chains converged.
Besides phylogenetic analyses, the alignment resulted from the ITS dataset was also subjected to distance estimation using MEGA version 726. The analysis preferences were as follows: p-distance substitution model including both transitions and transversions, uniform rates among sites, and pairwise deletion treatment.
Data availability
All data generated or analyzed during this study have been deposited in public databases as indicated.
References
Nobles, M. K. Conspecificity of Basidioradulum (Radulum) radula and Corticium hydnans. Mycologia 59, 192–211 (1967).
Ţura, D. A., Zmitrovich, I. V., Wasser, S. P., Spirin, W. A. & Nevo, E. Biodiversity of the Heterobasidiomycetes and non-gilled Hymenomycetes (former Aphyllophorales) of Israel (Gantner Verlag K.-G, 2011).
Larsson, K. H., Larsson, E. & Kõljalg, U. High phylogenetic diversity among corticioid homobasidiomycetes. Mycol. Res. 108, 983–1002 (2004).
Larsson, K. H. et al. Hymenochaetales: a molecular phylogeny for the hymenochaetoid clade. Mycologia 98, 926–936 (2006).
Zhou, L. W., Wang, X. W., Vlasák, J. & Ren, G. J. Resolution of phylogenetic position of Nigrofomitaceae within Hymenochaetales (Basidiomycota) and Nigrofomes sinomelanoporus sp. nov. (Nigrofomitaceae) from China. MycoKeys 29, 1–13 (2018).
Yurchenko, E. & Wu, S. H. A key to the species of Hyphodontia sensu lato. MycoKeys 12, 1–27 (2016).
Riebesehl, J. & Langer, E. Hyphodontia s.l. (Hymenochaetales, Basidiomycota): 35 new combinations and new keys to all 120 current species. Mycol. Prog. 16, 637–666 (2017).
Turland, N. J. et al. International Code of Nomenclature for algae, fungi, and plants (Shenzhen Code) adopted by the Nineteenth International Botanical Congress Shenzhen, China, July 2017. [Regnum Vegetabile no. 159.] (Koeltz Botanical Books, 2018).
Giraud, T., Refrégier, G., Le Gac, M., de Vienne, D. M. & Hood, M. E. Speciation in fungi. Fungal Genet. Biol. 45, 791–802 (2008).
Cai, L. et al. The evolution of species concepts and species recognition criteria in plant pathogenic fungi. Fungal Divers. 50, 121–133 (2011).
Steenkamp, E. T., Wingfield, M. J., McTaggart, A. R. & Wingfield, B. D. Fungal species and their boundaries matter - Definitions, mechanisms and practical implications. Fungal Biol. Rev. 32, 104–116 (2018).
Hibbett, D. S. et al. Progress in molecular and morphological taxon discovery in Fungi and options for formal classification of environmental sequences. Fungal Biol. Rev. 25, 38–47 (2011).
Tedersoo, L. et al. Global diversity and geography of soil fungi. Science 346, 1256688 (2014).
Petersen, J. H. Farvekort. The Danish Mycological Society’s colour chart. Greve: Foreningen til Svampekundskabens Fremme (1996).
Vilgalys, R. & Hester, M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4238–4246 (1990).
Gardes, M. & Bruns, T. D. ITS primers with enhanced specificity for Basidiomycetes: application to identification of mycorrhizae and rusts. Mol. Ecol. 2, 113–118 (1993).
White, T. J., Bruns, T., Lee, S. & Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics in PCR protocols: a guide to methods and application (eds. Innis, M. A., Gelfand, D. H., Sninsky, J. J. & White, T.J.) 315–322 (Academic Press, 1990).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Katoh, K., Kuma, K., Toh, H. & Miyata, T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33, 511–518 (2005).
Guindon, S. & Gascuel, O. A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Syst Biol 52, 696–704 (2003).
Posada, D. jModelTest: phylogenetic model averaging. Mol. Biol. Evol. 25, 1253–1256 (2008).
Silvestro, D. & Michalak, I. raxmlGUI: a graphical front end for RAxML. Org. Divers. Evol. 12, 335–337 (2012).
Stamatakis, A. RAxML-VI-HPC: maximum likelihood based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (2006).
Pattengale, N. D., Alipour, M., Bininda-Emonds, O. R. P., Moret, B. M. E. & Stamatakis, A. How many bootstrap replicates are necessary? J. Comput. Biol. 17, 337–354 (2010).
Ronquist, F. et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542 (2012).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Acknowledgements
Yu-Cheng Dai (BJFC, China), Tom May (MEL, Australia) and Chang-Lin Zhao (SWFC, China) are thanked for forwarding specimens as loans. Tom May is also thanked for improving language of the manuscript. The research was financed by the National Natural Science Foundation of China (Project Nos. 31970012 & 31570014) and Youth Innovation Promotion Association CAS (No. 2017240).
Author information
Authors and Affiliations
Contributions
X.W.W. and J.H.J. performed the molecular sequencing. X.W.W. and L.W.Z. performed the morphological examinations and wrote the manuscript. L.W.Z. analyzed the molecular data. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, XW., Jiang, JH. & Zhou, LW. Basidioradulum mayi and B. tasmanicum spp. nov. (Hymenochaetales, Basidiomycota) from both sides of Bass Strait, Australia. Sci Rep 10, 102 (2020). https://doi.org/10.1038/s41598-019-57061-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-57061-y
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.