Abstract
Olfactory receptors (ORs), encoded by the largest vertebrate multigene family, enable the detection of thousands of unique odorants in the environment and consequently play a critical role in species survival. Here, we advance our knowledge of OR gene evolution in procellariiform seabirds, an avian group which relies on the sense of olfaction for critical ecological functions. We built a cosmid library of Cory’s Shearwater (Calonectris borealis) genomic DNA, a model species for the study of olfaction-based navigation, and sequence OR gene-positive cosmid clones with a combination of sequencing technologies. We identified 220 OR open reading frames, 20 of which are full length, intact OR genes, and found a large ratio of partial and pseudogenes to intact OR genes (2:1), suggestive of a dynamic mode of evolution. Phylogenetic analyses revealed that while a few genes cluster with those of other sauropsid species in a γ (gamma) clade that predates the divergence of different avian lineages, most genes belong to an avian-specific γ-c clade, within which sequences cluster by species, suggesting frequent duplication and/or gene conversion events. We identified evidence of positive selection on full length γ-c clade genes. These patterns are consistent with a key role of adaptation in the functional diversification of olfactory receptor genes in a bird lineage that relies extensively on olfaction.
Similar content being viewed by others
Introduction
The sense of olfaction is used by vertebrates to process information about their surroundings, playing a critical role in survival and reproduction as it facilitates the recognition of suitable food, predators and prey, mates, and territories1,2. Odour perception begins with the binding of odorant molecules to G protein-coupled receptors (GPCRs) primarily expressed in the nasal olfactory epithelium3. These receptors, encoded by a complex multi-gene family containing large numbers of olfactory receptor (OR) genes4, allow discrimination of a vast array of unique odorants.
Phylogenetic analyses of olfactory receptor genes in model vertebrates enabled the identification of nine different monophyletic classes of receptors (α, β, δ, ε, γ, ξ, η, θ and κ) divided into Type I and Type II5,6 (Fig. 1). Type I is the most diverse, and includes Class I (α, β, δ, ε, ξ and θ groups) and Class II (γ). Particularly the α and γ groups have experienced extremely large copy number expansions and because they occur in tetrapods they are thought to mediate the recognition of airborne odorants5,7 (Fig. 1). The remaining Type I and Type II (which only includes group η) are mainly present in fish and amphibian genomes and are thought to detect water-soluble odorants5. The avian species surveyed in greater genomic depth (the chicken, Gallus gallus and the zebra finch Taeniopygia guttata) mostly have OR genes belonging to group γ, but also a few α and θ6,8,9.
Comparative studies have shown that OR gene copy number can vary by several orders of magnitude among vertebrate groups, and even among closely related lineages10,11,12. Although the proportion of OR pseudogenes is characteristically high (20–60%11), the relative numbers of intact, pseudo- (with an in-frame stop codon) and partial genes (without a start and/or stop codons) can also vary substantially among and within species11. Multi-gene families characterized by this diversity pattern follow a birth-and-death model of evolution13,14, according to which gene copy number variation emerges due to repeated bouts of duplication, deletion and gene conversion events14. While gene duplication occurs primarily due to unequal crossing over5, retention of duplicated genes may be facilitated by natural selection if a different and/or a larger number of genes confers a selective advantage by generating a more diverse receptor repertoire9,10,15,16,17. The striking variation in OR diversity patterns has been related to sensory trade-offs and lineage-specific ecological adaptation to new niches5,6,10,12,18,19,20,21. Despite this extraordinary gene diversity, OR gene structure is fairly conserved. Functional genes are approximately 1Kb long and generally lack introns. Based on patterns of variation and computational modeling in model organisms, the putative ligand binding sites are mainly located between the third and seventh transmembrane (TM) helical domains22,23.
Although it is now widely accepted that birds are able to recognize and act upon particular odours2,24, research on avian OR genes has been mostly based on G. gallus. The first survey in a taxonomically diverse group of nine bird species found a striking pattern of OR gene diversity, characterized by the presence of two distinct γ clades25. Genes clustered with putative orthologs from other species in one clade, suggesting that this γ sub-group was already present in the common ancestor of the different bird lineages25. In contrast, in the most abundant γ-c clade (also referred to as family 146), genes clustered by species, consistent with ubiquitous duplication events post-dating the divergence of different bird species. This expansion was later confirmed in the chicken and zebra finch genomes but found missing in the green anole and was thus hypothesized to be a unique feature of avian genomes8,9. A genomic survey of two falcon species found a small number of intact OR genes and a missing γ-c clade, possibly related to a foraging strategy more dependent on vision than olfaction26. A higher proportion of pseudogenes was found in penguins than in Procellariiformes, their sister group, a pattern that was also interpreted as resulting from a higher reliance on (underwater) vision for foraging27.
Seabirds in the avian order Procellariiformes, which includes albatrosses, petrels and shearwaters, have one of the largest relative olfactory bulb to brain size (OB) ratio of all birds28,29, and use the sense of smell in homing, foraging and navigation30,31,32. They forage day and night in the open ocean by building cognitive maps based on olfactory oceanic landscapes32,33. Despite having the largest OB ratio recorded in a group of nine species surveyed, the Snow petrel Pagodroma nivea was found to have amongst the fewest estimated total number of OR genes (n = 212), and among the surveyed genes, approximately 50% belong to the γ-c clade25. More recently, a survey of the genome of the Northern fulmar Fulmarus glacialis found 370 OR genes, of which 102 (only two intact) belong to the γ-c clade6. We focused on the Cory’s Shearwater Calonectris borealis, which is becoming a model seabird species for many ecological questions, including the importance of olfaction for homing and oceanic navigation31,32,33. Our goal was to extend the understanding of the evolution of OR genes to the Procellariiformes avian group, through their extensive characterization in C. borealis. Specifically, we aimed to determine 1) olfactory receptor gene diversity in C. borealis, 2) the ratio of full length to partial ORs and 3) whether there is evidence of lineage-specific expansion events in this species. Additionally, we aimed to find whether selection has been shaping the evolution of shearwater OR genes. This study will allow us to determine whether the evolutionary patterns observed are consistent with the hypothesis that Cory’s shearwaters rely heavily on their sense of smell for key functions such as foraging and homing, a critical contribution to the understanding of the genetic underpinnings of adaptation in this group.
Results
Genomic characterization of OR genes in Cory’s Shearwater
The characterization of the full genomic complement of OR gene families remains an important but challenging endeavour34,35. Multigene families can be reliably studied from high-quality genome references, but the combination of read lengths and the depth of coverage required make these costly and time consuming36,37. Conversely, short-read whole genome sequence data, while relatively inexpensive to generate, results in partial gene family sets with missing, truncated and possibly chimeric gene sequences, especially in families composed of genes with high pairwise sequence similarity or highly conserved sequence motifs38,39. Here, we opted for sequencing, at high depth of coverage and with a combination of sequencing technologies, a large number of long, OR- containing cosmids from a Cory’s shearwater genomic DNA library, a compromise on OR family completeness to achieve high-quality, representative gene sequences.
The most comprehensive hybrid assembly of the OR-containing cosmids had a cumulative length of 7.4 Mb. It consisted of 399 unique scaffolds with a cumulative length of 2.6 Mb and 22 K degenerate contigs totaling 4.8 Mb (Supplementary Info). The final set of EVm models supported by ORFs consisted of 1,501 genes encoding proteins with a mean length of 135 amino-acids (range of 75–1215 amino acids). Of these, 317 were found in the assembly scaffolds and the remaining in the degenerate contigs.
We identified 220 Cory’s shearwater full and partial length OR genes among the 1,501 gene models, ~ 60% of the OR gene number found in the genome assembly of the Northern fulmar, the only procellariidae with a draft genome available6,40, and 35% of the OR gene number found in the genomes of G. gallus and T. gutatta. Many additional small ORFs were discarded due to size (<75 amino acids), despite significant homology with other avian OR genes, so this is likely an underestimate of the total number of olfactory receptor genes in Cory’s shearwaters.
In total, 45 OR genes were in the main scaffolds, where ORFs ranged between 297–936 bp (with mean length ± standard deviation of 678.3 ± 249.3 bp). The remaining 175 OR genes were found in the degenerate contigs, and the ORFs ranged in length between 225–945 bp (mean ± SD 394.9 ± 118.6 bp).
Structure and proportion of potentially functional shearwater OR genes
Of all OR genes identified, only 56 (~ 25%) were not at the edge of a contig. Of these, 20 (36%) were complete genes, 15 (27%) were pseudogenes and 21 (38%) were partial sequences (missing a start and/or stop codon and no in-frame stop codons). In all except four cases there was only one OR locus per scaffold, even though almost 80 scaffolds were above 10 Kb (Fig. S1b). In the scaffolds with more than one OR locus, ORFs were between 44 and 1,517 bp apart, and at least one was always a pseudogene. Of the 220 OR genes, 85% were annotated as putatively belonging to the γ-c clade (family 146) (Table 1), 18 of which are complete. The remaining ORs belong to families already identified in birds and reptiles except for one gene, with highest sequence similarity to mammal genes (24-like family gene) (Table 1).
Full-length γ-c shearwater OR genes include a single exon with a length of 312 amino acids. The logo view of the alignment suggests that the distribution of genetic variation is not uniform along the length of these genes (Fig. 2). The characteristic, conserved olfactory receptor-specific motifs are present, such as the MAYDRYVAIC in the transmembrane domain (TM) 3 – intracellular domain (IC) 2 boundary, the FSTC(LP)H at the end of IC3, and the conserved cysteine residues in the extra-cellular (EC) 1 and EC2 domains and prolines at TM7 (Fig. 2), which are important for the conformation stability of the receptor41. The most variable positions are found in the TM 4 and TM5 and to a lesser extent in TM3 and TM6 (Fig. 2).
Sequence conservation in complete OR γ-c shearwater genes. The open reading frames of the aligned gene set (n = 17) was used to build a sequence logo. Putative location of transmembrane regions, (TM1-7), intra-cellular loops (IC1-3), extra-cellular loops (EC1-3), conserved motifs (□) and residues (✶) are shown.
Phylogenetics of shearwater OR gene sequences
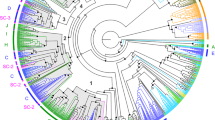
Phylogenetic analyses suggest that there are at least two distinct OR gene clades in the shearwater (Fig. 3a), in agreement with the functional annotation. A few OR genes cluster with those of the other sauropsid species in a γ clade that pre-dates the divergence of the different bird lineages. However, most chicken, zebra finch, fulmar and shearwater OR genes belong to the avian specific, well-supported γ-c clade, within which sequences cluster by avian family (Fig. 3a). The fulmar γ-c genes cluster with the shearwater genes in a paraphyletic Procellariidae clade, some clustering in a fulmar specific clade, suggesting that they are younger than the divergence of the two procellariids. When all 220 sequences are included in the analysis, some shearwater and fulmar γ-c sequences appear to be evolving at significantly faster rates than those from the other species, as revealed by the very long branch lengths (Fig. 3a). This pattern may be due to a large number of pseudogenes evolving free of selective constraints, as it does not persist in analysis including only intact OR genes (Fig. 3b). In the analysis with the intact genes, the monophyly of the avian γ-c clade remains well supported as is the clade including all shearwater genes (bootstrap value of 92%).
Maximum-likelihood phylogenetic analysis of sauropsid olfactory receptor genes. (a) Analysis includes the Cory’s Shearwater’s 220 OR genes aligned with 98 Northern fulmar genes from GenBank, 134 zebra finch genes, 214 chicken genes, and 112 green anole genes (from [8]). (b) Analysis using only the set of complete genes from the Cory’s Shearwater (n = 20), aligned with 44 full fulmar genes (from GenBank), 134 zebra finch genes, 214 chicken genes, and 112 green anole genes (from [8]). Both analyses are based on the JTT model as the best fitting substitution model. Outgroup sequences of the Adenosine receptor A2b are from GenBank. Nodal support values are shown for major clades.
Positive selection in full length γ-c shearwater OR genes
To determine the selection regimen governing the evolution of the intact γ-c shearwater genes, we considered two data partitions as GARD found evidence of one breakpoint at alignment position 246, which falls in the putative TM6 domain (Fig. 2). Based on the two-partition dataset of all full-length OR genes, the different methods found that the overall dN/dS was never significantly different from 1, varying between ω = 0.552 (SLAC) and ω = 1.044 (REL). However, positive selection was identified on individual site codons of the full-length shearwater γ-c genes, consistent with a key role of selection in the functional diversification of these genes in the shearwater (Fig. 4a, Table 2). The number of sites with evidence of positive selection varied with the method used, with amino-acid positions 109 and 274 (respectively in TM3 and TM7 domains) identified by all individual methods as well as by the integrative analysis (Table 2). In addition, positive selection was identified in amino-acid positions 111, 114, 209, 210 and 219 by at least two methods. These five sites are also located in TM domains (Fig. 4a). Critically, amino acids 109 and 274 have side chains oriented towards the center of the receptor (Fig. 4b), suggesting that, in this species, they may be part of the odorant binding pocket.
Secondary structure of a full-length γ-c OR gene from Cory’s shearwater. (a) Structure showing the α-helices of the trans-membrane domains in blue. The amino-acid sites estimated to be evolving under positive selection are plotted, showing two predicted by all methods to be under positive selection (in red) as well as those predicted by at least two methods to be under positive selection (in orange). (b). Detail of the secondary structure of the same OR receptor, showing the side chain of the two amino-acids with the strongest signal of positive selection, aa109 (TM3 domain) and aa274 (TM7 domain) (in red), orienting towards the center of the receptor, suggesting they might form part of the ligand binding pocket.
Discussion
This work provides one of the first detailed molecular insights into the evolution of chemoreception in an avian group for which olfaction has been shown to play a vital role, by specifically studying sequence diversity and evolution of olfactory receptors in the Cory’s shearwater genome.
Several studies suggest that mammalian and avian OR repertoires are influenced by life histories6,10,12,18,19,20. Procellariiform seabirds have anatomical characteristics that suggest a strong reliance on a functional olfactory system, such as one of the largest olfactory bulb size to brain size (OB) ratio among extant avian species and specialized tube-like nostrils42. It has been suggested that there is a positive correlation between OB ratio, OR gene repertoire and olfactory function10,15,21,25,28,29. In this context, one might expect shearwaters to have more OR genes than species with smaller OB ratio and less obvious reliance on olfaction for survival, such as the chicken and zebra finch, in which 674 and 688 isolated OR genes have been identified, respectively9. However, the procellariiform Snow petrel was found to have among the lowest estimated number of OR genes (n = 212) among nine surveyed avian species, despite its much larger OB ratio25. Likewise, the Northern fulmar was found to only have an average number of OR genes in a genomic survey that included 48 phylogenetic diverse avian lineages6. The evidence from Cory’s shearwater is inconclusive in this regard. While we identified with certainty 220 OR genes (similar to the numbers found in other petrels but about a third of the total number estimated in the chicken), our study was limited by the maximum number of cosmid clones considered for sequencing, and possibly by the specificity of the probes used to screen the library, which were restricted to known OR motifs. Furthermore, putative OR sequences mapping to the edge of assembly contigs and for which the translated sequence was shorter than 75 amino acids were discarded. Finally, the 220 shearwater OR genes were identified in only a very small (albeit biased toward OR-containing) fraction of the genome (7.4 Mb) compared with the other species for which the whole genome was analyzed. On the other hand, we found 17 intact γ-c genes in the shearwater, a larger number than the 2 previously found in the fulmar, despite possibly having missed characterization of the full repertoire. This raises the possibility that a smaller olfactory receptor repertoire is compensated by a larger number of intact genes, or possibly a broader receptor binding pocket specificity through the use of other sensory receptors 27,43. This issue should be revisited once a high quality assembled Cory’s shearwater genome becomes available.
A remarkable finding of this study is that the vast majority (85%) of the OR genes identified in the shearwater belong to the γ-c clade. If the proportion of γ-c genes among all identified shearwater OR genes is representative of their real frequency in the genome, then the olfactory receptor family in this species is mostly composed of young paralogous loci, reflecting a relatively recent expansion of highly similar genes putatively linked to the recognition of volatile odorants. Although the γ-c clade seems to be as abundant in the shearwater as in the chicken (63%) and the zebra finch (80%), it was found to be not as abundant in other avian species, including the Northern fulmar6, although low genome assembly quality may have compromised the ability to characterize large expansions of recently duplicated genes in some species44. In two falcon species the γ-c clade was found missing, possibly reflecting reliance on other sensory cues26. Nevertheless, the presence of this striking γ-c diversity in the shearwater genome, and its higher proportion of intact genes relative to what has been found in most species surveyed so far16, is an example of how ecological adaptation is likely contributing to an enhanced olfactory function in Procellariiformes.
Lineage-specific duplications and losses of olfactory receptor genes is characteristic of the evolution of the OR gene family in vertebrates and have resulted in a large range of OR gene repertoire size across lineages5,10,11,12,20. The co-occurrence of γ-c intact, partial and pseudogenes in the shearwater OR repertoire, sometimes in the same contig, agrees with the extreme dynamic nature of the evolutionary models proposed to govern the genes encoding OR and other vertebrate multi-gene families14,44,45. In the context of the olfactory receptor evolution, the presence of many paralogs facilitates the evolution of novel gene functions that allow the recognition of new odors, under a scenario of neo-functionalization, but may also result in large number of pseudogenes due to functional redundancy and consequent relaxation of selective constraints45,46,47.
A hallmark of the olfactory receptor structure is a highly variable pattern of amino acid sequence conservation determining the identity and specificity of odorants that are recognized by each receptor. Although it is possible that some ligand interactions will be mediated by conserved amino acids48,49, peaks of polymorphism have been consistently detected on the same transmembrane domains in different species, namely in the TM3, TM5, TM6 and TM716, similar with what was found in this study. If these domains do indeed form part of the odorant binding pocket in seabirds, as it has been shown in other vertebrate lineages22, it is not surprising to find evidence of diversifying selection at these sites, since allelic diversity would confer a selective advantage by allowing individuals to recognize a wider spectrum of odors.
Lineage-specific patterns of OR gene diversity have been found to be tightly linked to the habitat and foraging ecology of the different groups6,18,21. Procellariiform seabirds are pelagic species that forage over vast areas of a seemingly featureless sea expanse. However, these species rely on extremely fine tuned sensory cues to recognize heterogeneity in the marine environment, ensuring foraging efficiency over areas of patchy productivity. Exactly what odors are used to build an odoriferous seascape remains to be studied, but the large expansion of the γ-c clade, with its high levels of polymorphism and underlying patterns of adaptive evolution may contribute to the necessary discriminatory power to be used in a genetic mechanism linking the olfactory system with navigation, foraging and homing and perhaps even with kin recognition and other critical social interactions in this and other avian systems.
Much remains to be learnt about the structure and functioning of sensory receptors in birds6,11,12,16, and the isolation and characterization of olfactory receptor loci in the Cory’s shearwater is a significant step towards understanding the role of this fascinating multi-gene family in birds and, more generally, in the genomic underpinnings of ecological adaptation.
Materials and Methods
Sampling and OR gene isolation
A blood sample from a female Cory’s shearwater was collected in Selvagem Grande Island, and kept in 95% ethanol until laboratory analyses. Genomic DNA (~1500 μg) was extracted with a phenol-chloroform standard protocol and used to build a cosmid library (built with SuperCos1 Cosmid Vector Kit, Stratagene, La Jolla, CA, USA, outsourced to America Pharma Source LLC). The library had inserts that varied in size between 30–42 Kb and a titer equivalent to 4X of estimated genome size. Since there is no genome size estimates for Procellariiformes, we assumed 1,36 Gb based on the average of all avian genome sizes available50, a value similar to the genome assembly size for Northern fulmar of 1,135Gb40. Cosmid clones were screened using Southern Blotting with three probes, two targeting primarily sequences from the γ-c OR clade and one sequences from the non γ-c OR clade25. However, it is noteworthy that all probes targeted highly similar, evolutionarily conserved coding motifs that are present across all OR gene families, as implied by the cross-hybridization results below. Probes were generated as described elsewhere25. A ~ 500 bp fragment was amplified in the shearwater, which was cloned and sequenced to confirm olfactory receptor identity (Supplementary Info). We picked 96 positive clones, approximately a third were positive for all three probes, and the remaining for either one or two probes.
Sequencing, assembly and annotation of OR gene-positive cosmids
The sequencing strategy aimed at generating long assemblies while minimizing costs and avoiding chimeras. Sanger sequencing generated ~ 900 bp long reads from cosmid ends which anchored each cosmid sequence. Depth of coverage was obtained from an equi-molar pool of non-tagged cosmids, using Illumina HiSeq 2000. 34 million Illumina 100 bp-long reads were used to build multiple assemblies at different depth of coverage using Velvet51. The best Illumina data assembly, with an estimated 139X coverage, consisted of 5,180 contigs (125 bp to 38 Kb in length), with a total cumulative length of 3.02 Mb (Supplementary Info). Guided by the Illumina data, the 96 cosmid clones were pooled into ten subsets that minimized within pool clone similarity. For each pool, DNA was sheared to 3Kb and a MID 454 GS FLX Titanium Paired End library was built and sequenced. A total of 541,133 reads were obtained for the 10 pools, with an average read length of 311 bp, resulting in an additional 20X coverage of each of the 96 cosmids. Hybrid assemblies were built with the Celera Assembler v.752 using all 454 data and varying numbers of Illumina reads. Further analyses proceeded on the most comprehensive hybrid assembly.
All scaffold and degenerate contigs (the latter contain repetitive sequences that could not be unambiguously assembled with the main scaffolds) were processed through the Institute for Genome Sciences’eukaryotic structural annotation pipeline, which was configured to use a combination of tools, including ab initio gene finders, protein aligners and model combiners. Gene finders including GeneMark-ES (GM-ES)53, Augustus v2.754 and Maker55 were run using chicken evidence data, and GeneID v1.456 and GlimmerHMM v3.0.157 used human and zebra finch informant data, respectively. Exonerate’s protein2genome option58 was run with a score cutoff of 500 and a cutoff of 75% to identify homologs to known chicken (n = 37,258 protein models) and zebra finch (n = 17,773) proteins. Finally, Evidence Modeler (EVm) r2012-06-2559 was used to combine evidence from spliced protein alignments and ab initio gene predictions into weighed consensus gene models.
Proteins were searched with Hidden Markov Models (HMMs) generated with HMMbuild using default settings and calibrated with HMMcalibrate. One HMM was built from full length OR genes (mostly from G. gallus and T. guttata), and the second was based on an alignment of partial OR gene sequences from eight different avian species25. The HMMs were validated by searching UniProt (Universal Protein Resource) using HMMsearch (EMBL_EBI) with a cut off value of 1e−10. A set of open reading frames (ORFs) was generated with ENSEMBL’s getorf which was searched using the new HMMs, with default settings. ORFs and EVm models matching HMMs were compared and uniqued, and the assembly location of each of the resulting genes determined. Proteins generated by EVm were annotated with a custom functional annotation pipeline, implemented within a reusable Ergatis pipeline, where input proteins are searched using several tools in parallel including HMMer360 NCBI-BLASTP (cutoff 1e-6), SignalP61, ScanProsite62 and TMHMM63. The HMM database used for HMMer3 is custom-made with TIGRFAMs, PFAM, among other custom records. Proteins which match the HMMs above trusted cut off levels inherit these annotations, or else BLAST results were used.
Phylogeny of shearwater OR genes
Phylogenetic trees were estimated for two different amino acid (AA) alignments, one including all OR shearwater sequences of length > 75AA (n = 220) (Supplementary Dataset 1) and the other only the full-length ORs (n = 20) (Supplementary Dataset 2). The alignments included complete OR sequences from chicken, zebra finch and green anole (Anolis carolinensis) from9 and all non-redundant Northern fulmar (Fulmarus glacialis) proteins from Protein database of GenBank. We use a non-OR rhodopsin-like family GPCR gene (Adenosine receptor A2b from the chicken, zebra finch and emperor penguin (Aptenodytes forsteri), respective GenBank accession numbers NP_990418, XP_002198489, XP_009276449) as outgroup21. Sequences were aligned with MAFFT 7.1264, using E-INS-i parameters, and the alignments were manually curated in Mesquite v.2.7565. Approximate maximum-likelihood trees were reconstructed with FastTree v2.1.1066 under the JTT evolutionary model67, implemented in the CIPRES Science Gateway68. Nodal support was evaluated with Shimodaira-Hasegawa (SH) tests with 1,000 bootstrap replicates66.
Estimates of positive selection on clade γ-c
The ratio of non-synonymous to synonymous substitutions (ω = dN/dS) is used as an indicator of the strength of selection on protein-coding genes. The presence of recombination or gene conversion, frequent in multi-gene families, precludes the use of methods that estimate selection based on a single phylogenetic tree69,70. We estimated dN/dS per codon with four codon-based maximum likelihood methods available in HYPHY71, as implemented in the Datamonkey webserver72. We accounted for the possibility of recombination using the GARD module (also implemented in Datamonkey72,73 to infer breakpoints in the alignment and generate tree topologies for each of the non-recombinant data partitions. We then used SLAC, FEL, REL70, MEME74 methods, which differ in assumptions for the distribution of rates across sites or lineages, and an integrative approach, which considers all codon sites detected by each method, to infer selection within each partition. A significance level of P < 0.1 was used for SLAC, FEL and MEME, and a Bayes factor >50 for REL70.
WEBLOGO (http://weblogo.berkeley.edu/logo.cgi, 75) was used to generate a sequence logo with the amino acid alignment of all γ-c complete shearwater olfactory receptor sequences, to facilitate visualization of sequence variation for the putative transmembrane (TM), intracellular (IC) and extracellular (EC) domains. The height of each letter corresponds to the relative frequency of each amino acid at a given position, reflecting sequence conservation.
Chimera76 (http://www.cgl.ucsf.edu/chimera/index.html) was used to display the location of the polymorphic sites on the secondary structure of a shearwater complete γ-c OR.
Ethical statement
The volume of the blood sample collected from the female shearwater was 150 μl (approximately 0.2% vol/weight). Sampling guidelines were approved by the Instituto da Conservação da Natureza e da Biodiversidade (ICNB) and by the Parque Natural da Madeira (currently Instituto das Florestas e Conservação da Natureza), Portugal under permit: 2/2012S. Sampling further followed the requirements of the Directive 2010/63/EU of the European Parliament and of the council for the protection of animals used for scientific purposes.
Data availability
Cory’s Shearwater cosmid library shotgun Illumina HiSeq and 454 data as well as the assembled scaffold and degenerate contigs and OR gene sequences will be deposited in NCBI under BioSample accession number SAMN13721836. For additional information contact corresponding author (MCS).
References
Niimura, Y. & Nei, M. Evolutionary dynamics of olfactory and other chemosensory receptor genes in vertebrates. J. Hum. Genet. 51, 505–517 (2006).
Caro, S. P., Balthazart, J. & Bonadonna, F. The perfume of reproduction in birds: chemosignaling in avian social life. Horm. Behav. 68, 25–42 (2015).
Mombaerts, P. Genes and ligands for odorant, vomeronasal and taste receptors. Nature Rev. NeuroSci. 5, 263–278 (2004).
Buck, L. & Axel, R. A novel multi-gene family may encode odorant receptors: a molecular basis for odor recognition. Cell 65, 175–187 (1991).
Nei, M., Niimura, Y. & Nozawa, M. The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat. Rev. Genet. 9, 951–963 (2008).
Khan, I. et al. Olfactory receptor subgenomes linked with broad ecological adaptation in Sauropsida. Mol. Biol. Evol. 32, 2832–2843 (2015).
Freitag, J., Beck, A., Ludwig, G., von Buchholtz, L. & Breer, H. On the origin of the olfactory receptor family: receptor genes of the jawless fish (Lampetra fluviatilis). Gene 226, 165–174 (1999).
Niimura, Y. & Nei, M. Evolutionary dynamics of olfactory receptor genes in fishes and tetrapods. Proc. Natl. Acad. Sci. USA 102, 6039–6044 (2005).
Steiger, S. S., Kuryshev, V. Y., Stensmyr, M. C., Kempenaers, B. & Mueller, J. C. A comparison of reptilian and avian olfactory receptor gene repertoire: species-specific expansion of group γ genes in birds. BMC Genomics 10, 446 (2009).
Niimura, Y. & Nei, M. Extensive gains and losses of olfactory receptor genes in mammalian evolution. PLoS One 2, e708 (2007).
Niimura, Y. Olfactory receptor multigene family in vertebrates: from the viewpoint of evolutionary genomics. Curr. Genomics 13, 103–114 (2012).
Vandewege, M. W. et al. Contrasting patterns of evolutionary diversification in the olfactory repertoires of reptile and bird genomes. Genome Biol. Evol. 8, 470–480 (2016).
Nei, M., Gu, X. & Sitnikova, T. Evolution by the birth-and-death process in multigene families of the vertebrate immune system. Proc. Natl. Acad. Sci. USA 94, 7799–7806 (1997).
Nei, M. & Rooney, A. P. Concerted and birth-and-death evolution of multi-gene families. Ann. Rev. Genet. 39, 121–152 (2005).
Gilad, Y., Wiebe, V., Przeworski, M., Lancet, D. & Pääbo, S. Loss of olfactory receptor genes coincides with the acquisition of full trichromatic vision in primates. PLoS Biol. 2, E5 (2004).
Steiger, S. S., Fidler, A. E., Mueller, J. C. & Kempenaers, B. Evidence for adaptive evolution of olfactory receptor genes in 9 bird species. J. Hered. 101, 325–333 (2010).
Alioto, T. S. & Ngai, J. The odorant receptor repertoire of teleost fish. BMC Genomics 6, 173 (2005).
Hayden, S. et al. A cluster of olfactory receptor genes linked to frugivory in bats. Mol. Biol. Evol. 31, 917–927 (2014).
Dehara, Y. et al. Characterization of squamate olfactory receptor genes and their transcripts by the high-throughput sequencing approach. Genome Biol. Evol. 4, 602–616 (2012).
Rouquier, S., Blancher, A. & Giorgi, D. The olfactory receptor gene repertoire in primates and mouse: evidence for reduction of the functional fraction in primates. Proc. Natl. Acad. Sci. USA 97, 2870–2874 (2000).
Niimura, Y. On the origin and evolution of vertebrate olfactory receptor genes: comparative genome analysis among 23 chordate species. Genome Biol. Evol. 1, 34–44 (2009).
Floriano, W. B., Vaidehi, N. & Goddard, W. A. Making sense of olfaction through predictions of the 3D structure and function of olfactory receptors. Chem. Senses 29, 269–290 (2004).
Man, O., Gilad, Y. & Lancet, D. Prediction of the odorant binding site of olfactory receptor proteins by human-mouse comparisons. Protein Sci. 13, 240–254 (2004).
Hagelin, J. C., Jones, I. L. & Rasmussen, L. E. L. A tangerine-scented social odour in a monogamous seabird. Proc. Biol. Sci. 270, 1323–1329 (2007).
Steiger, S. S., Fidler, A. E., Valcu, M. & Kempenaers, B. Avian olfactory receptor gene repertoires: evidence for a well-developed sense of smell in birds? Proc. Biol. Sci. 275, 2309–2317 (2008).
Zhan, X. et al. Peregrine and saker falcon genome sequences provide insights into evolution of a predatory lifestyle. Nature Gen. 45, 563–568 (2014).
Lu, Q., Wang, K., Lei, F., Yu, D. & Zhao, H. Penguins reduced olfactory receptor genes common to other waterbirds. Sci. Rep. 6, 31671 (2016).
Bang, B. G. & Cobb, S. Size of olfactory bulb in 108 species of birds. Auk 85, 55–61 (1968).
Zelenitsky, D. K., Therrien, F., Ridgely, R. C., McGee, A. R. & Witmer, L. M. Evolution of olfaction in non-avian theropod dinosaurs and birds. Proc. Biol. Sci. 278, 3625–3634 (2011).
Nevitt, G. A. & Bonadonna, F. Sensitivity to dimethyl sulphide suggests a mechanism for olfactory navigation by seabirds. Biol. Lett. 1, 303–305 (2005).
Dell’Ariccia, G. & Bonadonna, F. Back home at night or out until morning? Nycthemeral variation in homing of anosmic Cory’s Shearwater in a diurnal colony. J. Exp. Biol. 216, 1430–1433 (2013).
Gagliardo, A. et al. Oceanic navigation in Cory’s Shearwaters: evidence for a crucial role of olfactory cues for homing after displacement. J. Exp. Biol. 261, 2798–2805 (2013).
Reynolds, A. M., Cecere, J. G., Paiva, V. H., Ramos, J. A. & Focardi, S. Pelagic seabird flight patterns are consistent with a reliance on olfactory maps for oceanic navigation. Proc. Biol. Sci. 282, 20150468 (2015).
Hughes, G. M., Finarelli, J. A., Murphy, W. J., Higgins, D. G. & Teeling, E. C. The birth and death of olfactory receptor gene families in mammalian niche adaptation. Mol. Bol. Evol. 35, 1390–1406 (2018).
Perry, B. W. et al. Molecular adaptations for sensing and securing prey and insight into amniote genome diversity from the Garter snake genome. Gen. Biol. Evol. 10, 2110–2129 (2018).
Worley, K. C., Richards, S. & Rogers, J. The value of new genome references. Exp. Cell Res. 358, 433–438 (2016).
Miller, J. R. et al. Hybrid assembly with long and short reads improves discovery of gene family expansions. BMC Genomics. 18, 541 (2017).
Treangen, T. J. & Salzberg, S. L. Repetitive DNA and next-generation sequencing: computational challenges and solutions. Nat. Rev. Genet. 13, 36–46 (2011).
Hughes, G. M., Gang, L., Murphy, W. J., Higgins, D. G. & Teeling, E. C. Using Illumina next generation sequencing technologies to sequence multigene families in the novo species. Mol. Ecol. Res. 13, 510–521 (2013).
Zhang, G., Li, B., Gilbert, M. T. P., Jarvis, E. D. & Wang, J. The Avian Genome Consortium. Comparative genomic data of the Avian Phylogenomics Project. GigaScience 3, 26 (2014).
Zhang, X. & Firestein, S. Genomics of olfactory receptors. Res. Probl. Cell Differ. 47, 25–36 (2008).
Bang, B. G. The olfactory apparatus of tubenosed birds. Acta Anat 65, 391–415 (1966).
Behrens, M. et al. ORA1, a zebrafish olfactory receptor ancestral to all mammalian V1R genes, recognizes 4-hydroxyphenylacetic acid, a putative reproductive hormone. J. Biol. Chem. 289, 19778–19788 (2014).
Jarvis, E. D. Perspectives from the Avian Phylogenomics Project: Questions that Can Be Answered with Sequencing All Genomes of a Vertebrate Class. Annu. Rev. Anim. Biosci. 4, 45–59 (2016).
Organ C. L., Rasmussen M., Baldwin M., Kellis M. & Edwards S. V. Phylogenomic approach to the evolutionary dynamics of gene duplication in birds in Evolution after gene duplication (eds. Dittmar K. & Liberles D.), 253–268 (John Wiley & Sons, Inc., 2010).
Lynch, M. & Conery, J. S. The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155 (2000).
Zhang, J. Evolution by gene duplication: an update. TREE 18, 292–298 (2003).
Pilpel, Y. & Lancet, D. The variable and conserved interfaces of modeled olfactory receptor proteins. Protein Sci. 8, 969–977 (1999).
Singer, M. S. & Sheperd, G. M. Molecular modeling of ligand-receptor interactions in the OR5 olfactory receptor. Neuroreport 5, 1297–1300 (1994).
Gregory, T. R. A bird’s-eye view of the C-value enigma: genome size, cell size and metabolic rate in the class Aves. Evolution 56, 121–130 (2002).
Zerbino, D. R. & Birney, E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18, 821–829 (2008).
Myers, E. W. et al. A whole-genome assembly of Drosophila. Science 287, 2196–2204 (2000).
Lukashin, A. V. & Borodosky, M. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 26, 1107–1115 (1998).
Stanke, M. & Waack, S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics 19, 215–225 (2003).
Cantarel, B. L. et al. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18, 188–196 (2008).
Blanco E., Parra G., Guigó, R Using geneid to identify genes in Protocols in Bioinformatics (ed. Baxevanis A Ed) Current Unit 4.3. (John Wiley & Sons Inc., 2002).
Majoros, W. H., Pertea, M. & Salzberg, S. L. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene finders. Bioinformatics 20, 2878–2879 (2004).
Slater, G. S. & Birney, E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics 6, 31 (2005).
Haas, B. J. et al. Automated eukaryotic gene structure annoatation using EvidenceModeler and the program to assemble spliced alignments. Genome Biol. 9, R7 (2008).
Mistry, J., Finn, R. D., Eddy, S. R., Bateman, A. & Punta, M. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 41, 1–10 (2013).
Petersen, T. N., Brunak, S., von Heijne, G. & Nielsen, H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature Meth. 8, 785–786 (2011).
De Castro E. et al. ScanProsite: detection of PROSITE signature matches and Pro-Rule-associated functional and structural residues in proteins. Nucleic Acid. Res. 34: W362-365 (2006).
Krogh, A., Larsson, B., von Heijne, G. & Sonnhammer, E. L. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 (2001).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Maddison, W. P. & Maddison, D. R. Mesquite: a modular system for evolutionary analysis. Version 2, 75 (2011).
Price, M. N., Dehal, P. S. & Arkin, A. P. FastTree2 – Approximately maximum-likelihood trees for large alignments. PLoS ONE 5, e9490 (2010).
Jones, D. T., Taylor, W. R. & Thornton, J. M. The rapid generation of mutation data matrices from protein sequences. Comp. Appl. Biosci. 8, 275–282 (1992).
Miller M.A., Pfeiffer W., Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In Proceedings of the Gateway Computing Environments Workshop (GCE), pp 1–8. New Orleans, LA (2010).
Anisimova, M., Nielsen, R. & Yang, Z. H. Effect of recombination on the accuracy of the likelihood method for detecting positive selection at amino acid sites. Genetics 164, 1229–1236 (2003).
Pond, S. L. K. & Frost, S. D. W. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol. Biol. Evol. 22, 1208–1222 (2005).
Pond, S. L. K., Frost, S. D. W. & Muse, S. V. HyPhy: hypothesis testing using phylogenies. Bioinformatics 21, 676–679 (2005).
Pond, S. L. K. & Frost, S. D. W. Datamonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21, 2531–2533 (2005).
Pond, S. L. K., Posada, D., Gravenor, M. B., Woelk, C. H. & Frost, S. D. W. GARD: a genetic algorithm for recombination detection. Bioinformatics 22, 3096–3098 (2006).
Murrell, B. et al. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 8, e1002764 (2012).
Crooks, G. E., Hon, G., Chandonia, J. M. & Brenner, S. E. WebLogo: A sequence logo generator. Genome Res. 14, 1188–1190 (2004).
Pettersen, E. F. et al. UCSF Chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Acknowledgements
We are indebted to P Catry and JP Granadeiro for collecting the Cory’s shearwater sample from the Selvagem Grande I.We thank João Moreno for his help with the protein handling software. We thank researchers from America Pharma Source, LLC for their advice during the building and screening of the cosmid library. This work was financed by Fundação para a Ciência e a Tecnologia (FCT), Portugal, through research project PTDC/BIA-BDE/100118/2008. MCS was supported by a post-doctoral research fellowship SFRH/BPD/85700/2012 from FCT, Portugal. JCS and JBM were supported in part by NSF’s Dimensions of Biodiversity program grant OCE-1046371.
Author information
Authors and Affiliations
Contributions
M.C.S. conceived and designed the study, carried out molecular work, analysed the data and drafted the manuscript. M.C. and S.D. carried out bioinformatics analyses, including cosmid assemblies and annotations. J.M. carried out data analysis and helped draft the manuscript. M.M.C. helped coordinate and draft the manuscript. J.C.S. contributed to study design, coordinated data collection and analyses and helped write the manuscript. All authors gave final approval for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
C. Silva, M., Chibucos, M., Munro, J.B. et al. Signature of adaptive evolution in olfactory receptor genes in Cory’s Shearwater supports molecular basis for smell in procellariiform seabirds. Sci Rep 10, 543 (2020). https://doi.org/10.1038/s41598-019-56950-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-56950-6
This article is cited by
-
The role of the odorant receptors in the formation of the sensory map
BMC Biology (2021)
-
A unified nomenclature for vertebrate olfactory receptors
BMC Evolutionary Biology (2020)
-
Genome-wide selective sweep analysis of the high-altitude adaptability of yaks by using the copy number variant
3 Biotech (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.