Abstract
Invasive aspergillosis (IA) is a life-threatening complication among allogeneic hematopoietic stem cell transplant (alloSCT) recipients. Despite well known risk factors and different available assays, diagnosis of invasive aspergillosis remains challenging. 103 clinical variables from patients with hematological malignancies and subsequent alloSCT were collected. Associations between collected variables and patients with (n = 36) and without IA (n = 36) were investigated by applying univariate and multivariable logistic regression. The predictive power of the final model was tested in an independent patient cohort (23 IA cases and 25 control patients). Findings were investigated further by in vitro studies, which analysed the effect of etanercept on A. fumigatus-stimulated macrophages at the gene expression and cytokine secretion. Additionally, the release of C-X-C motif chemokine ligand 10 (CXCL10) in patient sera was studied. Low monocyte concentration (p = 4.8 × 10−06), severe GvHD of the gut (grade 2–4) (p = 1.08 × 10−02) and etanercept treatment of GvHD (p = 3.5 × 10−03) were significantly associated with IA. Our studies showed that etanercept lowers CXCL10 concentrations in vitro and ex vivo and down-regulates genes involved in immune responses and TNF-alpha signaling. Our study offers clinicians new information regarding risk factors for IA including low monocyte counts and administration of etanercept. After necessary validation, such information may be used for decision making regarding antifungal prophylaxis or closely monitoring patients at risk.
Similar content being viewed by others
Introduction
Invasive aspergillosis (IA) is the most relevant mould infection in immunocompromised patients. It is associated with significant mortality in patients with haematological malignancies and those receiving allogeneic stem cell (alloSCT) or solid organ transplants1,2,3. Diagnosis of IA is challenging as clinical symptoms are often non-specific and conventional diagnosis is poor4,5. Methods such as high resolution computed tomography scans6 only show typical signs once infection is established, and even then, specific signs can be transient and could be associated with manifestations other than IA4,7. Serological tests detecting galactomannan (GM) and β-D-glucan have low positive predictive values and are better used for exclusion rather than for diagnosis of IA4,5,7. PCR-based assays to detect fungal DNA in patient specimens operate at the very limit of detection, due to the small amounts of fungal DNA recovered from blood samples and positivity occurs only transiently8. Although the testing of bronchoalveolar lavage fluid is likely associated with increased quantities of biomarker, sampling is often contra-indicated by the patient’s underlying clinical condition. As IA is a multi-factorial disease, it seems promising to combine different biomarkers, such as the use of GM and PCR assays to improve diagnosis9,10,11.
While there is limited evidence of current diagnostic assays being used to monitor the fungal load for prognostic purposes or during antifungal therapy, host biomarkers might better reflect the clinical course of IA. Furthermore, host biomarkers might allow individual pre-emptive risk stratification for IA. These markers include genetic (single nucleotide polymorphisms (SNPs)), clinical, environmental, and personal factors, which determine the individuals risk for IA and potentially the course and outcome of IA12,13. They include among others advanced age, the type of underlying disease, the dose and duration of corticosteroid therapy, viral infection (e.g. cytomegalovirus (CMV) or respiratory virus infection), lymphopenia, and neutropenia13,14. Although many of these factors have been individually associated with IA, defining patient-specific risk profiles for IA remains extremely challenging, as knowledge about the combination of risk factors with and without fungal biomarkers remains scarce.
The aim of this study was to identify distinct new risk factors, which in combination with known risk factors and fungal biomarkers might allow risk stratification and improve diagnosis of IA in patients that underwent alloSCT. Therefore, our approach is to identify significant associations between a large number of individual clinical variables and the occurrence of IA in parallel to identify new significant risk factors for the development of IA after alloSCT. Such risk factors might in future allow the definition of personalized stratification protocols for alloSCT patients according to their individual risk of developing IA.
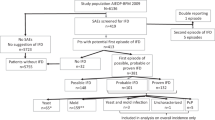
We present a longitudinal case-control study, where post stem cell transplantation clinical variables were collected twice weekly, when feasible (Fig. 1). We compared 36 alloSCT patients with probable IA and 36 alloSCT controls with no evidence of fungal infection. The relevance of variables identified in this study as significantly associated with IA was validated in a confirmatory study of 23 cases of probable IA and 25 control alloSCT patients.
Overall study design and workflow. Clinical variables from allogeneic hematopoietic stem cell transplant (alloSCT) recipients were collected after stem cell transplantation twice weekly, whenever feasible. Monocyte count and treatment with etanercept were confirmed in validation cohort and therefore studied further.
To support and interpret our findings, the function of the Tumor Necrosis Factor-alpha (TNF-alpha) blocker etanercept, which has been also used to treat chronic and acute graft-versus-host disease (GvHD), was studied on cytokine secretion and gene expression profiles were investigated in vitro in monocyte-derived macrophages stimulated with Aspergillus fumigatus. Furthermore, CXCL10 release in serum samples from probable IA patients with or without etanercept treatment was investigated.
Results
We performed a case-control study, consisting of 36 patients suffering from probable IA and 36 control patients without any signs of fungal infection after alloSCT. Detailed information regarding patient demographics is shown in Materials and Methods (paragraph “selection of patients”) and in Table 1. From these patients, we collected time-resolved data related to 96 binary clinical and seven continuous variables.
Among them, seven binary variables were significantly associated with IA (p < 0.05, Table 2). However, positive Galactomannan (GM) ELISA and Aspergillus PCR test results in patients with probable IA do not represent independent variables as they are linked to the diagnosis of IA6,15. The use of nitroimidazole and dexamethasone and the presence of Enterococcus faecalis in the blood culture could not be confirmed in the validation cohort as an independent risk factor. Thus, we excluded these variables from the following studies and focused our further analyses and functional studies on the two remaining significant variables, more precisely the administration of etanercept and GvHD of the gut (grade ≥ 2). In the next step, we screened the specified continuous variables for association with IA.
Low monocyte concentration is a quantitative risk factor for IA after alloSCT
Temporally continuous variables, including the blood counts of various types of leukocytes were investigated over a period of 4 weeks prior to the diagnosis of IA. One week before diagnosis the concentration of total leukocytes, monocytes and granulocytes were all significantly associated with the diagnosis of IA (Table 3). The total leukocyte concentration was significantly reduced to 611 cells/µl in cases in contrast to 1101 cells/µl in controls (odds ratio (OR) = 1.14, 95% confidence interval (CI) = 1.1 to 1.3, p = 1.4 × 10−3), while granulocyte concentration reached levels of 354 cells/µl (cases) and 648 cells/µl (controls) (OR = 1.33, 95% CI = 1.1 to 1.7, p = 4.1 × 10−03) (Table 3).
In particular, monocyte concentrations were relatively lowered in cases to 44 cells/µl compared to controls 162 cells/µl (OR = 13.26, 95% CI = 3.3 to 80.2, p = 4.8 × 10−06) (Fig. 2). The earlier weeks (up to four weeks prior to the diagnosis) exhibited a similar significant reduction of the monocyte concentration (Fig. 2). In validation data, we observe significantly lowered monocytes, but only starting at second week before diagnosis (OR = 4.97, 95% CI = 1.23 to 32.19, p = 0.018), whereas the first week before diagnosis shows no changes in monocyte concentration.
Boxplot showing concentrations of monocytes up to four weeks before IA diagnosis for cases. Control data (white boxplots) was matched to the time point of alloSCT of all cases (blue boxplots) per week. Differences were determined by the Likelihood-Ratio test. Nominally significant differences (i.e. p ≤ 0.05) were observed for all 4 weeks.
Graft versus Host Disease of the gut (grade ≥ 2) is a risk factor for IA
Severe GvHD of the gut (grade 2–4) was significantly associated with IA (OR = 4.52, 95% CI = 1.4 to 17.71, p = 1.08 × 10−02). Case patients developed GvHD with a risk of 36% (13/36), while controls exhibited a lower risk of 11% (4/36). To verify this result, we also tested the significance of GvHD in our validation cohort result (OR = 6.92, 95% CI = 1.51 to 50.24, p = 1.1 × 10−02). In comparison, a general GvHD variable which also included less severe types of other organs (e.g. skin) was not significantly associated with IA.
Etanercept is a binary risk factor for IA after alloSCT
The TNF-alpha blocker etanercept, which is frequently used for treatment of autoimmune inflammatory diseases, is also used to treat severe refractory GvHD.
From the group of drug-linked variables, etanercept was significantly associated with IA (OR = 6.22, 95% CI = 1.8 to 29.4, p = 3.5 × 10−03). In fact, the probability of getting IA after alloSCT under etanercept treatment among cases was 36% (13/36) in contrast to 8% (3/36) in controls. To validate etanercept as a risk factor for IA after alloSCT, we performed our statistical analyses in the validation cohort, which showed that 80% (8/10) of the patients who received etanercept treatment developed IA (OR = 6.13, 95% CI = 1.32 to 44.5, p < 0.019). Interestingly, both, severe GvHD of the gut (grade 2–4) and the use of etanercept were significantly associated with IA in univariate models. Therefore, we tested the effect of etanercept, when added as a new variable to the GvHD model. Notably, the predictive power of the model was significantly increased (p = 0.0152). In fact, patients with etanercept treatment of severe GvHD showed the highest risk to develop IA (10/10, 100%), compared to a lower risk (3/7, 43%) for patients developing GvHD of the gut (grade ≥ 2), which were not treated with etanercept (Table 2).
Combination of risk factors
A multiple logistic regression model combined the different selected variables into a single model. This allowed the prediction of disease status based on knowledge about GvHD, etanercept treatment in combination with the appearance of low monocyte counts.
As monocyte concentration significantly varied over time, we generated a model based on all data up to four weeks prior to the diagnosis of IA. For model evaluation, we applied the model subsequently on the validation data. Receiver operating characteristic (ROC) analysis was used to quantify the performance of the model. In Fig. 3, we show the resulting ROC curves for each week. The predicted diagnostic model performs exceptionally well on the validation data within week 2 (AUC = 0.85, 95% CI = 0.73 to 1) and 3 (AUC = 0.78, 95% CI = 0.54 to 1), while in week 1 (AUC = 0.62, 95% CI = 0.31 to 0.88) and week 4 (AUC = 0.68, 95% CI = 0.36 to 0.89) the model still provides a good classification. One explanation is that several cases appear to have elevated monocyte counts in week 1, which leads to lowered model sensitivity.
The effect of etanercept on CXCL10 secretion
To identify and interpret any potential influence of etanercept on the immune response against A. fumigatus, we performed a variety of functional in vitro assays. Human primary MDM were selected, as they are among the most important cells involved in defense against IA. MDM were co-cultured with live A. fumigatus germ tubes in presence or absence of etanercept.
As determined by multiplex ELISA analysis of the culture supernatants, confrontation of MDM with A. fumigatus induced a strong inflammatory response characterized by significant up-regulation of ten cytokines (Supplementary Table S2a). Treatment of unstimulated MDM with etanercept led to low secretion of IL-1RA and MCP-1 (Supplementary Table S2b). As expected, we detected lower TNF-alpha levels (p = 0.0104) after stimulation of MDM with A. fumigatus germ tubes in the presence of etanercept than compared to MDM which were infected with A. fumigatus germ tubes only (Fig. 4, Supplementary Table S2c). Interestingly, our data also showed a strong reduction of CXCL10 (IP-10) release (p = 0.0004) in etanercept-treated MDM stimulated with A. fumigatus germ tubes (Fig. 4).
Comparison of cytokine release in monocyte-derived macrophages (MDM) stimulated with A. fumigatus in presence or absence of etanercept. Differences were determined by the Wilcoxon-Mann-Whitney test (p ≤ 0.05 are highlighted by *). (a) Secretion of cytokines TNF-alpha (p = 0.0104) and CXCL10 (p = 0.0004) is significantly downregulated in MDM stimulated with A. fumigatus under etanercept treatment (AF_ET) compared MDM stimulated with A. fumigatus only (AF). (b) CXCL10 release in serum samples from eight IA patients without etanercept treatment (untreated) and IA patients under etanercept treatment (treated).
To validate this finding, CXCL10 levels in serum samples from 8 selected patients with probable IA were performed. Our data showed that patients with probable IA and etanercept treatment had significantly lower CXCL10 serum concentrations (p = 0.017) than patients without etanercept treatment (Fig. 4).
Furthermore, to achieve a more holistic view on the influence of etanercept on the macrophage – A. fumigatus interaction in vitro, we performed RNA-sequencing and compared gene expression of MDMs treated with etanercept and confronted with A. fumigatus germ tubes to gene expression of MDM confronted with A. fumigatus germ tubes only. Non-unstimulated MDM treated with etanercept served as a control. Confrontation of MDM with A. fumigatus germ tubes led to a significant induction of the expression of 382 genes while only 6 genes were repressed. A. fumigatus induces a fast and strong induction of genes involved in immune response and inflammation. Our data showed a more than 80-fold induction of the genes encoding for the chemokines CXCL2, CXCL3, CCL4 and CXCL10, respectively. Interestingly, etanercept treatment led to only minor transcriptional changes in unstimulated MDM. The drug did not significantly induce the expression of any gene while it repressed the expression of six genes encoding metallothioneins (MT2A, MT1E, MT1X, MT1F, MT1H, MT1M).
Next, we compared gene expression profiles of MDM treated with etanercept and A. fumigatus germ tubes to gene expression profiles of MDM confronted with the fungus only. Among the 18 differentially expressed genes which were all down-regulated in MDM treated with etanercept and A. fumigatus, were genes encoding for metallothioneins (mentioned above) and genes encoding CXCL10, DRAK1, ICAM1, BID, FUT4, RELB, RASGRP1, RNF144B, POU2F2, BIRC3, SLC2A6 and BCL3 (Fig. 5a, Supplementary Table S3). Although TNF-alpha expression was not significantly altered after etanercept treatment, genes located downstream of TNF-alpha including the transcription factor RELB, the intercellular adhesion molecule 1 (ICAM1) and the apoptotic molecules BCL3 and BIRC3 were significantly downregulated in the presence of A. fumigatus and etanercept, compared to MDM stimulated with A. fumigatus only (Fig. 5).
Regulation of NF-kappaB target genes which are differentially expressed in monocyte-derived macrophages treated with A. fumigatus and etanercept (AF + ET) compared to A. fumigatus (AF) only. Down-regulated genes are marked with blue color. (a) The heatmap displays downregulation of genes after addition of etanercept and the regulatory network (b) summarizes the effect of etanercept on the TNF-alpha signaling pathway.
Discussion
Among opportunistic infections in immunocompromised patients, IA remains most challenging. The detection of fungal biomarkers in blood is imperfect due to variable sensitivity and specificity of the available diagnostic assays, especially in non-neutropenic patients or those receiving antifungal therapy or mold-active prophylaxis. Furthermore, positivity often occurs transiently as many diagnostic assays are run at the very limit of detection, due to the small amounts of fungal biomarkers. Consequently, and in contrast to many other infectious agents, following the fungal load for prognostic purposes or antifungal therapy monitoring is limited with the current diagnostic tools. Thus, the discovery and validation of new biomarkers are mandatory. This includes host biomarkers, which might better reflect the clinical course of IA and allow monitoring the response to antifungal therapy. Furthermore, host biomarkers might also allow risk stratification leading to an earlier start of antifungal treatment in afebrile patients thus resulting in more comprehensive pre-emptive treatment strategies.
Among the biomarkers described in this study, low monocyte counts up to four weeks prior to the diagnosis of probable IA was significantly associated with disease. Monocytes serve three main functions, including phagocytosis of microbes, the presentation of antigens to cells of the adaptive immune system, and the production of inflammatory cytokines and chemokines16. Moreover, monocytes also act as precursors for other inflammatory immune cells17. Therefore, reduction in monocyte levels might lead to a disrupted immune response. Our study revealed that monocytopenia occurred in patients with probable IA to a mean concentration of 44 cells/µl blood, compared to 162 cells/µl in control patients one week before diagnosis of IA (Fig. 2). These results imply that monocytopenia should be regarded as a risk factor for IA after alloSCT.
The high classification performance of the multivariable model on the validation data (validation cohort) underlines the predictive power of our variables (monocytes, GvHD and etanercept), while the temporal variation of the AUC performance remains a challenge.
Etanercept is a TNF-alpha blocker that antagonizes ligand binding and activation of tumor necrosis factor receptor 2 (TNFR2) and consequently inhibits downstream signaling including NF-κB activation. It can bind both, the soluble and the membrane form of TNF-alpha molecules removing them from the circulation18. TNF-alpha is a key cytokine involved in inflammatory and immune responses and has been shown to play a crucial role during fungal infections19. Thus, the use of drugs that block host immunity against microorganisms likely increases the risk of infection19,20,21,22. Since etanercept is most commonly used for the treatment of autoimmune diseases, many studies have focused on investigating mechanisms of action and side-effects of this drug among rheumatoid arthritis (RA) patients22,23. Etanercept has been already associated with an increased incidence of opportunistic infections in RA patients19, including bacterial and viral infections20,21,22,24.
Salvage therapy with high-dose corticosteroids and etanercept is used only in steroid-refractory GvHD with gastrointestinal involvement25,26,27. Until now, data on fungal infections associated with TNF-alpha blockade were limited to isolated cases or small number of patients. In a previous study from Tsiodras and colleagues21 it was shown that the most prevalent invasive fungal infections associated with TNF-alpha antagonist treatment were histoplasmosis (30%), candidiasis (23%) and aspergillosis (23%). This study focused on fungal infections associated with TNF-alpha blockade from 1966 to 2007 and revealed that among different patient groups receiving TNF-alpha blocker therapy, the GvHD group showed the worst clinical picture21. Although etanercept is available for the treatment of severe GvHD, little is known about its mechanism of action in association with Aspergillus spp. infections.
In vitro studies allow the reproduction of results observed in vivo by using highly controlled and standardized conditions. MDM represent a major TNF-alpha producing cell population of the innate immune system and play a major role in the immune defence against A. fumigatus. Confrontation of MDM with etanercept in vitro resulted in down-regulation of 6 genes that encode metallothioneins which are involved in the maintenance of zinc homeostasis28. By binding and releasing zinc, metallothioneins regulate zinc levels within the body, which influences the activation and binding of a variety of transcription factors. In addition, and as expected, stimulation of immune cells with A. fumigatus resulted in strong induction of genes involved in inflammatory responses29,30. Scicluna and colleagues31 reported that etanercept pre-treatment leads to a decreased transcriptional response to LPS. Furthermore, Tan and colleagues23 showed regulation of multiple inflammatory pathways in patients with psoriasis treated with etanercept. Data from our study showed the down-regulation of genes involved in immune responses and TNF-alpha signalling (Fig. 5). Significantly, down-regulated genes were involved in NFkB signalling, antimicrobial humoral response and apoptotic processes (ICAM1, BCL3, BID, BIRC3, RELB, DRAK1 and FUT4) (Supplementary Table S3). For example, ICAM1 is an important adhesion molecule, expressed in immune cells32 and plays a major role in the immune response in association with T-cells33. BCL3 (B-cell lymphoma 3) is a regulator of the NFkB activity and essential for the proper development, survival and activity of adaptive immune cells34. BIRC3 (Baculoviral IAP repeat-containing protein3) encodes cIAP2, which is an element of the TNFR2 receptor complex and therefore plays an important role in the TNF-alpha signalling pathway35. RELB (RELB Proto-Oncogene, NF-KB Subunit) is one of the members of the NFkB family and is activated by Dectin-1, which recognizes specific morphologies of A. fumigatus36,37.
Our in vitro studies showed that the presence of etanercept significantly decreases the secretion of the chemokine CXCL10 from MDM stimulated with A. fumigatus (p = 0.0004, Fig. 4). Furthermore, our data suggest that patients with IA being treated etanercept have lower CXCL10 serum concentrations compared to IA patients without treatment (Fig. 4). All patients analysed suffered from IA relatively late after alloSCT (range 108–219 days) and IA occurred during or immediately after administration of etanercept.
It is well established that high levels of TNF-alpha activate CXCL10 secretion via the STAT1 – NFkB1 (p50) – RelA (p65) pathway38. CXCL10 in blood and tissue exhibits strong chemotactic properties for monocytes, Natural Killer cells and CD4 + T cells leading to the conclusion that decreased CXCL10 levels prevent the presence of a variety of immune cell populations important for successful defense of IA39,40. Furthermore, Zhao and colleagues recently demonstrated a direct role of CXCL10 in IFN-γ-primed human monocytes to induce a robust production of proinflammatory cytokines, such as IL-12 and IL-2341. This leads to the conclusion that reduced concentrations of CXCL10 (in addition to already markedly reduced TNF-alpha concentrations due to etanercept treatment) might multimodally hamper a successful defense against A. fumigatus in patients treated with etanercept.
In this context, our own group demonstrated in 2008 that healthy individuals have lower CXCL10 concentrations in sera in comparison to patients after alloSCT. Furthermore, alloSCT patients with no fungal infection showed lower CXCL10 concentrations in comparison to patients with proven or probable IA, being highest in those patients who survived IA42. Moreover, SNPs in CXCL10 were described to be associated with a reduced expression of CXCL10 and an increased risk of IA42, which again shows the comprehensive functional relevance of this chemokine during Aspergillus infections.
Taken together, our in vitro studies show the ability of etanercept to inhibit immune responses through modulation of NFkB activity, TNF-alpha activity and CXCL10 release, suggesting that etanercept treatment is an important risk factor for the development of IA.
In conclusion, our study provides new and valuable information on potential factors predicting the risk for IA in alloSCT recipients. They also provide mechanistic and functional details on how etanercept treatment lowers CXCL10 concentrations in vitro and in vivo, thereby multiplying the effect of already diminished TNF-alpha concentrations. This study should direct clinicians to the fact that low monocyte counts and the administration of etanercept necessitate special caveats, such as continuous monitoring of patients with highly sensitive diagnostic assays and the use of anti-Aspergillus prophylaxis. Further large multi-centre studies will be necessary to identify new biomarkers and to evaluate existing markers on a broader basis considering multi-factorial and multimodal analysis strategies.
Materials and Methods
Characterization of clinical variables
A collection of 96 static and seven dynamic, i.e. temporal, variables were defined previously (Supplementary Table 1) to acquire patient characteristics in the frame of the study “Aspergillosis – Individual Risk Estimation (ASPIRE, unpublished data). The static variables included age, gender, underlying disease, stem cell type, treatment variables such as the use of antibiotics, antifungal drugs, and drugs for immune suppression. Temporal variables comprised blood cell counts including total leukocytes, monocytes, granulocytes and lymphocytes.
Selection of patients
Between 2007 and 2013, patients undergoing alloSCT at the University Hospital of Würzburg were followed and categorized according to the first revision of the EORTC/MSG definitions6. Among them, a total of 36 patients with probable IA yielded complete clinical data records containing all 103 hitherto defined variables mentioned above. Thus, these patients qualified for inclusion into the current study.
Cases (with IA) and respective controls without signs of IA (n = 36, respectively) were balanced according to gender, age and underlying diseases (Table 1). All study patients suffered from defined haematological malignancies and subsequently underwent an alloSCT at the University Hospital Wuerzburg43.
The main underlying disease was acute myeloid leukemia (AML, n = 20) followed by multiple myeloma (n = 15) (patient demographics are shown in Table 1). The total patient cohort consisted of 25 females and 47 males with an average age of 55 years (range: 20–72 years). In female patients, AML was the main underlying disease in both cases (n = 5) and controls (n = 4), followed by other diseases (cases, n = 4; controls, n = 3), multiple myeloma (cases, n = 2; controls, n = 2), myelodysplastic syndrome (cases, n = 1; controls, n = 2), and acute lymphoblastic leukemia (cases, n = 1; controls, n = 1). In male patients, multiple myeloma (controls, n = 9) and AML (cases, n = 8) were the main underlying diseases, followed by myelodysplastic syndrome (cases, n = 3; controls, n = 2), chronic lymphocytic leukemia (cases, n = 3; controls, n = 2), acute lymphoblastic leukemia (cases, n = 4; controls, n = 1), and other diseases (cases, n = 3; controls, n = 7).
Controls did not show any signs of fungal infection. To study temporal changes after alloSCT, we collected all variables (Supplementary Table S3) twice weekly (when available) starting directly after SCT (day + 0) until the patient was discharged from the hospital or died. Patients developed IA on average 105 days after alloSCT.
To confirm the most relevant results of this study, a second, independent validation cohort was defined. For this second patient cohort, only information about significant variables identified by the first cohort was collected. Patients, within complete data for103 variables and consequently excluded from the first cohort were included in the validation cohort. Again, patients who suffered from hematological malignancies and received alloSCT between 2007 and 2015 were considered. This cohort consisted of 23 cases (with probable IA) and 25 controls, with no evidence of fungal infection. The cohort comprised 25 females and 23 males with an average age of 56 years (range: 26–72 years). The main underlying disease was AML (n = 28) followed by multiple myeloma (n = 5). The average time from alloSCT to IA diagnosis was 101 days.
Blood cell counts
Quantitative data on blood cell counts from cases were obtained weekly over a four week window prior to the diagnosis of IA, using the date of diagnosis of IA as a reference time point. Suitable controls were matched considering only samples with a similar temporal distance to the date of alloSCT as a reference.
In vitro generation of human monocyte-derived macrophages (MDM)
Peripheral blood mononuclear cells (PBMC) were isolated from three healthy anonymous volunteers (Institute of Transfusion Medicine and Haemotherapy, University Hospital Würzburg) by density gradient centrifugation using Biocoll separating solution (Biochrom). Monocytes were purified from PBMCs by magnetic cell sorting using CD14 microbeads (Miltenyi Biotech) and incubated at 37 °C and 5% CO2 atmosphere in RPMI-1640 supplemented with 10% heat-inactivated FCS, 120 µg/ml Refobacin (Merck) and 25 ng/ml GM-CSF (Sanofi-Aventis). MDM were used at day six for further analyses.
Aspergillus fumigatus culture
Aspergillus fumigatus ATCC 46645 (American Type Culture Collection) was used as a stimulus for all experiments. Cultivation of A. fumigatus and generation of germ tubes was according protocol described by Hefter and collegues44. After overnight incubation, germ tubes were centrifuged for 5 minutes at 5000xg and resuspended in fresh medium. MDM were incubated for 6 hours with live A. fumigatus germ tubes at a multiplicity of infection (MOI) of 0.5 at 37 °C and 5% CO2 atmosphere.
Etanercept
The effects of etanercept (Enbrel®, Pfizer, Germany) on macrophage gene expression profiles and cytokine response were evaluated at a concentration of 2 µg/ml, which is comparable with concentrations found in plasma of arthritic patients45,46. MDM were challenged with etanercept for 6 hours at 37 °C in a 5% CO2 atmosphere.
RNA extraction, RNA sequencing and RNAseq data processing
The cell pellet was resuspended in RNAprotect (Qiagen) and RNA was extracted using the RNeasy mini kit (Qiagen) according to the manufacturer’s protocol. RNA concentration and purity were determined in a NanoDrop ND-1000 spectral photometer (Thermo Fisher Scientific). The integrity of all samples was analysed in a 2100 Bioanalyzer (Agilent Technologies, Germany) using the RNA 6000 Nano LabChip Kits according to the manufacturer’s instructions (Agilent Technologies). All samples had RIN values above 8.5 and were sent for library preparation, rRNA removal, and 75 bp single-end sequencing on an Illumina Next Seq 500 to IMGM Laboratories GmbH (Munich, Germany). Raw sequencing reads were deposited at Gene Expression Omnibus (GSE113254). Reads were quality checked using FastQC and trimmed using Trimmomatic (v 0.36, window size 15 average quality 25, MINLEN:30)47. Quality trimmed reads were mapped onto the human reference genome (GRCh38) using HiSat2 (v 2.1.0) by supplying GFF file48. A number of reads were counted using the Rsubread package49 together with the gene structural annotation (GRCh38-GFF file). Raw counts were normalized using RPKM values. Differentially expressed genes were detected using DESeq250 with an adjusted p-value of ≤ 0.05 combined with an absolute log2-fold-change cutoff of at least 1.5. Enriched Gene Ontology categories were detected using the tool FungiFun251. The following genes were further analysed in a gene regulatory network: BCL3 (ENSG00000069399), BIRC3 (ENSG00000023445), CXCL10 (ENSG00000169245), ICAM1 (ENSG00000090339), NFKB1 (ENSG00000109320), and RELB (ENSG00000104856). Network interactions were derived from the database of Pathway Studio V9, which contains automatically extracted information from PubMed publications52.
Cytokine quantification
The abundance of cytokines secreted by MDM was measured using a Bio-Plex Pro Human Cytokine 27-plex Assay (Bio-Rad). CXCL10 concentration in serum samples (n = 80) from IA patients (n = 8) with or without etanercept treatment was measured with the ELISA MAXTM - human CXCL10 (IP-10) assay (BioLegend) according to the manufacturer’s instructions. Preprocessing of the data was performed according to the instructions from the manufacturer. Differentially secreted cytokines were detected using Wilcoxon-Mann-Whitney tests applying a p-value ≤ 0.05.
Statistical methods
Univariate logistic regression models were employed to find associations between all variables and the diagnosis of IA (case-control status) in the training data set of 36 patients suffering from probable IA and 36 control patients. For temporally dynamic variables, models were generated separately for each week and the first 4 weeks before diagnosis. Since blood cell counts can vary considerably over time, we performed time-dependent case-control matching. For example, the monocyte concentration of a case suffering from IA at day 60 after alloSCT was compared with a matched monocyte concentration of a control patient also (or closest to) 60 days after alloSCT. Each model and thus the contribution of each variable were evaluated by a likelihood ratio test (LRT) against an intercept-only model. We explored the univariate models and pursued variables if their univariate screening p-value was ≤ 0.05. Variables with too little variability were not considered further. Afterwards, multiple logistic regression models were built to test the joint combined effect of the selected variables from the univariate models. Additional variables were integrated and tested with LRT for improvement of the prediction power of the model.
The predictive power of our final model was assessed using an independent, similarly ascertained, validation sample of 23 IA cases and 25 control patients. We report statistical estimates of the area under ROC curve (AUC) for all models to allow for evaluation of the model. All statistical analyses were done in the statistical environment R (version 3.3).
Ethics approval
All methods were performed in accordance with the relevant guidelines and regulations. Informed consent has been obtained for study participation and for publication of information, as requested by the Ethics Committee of the University Hospital of Wuerzburg (permit #173/11).
References
Girmenia, C. et al. Prophylaxis and treatment of invasive fungal diseases in allogeneic stem cell transplantation: results of a consensus process by Gruppo Italiano Trapianto di Midollo Osseo (GITMO). Clin Infect Dis 49, 1226–1236, https://doi.org/10.1086/605665 (2009).
Garcia-Vidal, C., Upton, A., Kirby, K. A. & Marr, K. A. Epidemiology of invasive mold infections in allogeneic stem cell transplant recipients: biological risk factors for infection according to time after transplantation. Clin Infect Dis 47, 1041–1050, https://doi.org/10.1086/591969 (2008).
Chakrabarti, A., Chatterjee, S. S., Das, A. & Shivaprakash, M. R. Invasive aspergillosis in developing countries. Med Mycol 49(Suppl 1), S35–47, https://doi.org/10.3109/13693786.2010.505206 (2011).
Hope, W. W. & Denning, D. W. Invasive aspergillosis: current and future challenges in diagnosis and therapy. Clin Microbiol Infect 10, 2–4 (2004).
Alanio, A. & Bretagne, S. Challenges in microbiological diagnosis of invasive Aspergillus infections. F1000Res 6, https://doi.org/10.12688/f1000research.10216.1 (2017).
De Pauw, B. et al. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin Infect Dis 46, 1813–1821, https://doi.org/10.1086/588660 (2008).
Barton, R. C. Laboratory diagnosis of invasive aspergillosis: from diagnosis to prediction of outcome. Scientifica (Cairo) 2013, 459405, https://doi.org/10.1155/2013/459405 (2013).
White, P. L. et al. Clinical Performance of Aspergillus PCR for Testing Serum and Plasma: a Study by the European Aspergillus PCR Initiative. Journal of Clinical Microbiology 53, 2832–2837, https://doi.org/10.1128/Jcm.00905-15 (2015).
Loeffler, J. et al. Prospective Biomarker Screening for Diagnosis of Invasive Aspergillosis in High-Risk Pediatric Patients. J Clin Microbiol 55, 101–109, https://doi.org/10.1128/JCM.01682-16 (2017).
Springer, J. et al. Multicenter comparison of serum and whole-blood specimens for detection of Aspergillus DNA in high-risk hematological patients. J Clin Microbiol 51, 1445–1450, https://doi.org/10.1128/JCM.03322-12 (2013).
Rogers, T. R. et al. Combined real-time PCR and galactomannan surveillance improves diagnosis of invasive aspergillosis in high risk patients with haematological malignancies. Br J Haematol 161, 517–524, https://doi.org/10.1111/bjh.12285 (2013).
White, P. L., Parr, C. & Barnes, R. A. Predicting Invasive Aspergillosis in Hematology Patients by Combining Clinical and Genetic Risk Factors with Early Diagnostic Biomarkers. J Clin Microbiol 56, https://doi.org/10.1128/JCM.01122-17 (2018).
Singh, N. & Paterson, D. L. Aspergillus infections in transplant recipients. Clin Microbiol Rev 18, 44–69, https://doi.org/10.1128/CMR.18.1.44-69.2005 (2005).
Baddley, J. W. Clinical risk factors for invasive aspergillosis. Med Mycol 49(Suppl 1), S7–S12, https://doi.org/10.3109/13693786.2010.505204 (2011).
Mengoli, C., Cruciani, M., Barnes, R. A., Loeffler, J. & Donnelly, P. Use of PCR for diagnosis of invasive aspergillosis: systematic review and meta-analysis. Lancet Infectious Diseases 9, 89–96, https://doi.org/10.1016/S1473-3099(09)70019-2 (2009).
Jakubzick, C. V., Randolph, G. J. & Henson, P. M. Monocyte differentiation and antigen-presenting functions. Nat Rev Immunol 17, 349–362, https://doi.org/10.1038/nri.2017.28 (2017).
Randolph, G. J., Jakubzick, C. & Qu, C. Antigen presentation by monocytes and monocyte-derived cells. Curr Opin Immunol 20, 52–60, https://doi.org/10.1016/j.coi.2007.10.010 (2008).
Sedger, L. M. & McDermott, M. F. TNF and TNF-receptors: From mediators of cell death and inflammation to therapeutic giants - past, present and future. Cytokine Growth Factor Rev 25, 453–472, https://doi.org/10.1016/j.cytogfr.2014.07.016 (2014).
Filler, S. G., Yeaman, M. R. & Sheppard, D. C. Tumor necrosis factor inhibition and invasive fungal infections. Clin Infect Dis 41(Suppl 3), S208–212, https://doi.org/10.1086/430000 (2005).
Downey, C. Serious infection during etanercept, infliximab and adalimumab therapy for rheumatoid arthritis: A literature review. Int J Rheum Dis 19, 536–550, https://doi.org/10.1111/1756-185X.12659 (2016).
Tsiodras, S., Samonis, G., Boumpas, D. T. & Kontoyiannis, D. P. Fungal infections complicating tumor necrosis factor alpha blockade therapy. Mayo Clin Proc 83, 181–194 (2008).
Nedel, W. L., Kontoyiannis, D. P. & Pasqualotto, A. C. Aspergillosis in patients treated with monoclonal antibodies. Rev Iberoam Micol 26, 175–183, https://doi.org/10.1016/j.riam.2009.04.001 (2009).
Tan, J. K., Aphale, A., Malaviya, R., Sun, Y. & Gottlieb, A. B. Mechanisms of action of etanercept in psoriasis. J Investig Dermatol Symp Proc 12, 38–45, https://doi.org/10.1038/sj.jidsymp.5650037 (2007).
Gottlieb, A. B. Tumor necrosis factor blockade: mechanism of action. J Investig Dermatol Symp Proc 12, 1–4, https://doi.org/10.1038/sj.jidsymp.5650029 (2007).
Garcia-Cadenas, I. et al. Patterns of infection and infection-related mortality in patients with steroid-refractory acute graft versus host disease. Bone Marrow Transplant 52, 107–113, https://doi.org/10.1038/bmt.2016.225 (2017).
Park, J. H. et al. Etanercept for steroid-refractory acute graft versus host disease following allogeneic hematopoietic stem cell transplantation. Korean J Intern Med 29, 630–636, https://doi.org/10.3904/kjim.2014.29.5.630 (2014).
De Jong, C. N. et al. Etanercept for steroid-refractory acute graft-versus-host disease: A single center experience. PLoS One 12, e0187184, https://doi.org/10.1371/journal.pone.0187184 (2017).
Subramanian Vignesh, K. & Deepe, G. S., Jr. Metallothioneins: Emerging Modulators in Immunity and Infection. Int J Mol Sci 18, https://doi.org/10.3390/ijms18102197 (2017).
Rodland, E. K. et al. Expression of genes in normal human monocytes in response to Aspergillus fumigatus. Med Mycol 46, 327–336, https://doi.org/10.1080/13693780701874507 (2008).
Rivera, A., Hohl, T. & Pamer, E. G. Immune responses to Aspergillus fumigatus infections. Biol Blood Marrow Transplant 12, 47–49, https://doi.org/10.1016/j.bbmt.2005.09.007 (2006).
Scicluna, B. P. et al. Role of tumor necrosis factor-alpha in the human systemic endotoxin-induced transcriptome. PLoS One 8, e79051, https://doi.org/10.1371/journal.pone.0079051 (2013).
Loeffler, J. et al. Interaction analyses of human monocytes co-cultured with different forms of Aspergillus fumigatus. J Med Microbiol 58, 49–58, https://doi.org/10.1099/jmm.0.003293-0 (2009).
Holland, J. & Owens, T. Signaling through intercellular adhesion molecule 1 (ICAM-1) in a B cell lymphoma line. The activation of Lyn tyrosine kinase and the mitogen-activated protein kinase pathway. J Biol Chem 272, 9108–9112 (1997).
Herrington, F. D. & Nibbs, R. J. Regulation of the Adaptive Immune Response by the IkappaB Family Protein Bcl-3. Cells 5, https://doi.org/10.3390/cells5020014 (2016).
Conte, D. et al. Inhibitor of apoptosis protein cIAP2 is essential for lipopolysaccharide-induced macrophage survival. Mol Cell Biol 26, 699–708, https://doi.org/10.1128/MCB.26.2.699-708.2006 (2006).
Bren, G. D. et al. Transcription of the RelB gene is regulated by NF-kappaB. Oncogene 20, 7722–7733, https://doi.org/10.1038/sj.onc.1204868 (2001).
Steele, C. et al. The beta-glucan receptor dectin-1 recognizes specific morphologies of Aspergillus fumigatus. PLoS Pathog 1, e42, https://doi.org/10.1371/journal.ppat.0010042 (2005).
Gattass, C. R., King, L. B., Luster, A. D. & Ashwell, J. D. Constitutive expression of interferon gamma-inducible protein 10 in lymphoid organs and inducible expression in T cells and thymocytes. J Exp Med 179, 1373–1378 (1994).
Taub, D. D., Key, M. L., Clark, D. & Turcovski-Corrales, S. M. Chemotaxis of T lymphocytes on extracellular matrix proteins. Analysis of the in vitro method to quantitate chemotaxis of human T cells. J Immunol Methods 184, 187–198 (1995).
Taub, D. D., Sayers, T. J., Carter, C. R. & Ortaldo, J. R. Alpha and beta chemokines induce NK cell migration and enhance NK-mediated cytolysis. J Immunol 155, 3877–3888 (1995).
Zhao, Q. et al. A novel function of CXCL10 in mediating monocyte production of proinflammatory cytokines. J Leukoc Biol 102, 1271–1280, https://doi.org/10.1189/jlb.5A0717-302 (2017).
Mezger, M. et al. Polymorphisms in the chemokine (C-X-C motif) ligand 10 are associated with invasive aspergillosis after allogeneic stem-cell transplantation and influence CXCL10 expression in monocyte-derived dendritic cells. Blood 111, 534–536, https://doi.org/10.1182/blood-2007-05-090928 (2008).
The International Agency for Research on Cancer. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. (eds Swerdlow S., Campo E., Lee Harris N., Jaffe, Pileri S. A., Stein H., Thiele J., Vardiman J. W.) 439 (World Health Organization, 2008).
Hefter, M. et al. Human primary myeloid dendritic cells interact with the opportunistic fungal pathogen Aspergillus fumigatus via the C-type lectin receptor Dectin-1. Medical mycology 55, 573–578, https://doi.org/10.1093/mmy/myw105 (2017).
Keystone, E. C. et al. Once-weekly administration of 50 mg etanercept in patients with active rheumatoid arthritis - Results of a multicenter, randomized, double-blind, placebo-controlled trial. Arthritis Rheum 50, 353–363, https://doi.org/10.1002/art.20019 (2004).
Obeng, J. A. et al. Modulation of human monocyte/macrophage activity by tocilizumab, abatacept and etanercept: An in vitro study. Eur J Pharmacol 780, 33–37, https://doi.org/10.1016/j.ejphar.2016.03.028 (2016).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120, https://doi.org/10.1093/bioinformatics/btu170 (2014).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12, 357–360, https://doi.org/10.1038/nmeth.3317 (2015).
Liao, Y., Smyth, G. K. & Shi, W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 41, e108, https://doi.org/10.1093/nar/gkt214 (2013).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550, https://doi.org/10.1186/s13059-014-0550-8 (2014).
Priebe, S., Kreisel, C., Horn, F., Guthke, R. & Linde, J. FungiFun2: a comprehensive online resource for systematic analysis of gene lists from fungal species. Bioinformatics 31, 445–446, https://doi.org/10.1093/bioinformatics/btu627 (2015).
Yuryev, A. et al. Automatic pathway building in biological association networks. BMC Bioinformatics 7, 171, https://doi.org/10.1186/1471-2105-7-171 (2006).
Acknowledgements
This work was supported by the Deutsche Forschungsgemeinschaft (Collaborative Research Center/Transregio 124 “Pathogenic fungi and their human host: Networks of interaction – FungiNet”, [project A2 to H.E. and Ju.L., project C3 to O.K., project INF to Jö.L.]) and by the Free State of Thuringia and the European Regional Development Fund (T.Z., Jö.L., S.S. and B.S.).
Author information
Authors and Affiliations
Contributions
T.Z. and M.W. contributed to the study design, experimental work, analysis, and writing of the manuscript. J.S. contributed to the design of experiments, data analysis, and preparation of the manuscript. J.B., Aa.S., C.L., B.S. and Ae.S. contributed to the data analysis. K.H. contributed to the experimental work, data analysis, and preparation of the manuscript. P.L.W. and O.K. contributed to the data analysis and to the writing the manuscript. S.S., C.O.M. and H.E. contributed to the writing the manuscript. Ju.L. and Jö.L. contributed to the study design, data analysis, and the writing the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zoran, T., Weber, M., Springer, J. et al. Treatment with etanercept and low monocyte concentration contribute to the risk of invasive aspergillosis in patients post allogeneic stem cell transplantation. Sci Rep 9, 17231 (2019). https://doi.org/10.1038/s41598-019-53504-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-53504-8
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.