Abstract
Nile tilapia (Oreochromis niloticus) is a globally significant aquaculture species rapidly gaining status as a farmed commodity. In West Africa, wild Nile tilapia genetic resources are abundant yet knowledge of fine-scale population structure and patterns of natural genetic variation are limited. Coinciding with this is a burgeoning growth in tilapia aquaculture in Ghana and other countries within the region underpinned by locally available genetic resources. Using 192 single nucleotide polymorphism (SNP) markers this study conducted a genetic survey of Nile tilapia throughout West Africa, sampling 23 wild populations across eight countries (Benin, Burkina Faso, Côte d’Ivoire, Ghana, Togo, Mali, Gambia and Senegal), representing the major catchments of the Volta, Niger, Senegal and Gambia River basins. A pattern of isolation-by-distance and significant spatial genetic structure was identified throughout West Africa (Global FST = 0.144), which largely corresponds to major river basins and, to a lesser extent, sub-basins. Two populations from the Gambia River (Kudang and Walekounda), one from the western Niger River (Lake Sélingué) and one from the upper Red Volta River (Kongoussi) showed markedly lower levels of diversity and high genetic differentiation compared to all other populations, suggesting genetically isolated populations occurring across the region. Genetic structure within the Volta Basin did not always follow the pattern expected for sub-river basins. This study identifies clear genetic structuring and differentiation amongst West African Nile tilapia populations, which concur with broad patterns found in previous studies. In addition, we provide new evidence for fine-scale genetic structuring within the Volta Basin and previously unidentified genetic differences of populations in Gambia. The 192 SNP marker suite used in this study is a useful tool for differentiating tilapia populations and we recommend incorporating this marker suite into future population screening of O. niloticus. Our results form the basis of a solid platform for future research on wild tilapia genetic resources in West Africa, and the identification of potentially valuable germplasm for use in ongoing breeding programs for aquaculture.
Similar content being viewed by others
Introduction
The Oreochromines are a sub-family of the Cichlidae family of fishes native to Africa and parts of the Levant that have large commercial significance for aquaculture and capture fisheries1. The Nile tilapia, Oreochromis niloticus, has a broad natural distribution spanning from the Nile River basin southwards through the Eastern and Western Rift Valley lakes in East Africa, and westwards through the basins of Lake Chad, Niger, Benue, Volta, Gambia and Senegal rivers1. The species has been well studied from a taxonomic perspective. A complex sub-species structure and distribution of O. niloticus has been documented, especially across East Africa where a network of geographically and biologically diverse lakes have shaped its evolution. Seven sub-species of O. niloticus have been identified based on traditional taxonomic methods; all except one – O. niloticus niloticus - are distributed amongst the Rift Valley lakes of East Africa1.
Molecular studies have identified clear differentiation between west and east African populations of O. niloticus2,3,4,5, which can be generalized into three main groups: (i) Sudano-Sahelian populations, covering West Africa, (ii) Ethiopian Rift valley populations and (iii) Nile drainage and Kenyan Rift Valley populations5,6. A hierarchical pattern of genetic differentiation has been observed in O. niloticus, whereby the effects of major paleo-geographic and climatic events are the predominant factor at the macro-geographic scale, and present-day river systems are influential on genetic connectivity and patterns of genetic structure at smaller geographic scales5. The same authors have also suggested limited dispersal capabilities combined with non-random mating caused by social behaviours may also influence small scale population structure. Other studies indicate that isolated and previously undescribed O. niloticus populations in Kenya can show strong genetic differentiation from other nearby populations, and may constitute new wild genetic resources with potential importance for exploitation in commercial aquaculture activities7. Furthermore, studies of patterns of mitochondrial DNA variation have revealed additional complexities amongst O. niloticus populations, with evidence of historical hybridization occurring between closely related O. aureus8 and O. leucosticus9 species occurring in the western and eastern regions of Africa, respectively.
Although much attention has been given to the population characterization and genetic structure of O. niloticus in East Africa, relatively few studies have focused on populations throughout West Africa with high resolution. This is despite West Africa being the largest ichthyoregion naturally occupied by O. niloticus. Earlier studies have indicated genetic differentiation at broad scales (i.e. 1000’s of km)3,4,5. However, spatial coverage of these studies is limited and consequently knowledge of finer scale patterns of genetic structure of O. niloticus in West Africa remains uncertain.
Oreochromis niloticus is by far the most prominent amongst tilapia species produced by aquaculture, being the most farmed tropical fish species globally. Approximately 4.1 million tonnes of Nile tilapia were produced by aquaculture in 2017, having a value of US$7.6 billion10. West African O. niloticus populations have made a significant contribution towards the development of farmed genetic resources that underpin much of global production. For example, at least four of the eight founder populations (and likely to be five) of the highly successful Genetically Improved Farmed Tilapia (GIFT) strain of O. niloticus were either sourced directly from wild populations in Ghana and Senegal or cultured populations derived from stocks of Ghanaian origin11. The GIFT strain has undergone continuous selective breeding in Southeast Asia since the early 1990’s, and has been widely recognized as having substantial impact on tilapia aquaculture development across the region12,13. Additionally, a selective breeding program based on locally sourced wild O. niloticus is rapidly gaining popularity by aquaculture farmers in Ghana and bringing economic benefits and boosting primary productivity for the country14,15,16. Such successes demonstrate the potential value that can be derived from these wild genetic resources provided the appropriate management and technological approaches are applied.
Tilapia aquaculture is poised for rapid growth throughout Sub-Saharan Africa in the coming years. As tilapia aquaculture increases throughout Sub-Saharan Africa, an impetus to better understand the distribution of wild genetic resources and patterns of population structure throughout the natural distribution of O. niloticus is similarly increasing6,17. The historic popularity of Nile tilapia as a capture fishery and for aquaculture production has seen its repeated translocation and deliberate introduction to many places where it was not originally found. The genetic origin of such introductions are often vague or unknown, yet may have an important impact on the genetic composition of wild populations.
The advantages of investigating molecular genetic characteristics of wild Nile tilapia are two-fold. Firstly, understanding wild population structure can assist the management or safeguarding of wild genetic resources, particularly in the context of the potential impacts caused by aquaculture escapes. Secondly, the identification of genetically distinct and diverse wild populations is of potential benefit to aquaculture breeding programs, which require a broad genetic diversity in their founder populations to achieve sustained productivity improvements and avoid problems of inbreeding accumulation. By providing insights to both the management and use of tilapia genetic resources, molecular genetic data offers better understanding in a climate where large-scale aquaculture development in West Africa is growing and proper resource usage is critical.
This study presents the opportunity to understand patterns of genetic structure and factors affecting diversity across the region, to better document the natural Nile tilapia genetic resources in West Africa, and further build on previous research to understand the potential macro or micro geographic factors influencing their diversity. Using single nucleotide polymorphism (SNP) markers, we studied 23 populations across five river basins in West Africa to assess spatial patterns of genetic diversity and genetic differentiation of indigenous wild Nile tilapia populations. The specific aims of this study are to: (1) Characterize genetic diversity of Nile tilapia populations throughout West Africa using SNP markers; (2) Quantify the relative differences of genetic structure in Nile tilapia populations within versus among watershed basins throughout West Africa; (3) Explore the genetic characteristics of Nile tilapia populations not recognized as part of its natural distribution but are known locally to be sources of the species; and (4) Identify potential factors contributing to patterns of genetic diversity and differentiation in West African Nile tilapia populations. We anticipate this will provide a clearer insight towards what populations may be of potential value for conservation or further use in ongoing aquaculture expansion plans for the region.
Material and Methods
Origin and location of the sampling populations
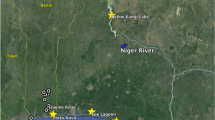
Twenty three wild O. niloticus populations were sampled from across eight countries (Benin, Burkina Faso, Côte d’Ivoire, Gambia, Ghana, Mali, Senegal and Togo) of West Africa, representing the major catchments of the Volta, Niger, Senegal and Gambia River basins (Fig. 1). Fine-scale patterns of genetic differentiation were targeted within the Volta Basin. Two to five population samples were taken from within several sub-basins, including Lake Volta, a large reservoir formed by the Akosombo Dam constructed in 1965, and each of its four main tributaries, namely, the Black Volta River (also known as the Mouhoun), the Red (Nazinon) and White Volta (Nakambé) Rivers and the Oti River (Table 1). One site from the Comoe Basin (Burkina Faso) and the Kloukpa River (Ghana) were also sampled, which are not recognized as being part of the natural distribution of O. niloticus1 but are known sources of the species locally.
Sample collection
A total of 1001 individual fish were sampled from 2009 to 2011. Fish were collected directly from commercial fishing boats or at the landing sites of known fishing areas. A caudal finclip approximately 1 cm × 2 cm was cut using forceps and a sharp, clean pair of scissors, and preserved in individually labeled vials using 95% ethanol. All the samples were stored at room temperature until later processing. Each sample site was geo-referenced using a handheld GPS receiver. Between 24 and 50 individuals were collected from each site, depending on availability. Sex of each fish was indeterminable from external examination at sampling, and it is assumed that an approximately even sex ratio of males to females were sampled. A summary of each sampling site, including geo-reference details, is described in Table 1.
Ethics statement
Live experimental animals were not used for this study; thus the requirement for animal ethics approval was not applicable. Finclips were taken from dead fish caught by small-scale commercial fishers using standard gear and practices. Fishers were not commissioned by researchers of this study and study animals were purchased whole or sampled at the point of sale to the public on the day of catch.
DNA extraction protocol
After collection, DNA extraction was performed using a commercially available kit (Promega WIZARD® Genomic DNA Purification Kit) as per the manufacturer’s instructions, and stored at −20 °C until later processing. DNA extraction was conducted at the laboratory of the International Centre for Research and Development on Livestock in Subhumid Zones (CIRDES), Burkina Faso. Samples were standardized to a DNA concentration of 50 ng ul−1 and shipped with dry ice by air to Wageningen University for genotyping.
Single nucleotide polymorphism (SNP) genotyping
Multi-locus genotypes were generated from 192 SNP markers, selected from a suite of 384 SNP markers developed for O. niloticus18. Genotyping was performed in a 192 SNP multiplex assay using the Golden Gate Assay (Illumina), and deployed on a BeadXpress platform using Veracode technology. A total of 15 SNPs on the assay were located on chromosomes/linkage groups where there are known associations with sex determination (LG1, LG20, LG23) in Nile tilapia19,20, however, these SNPs used were not located in the vicinity of these sex-linked regions (data not shown) and therefore have minimal risk of biasing results of the study. A minimum call rate of 0.8 was used to score genotypes. A full list of the SNP markers used, the linkage group according to Guyon et al.21 and the dbSNP accession number of each SNP are given in the Supplementary Materials section.
Statistical analyses
Genetic diversity
Number of polymorphic loci, observed heterozygosity and expected heterozygosity were calculated for each population using the R package adegenet22. Allelic richness of each population was calculated using the R package hierfstat23 based on a minimum sample size of 15 individuals, which accounts for differences in sample size and genotype failure. Tests for Hardy-Weinberg Equilibrium (HWE) were performed at each locus for each population using the HWE.test.genind() function of adegenet, based on the Chi-squared statistic. P-values of each test were adjusted within population according to the false discovery rate method of Benjamini and Hochberg24 using the p.adjust() function in R25, to an equivalent significance level of α = 0.05.
Effective population size (Ne)
Effective population size and its 95% confidence interval were estimated for each population using the linkage disequilibrium method of Waples26, as implemented in NeESTIMATOR v2.0127. A random mating model was applied, using a PCRIT (lowest allele frequency) of 0.05.
Population differentiation and genetic distances
Wright’s F-statistics (FST, FIS and FIT) were estimated using ARLECORE, the console version of ARLEQUIN v3.528, to determine the proportion of genetic variation partitioned within and among various levels of population sub-structure (i.e. individual, sub-population and total population). Pairwise FST between populations were also calculated using ARLECORE, with the significance of pairwise values tested using a non-parametric Monte Carlo approach (1000 permutations). A Bonferroni correction was applied to P-values using the p.adjust() function in R25 to control the likelihood of type I errors associated with multiple comparisons. To investigate the significance of major geographical factors such as river basins (watersheds) on genetic structuring in Nile tilapia, an hierarchical Analysis of Molecular Variation (AMOVA) was implemented using ARLEQUIN. Several models were tested based on different groupings of populations representing reasonable explanatory drivers of genetic differentiation. The aim here is to explore what groupings could better explain the observed genetic variation when partitioned amongst groups (FCT), amongst populations within groups (FSC) and amongst populations (FST). The population were grouped by (1) major river basins; (2) major river basins and sub-basins (where present); and (3) climatic zones.
Isolation by distance test
To test if genetic differentiation patterns followed an isolation-by-distance model of population divergence, Mantel’s test for correlation between geographic distances (km) and genetic distances (FST) was done based on 10 000 permutations using the mantel.randtest() function in adegenet. Geographic distances used are straight line distances between population sample sites calculated from longitude and latitude coordinates using the earth.dist() function of the R package fossil29.
Spatial principal component analysis (sPCA)
Spatial patterns of genetic variability in O. niloticus were further investigated through a spatial principal component analysis (sPCA) of population allelic frequencies30. This method allows the analysis of both variability in allele frequencies and spatial autocorrelation amongst populations. sPCA was conducted using the spca() function of adegenet22. Spatial information was incorporated into the analysis based on a Gabriel graph connection network of population sample sites. Monte-Carlo tests for the presence of significant global and local spatial structure were performed using the global.rtest() and local.rtest() functions of adegenet, respectively, based on 9999 permutations. Here, global structure refers to positive spatial autocorrelation (i.e. populations closer to each other are more similar) and local structure refers to negative spatial autocorrelation (closer populations are more dissimilar). Because of the potential sampling biases in allele frequencies caused by the presence of rare variants, the dataset was trimmed to use only those loci that had a minor allele frequency (MAF) greater than 0.01 across all individuals.
Genetic clustering - Bayesian approach
The Bayesian clustering approach implemented in the software STRUCTURE v2.3 was used to partition genotype data according a predefined number of groups (K) that conform to expectations of Hardy-Weinberg equilibrium and linkage equilibrium between loci31,32. An admixture model with correlated allele frequencies was chosen to estimate the group membership probability (Q) of each individual for K = 2 to K = 20. For each level of K ten replicates of 300 000 Markov-Chain Monte Carlo iterations were run after a 150 000 iteration burn-in period. Default parameters were used otherwise. To identify the most likely number of clusters the Evanno method33 was implemented using STRUCTURE HARVESTER34. CLUMPP35 was used to align clusters across replicate runs and calculate a mean Q estimate for each individual, which were visualized using DISTRUCT36.
Genetic clustering - Multivariate approach
An alternative approach to identifying genetically distinct clusters is through Discriminant Analysis of Principal Components (DAPC)37. DAPC creates synthetic variables (discriminant functions) that attempt to maximize differences between groups or genetic clusters whilst minimizing allele frequency variance within a cluster. It has a major advantage in that no population genetic assumptions (such as Hardy-Weinberg expectations) are required to describe patterns of genetic variation across individuals and clusters. Prior definitions of groups are required for DAPC, which were done using the find.clusters() function of adegenet to implement a k-means clustering algorithm. The function also generates a Bayesian Information Criterion (BIC) for each value of k, which was then used to identify the optimal value of k. Ideally the BIC will decrease until an optimal k is reached and then subsequently increase for greater values of k. DAPC was performed for values of k increasing from 2 to 20 using the dapc() function of adegenet, retaining 75 principal components and 6 discriminant functions for each value of k. The trimmed dataset containing loci with a MAF greater than 0.01 was also used for this analysis, for the same reasons described earlier.
Results
Dataset cleaning
A total of 192 SNPs were genotyped for all 1001 samples. Four samples were removed from the final dataset due to genotyping failure. Eight SNPs were monomorphic and seven SNPs had ≥15% missing genotypes, and were subsequently removed from the final dataset. A total of 177 SNP markers were used for the final analyses, unless otherwise stated. Fifty four SNP markers had a minor allele frequency (MAF) < 0.01.
Summary statistics and genetic diversity
Genetic diversity was lowest in the Kudang, Walekounda (both located in the Gambia River basin), Lake Sélingué (western Niger basin) and Kongoussi (Red/White Volta basin) populations. Populations that showed the greatest level of genetic variation were from Malanville (eastern Niger basin) and two populations from the southern region of Ghana (Site VL2 from Lake Volta and the Afife Reservoir in southern Ghana). Total number of alleles within each population ranged from 257 to 345, which translated to an overall percentage of polymorphic loci by population ranging from 33.8 to 79.7 percent. Allelic richness (Rs), a measure of the average number of alleles per locus taking into account potential biases due to differences in sample sizes, ranged from 1.30 to 1.62. Observed heterozygosity was very similar to expected heterozygosity in all populations when averaged across loci (Table 2), which indicates a general conformation to expectations of Hardy-Weinberg Equilibrium (HWE). This is supported by HWE tests by locus, whereby less than 5 percent of loci within each population showed a significant departure from Hardy-Weinberg expectations at all sites (P < 0.05), with the exception of Nawuni in northern Ghana (5.2 percent loci not in HWE) and the Afife Reservoir (15.1 percent).
Effective population size
Effective population sizes (Ne) was in general positively correlated with genetic diversity measures. Over 50 percent of populations sampled had Ne in excess of 100 and none below 50. An exception to this pattern was the Kongoussi population in northern Burkina Faso, which showed the greatest Ne (351.6) yet had comparatively low heterozygosity and allelic richness (Table 2). Full details of the summary and genetic diversity statistics of each population are outlined in Table 2.
Population differentiation
Significant population differentiation was observed at the global level (p < 0.001), with 14.4% of overall genotypic variation attributable to differences among populations (i.e. FST.) when no hierarchical sub-groupings were considered other than the individual and population level (Table 3). When populations were grouped based on river basins using hierarchical AMOVA, regional patterns of genetic structure according to river basins or sub-basins begin to emerge. In some instances differences amongst regional grouping (FCT) accounts for up to 13.2 per cent of overall variation (Table 3). Grouping populations based on their climate zones could explain only 1.6% of overall variation. Population pairwise FST show significant differences amongst all populations with the exception of several groups of closely situated populations in the southern areas of Lake Volta (VAk, VL1 and VL2; and VL3 and VLk), two populations from the Gambia River (GKu and GWa), and two populations from the Oti River in the Volta Basin (VOt and VPj), which showed no significant differences to each other after Bonferroni correction. Clear patterns of genetic differentiation are observed between the Gambia River populations and all other populations sampled (Fig. 2). Populations from Lake Sélingué (western Niger River) and Koungoussi (upper Red/White Volta River) also showed high to moderate differences with all other sites, with pairwise FST ranging from 0.2 to 0.49 (Fig. 2).
Spatial genetic patterns
Mantel’s test showed a significant, positive relationship between pairwise genetic distances (FST) and geographic distances (see Supplementary Materials), indicating isolation-by-distance is a moderate influence on genetic differentiation at the regional scale (r = 0.521, p = 0.001). Consistent with this, sPCA revealed significant global spatial structure (p = 0.03) and non-significant local structure (p = 0.97) overall, indicating the genetic similarity of populations are positively correlated with their geographic proximity to each other. Within the Volta Basin, however, a significant yet much weaker relationship between genetic and geographic distances was present (r = 0.196, p = 0.029), indicating isolation-by-distance is a relatively minor factor shaping genetic differentiation amongst populations. Patterns of genetic differences and similarity across West Africa are evident when the lagged scores of the first three principal components of the sPCA are spatially plotted (Fig. 3). A “colorplot”, representing the lagged scores of the first three principal components of each population on the Red-Green-Blue colour space, highlights a genetic similarity within the Niger River basin populations, the Oti River and Lake Volta populations, the Red/White Volta and the Black Volta River populations, and the Gambia River populations (Fig. 3).
Spatial principal components analysis (sPCA) of Nile tilapia populations. The first two global principal components (PCs) are shown separately, where each square represents a population loading score. A combined illustration of all three PCs is shown through a colorplot, whereby the three loading scores are represented by colors according to the red, blue and green channels of the RGB color system.
Individual genetic clustering
Bayesian (STRUCTURE) and multivariate (DAPC) based approaches revealed highly consistent patterns of individual genetic clustering for differing values of k ranging from 2 to 15. For clarity, only DAPC results are shown. Plots of individual posterior probabilities for assignment to a given cluster show that patterns of genetic differentiation based on river basin are apparent at various values of k (Fig. 4). The most likely number of clusters (k) for the Bayesian approach was four, based on the Delta K method33, although a secondary peak in Delta K is also seen for k = 13 which could indicate a more fine scale genetic structuring. Changes in BIC showed that an optimum value of k to use for DAPC is likely to fall between 10 and 15. Detailed changes in BIC and Delta K, and the comparison between DAPC and STRUCTURE results can be found in the Supplementary Materials. Clustering at the lowest level (k = 2) shows a distinction between the Volta Basin populations and all others, particularly for the DAPC analysis (Fig. 4). As the number of clusters (k) increases, individual clustering follows closely according to river basin. Within the Volta Basin, four genetic clusters can be observed and can be described as a Lake Volta cluster (including Nawuni, which lies on the White Volta River), an Oti River cluster, a Black Volta cluster and a Red/White Volta cluster. The relative differences amongst clusters identified with DAPC is visualized for k = 10 in Fig. 5. It highlights the largest genetic differences among all clusters are from two clusters comprised of individuals that are from Lake Sélingué (Niger River) and from the two populations of the Gambia River.
Bar plots representing individual-based clustering analyses of Nile tilapia throughout West Africa performed for differing number of clusters (k) using discriminant analysis of principal components (DAPC). Each vertical line represents an individual, and each color represents the membership probability of an individual to a cluster for a given value of k. Black bars separate different populations. Upper labels refer to population code and lower labels indicate river basin (or sub-basin).
DAPC scatterplot of Nile tilapia SNP genotypes displaying (A) principal components 1 and 2; (B) principal components 2 and 3; and (C) principal components 1 and 3, for k = 10 clusters. Clusters are represented by colours and inertia ellipses, and individuals are shown as dots. Colours used for each cluster correspond to the same colours as those used for clusters in Fig. 4. (D) Number of individuals of each population assigned to different cluster groups. Population codes are listed in Table 1.
Discussion
The multiple analyses conducted in this study all corroborate a pattern of spatial genetic structure and differentiation of Nile tilapia populations across West Africa. The broad scale isolation-by-distance patterns are consistent with the significant spatial correlations of population allele frequencies identified by sPCA. Genetic differentiation of O. niloticus across West Africa can generally be explained by river basins, and to some extent, sub-basins, with gene flow amongst populations following patterns of isolation-by-distance. However, genetic similarity in populations that are over 1400 km apart connected by the Niger River (e.g. Malanville and Mopti) provides evidence that sufficiently high gene flow along a large, continuous waterway can occur over genetically relevant timescales. In contrast, within a much shorter distance around the Red and White Volta River regions we see genetic heterogeneity among several populations (Bazega, Lake Bagre and Nawuni), indicating population differences can persist across relatively nearby geographies.
Our study reveals additional population complexity that has not been documented previously. The most divergent populations amongst those sampled were from Lake Sélingué in the Niger Basin and from the Gambia River, which showed strong differentiation to all other populations. Unlike Rognon & Guyomard8, who observed O. niloticus populations from Senegal to be most divergent from other West African populations, we observe a genetic similarity between populations from Senegal and those from Upper and mid-Niger River. This is consistent with the belief that following the last pluvial, around 12,000 to 7,500 years ago, the Senegal and Gambia Rivers were recolonized by freshwater fauna from the Niger Basin38. Similarly, the Black Volta and the Pendjari Rivers are likely to have been tributaries of the Niger River in the past38, which may explain the moderate genetic differentiation observed among sampling sites covering the three main tributaries of the Volta Basin.
We find genetic diversity using SNPs to be similar across the majority of populations sampled, with the exception of several populations exhibiting lower genetic diversity than others. Most populations showed greater SNP heterozygosity than previously reported for the Volta Basin18 and in two selectively bred tilapia populations39, albeit using SNP arrays not necessarily optimized for informativeness in West Africa. Lower genetic diversity in the Gambia River and the far western region of the Niger River is consistent with patterns often seen at a species range limit or isolated populations. Rognon & Gouyomard8 found decreased enzyme diversity in Nile tilapia samples from the Senegal River and cited founder effects at the margins of the distribution as the likely explanation. Founder effects may explain the pattern of genetic differentiation and reduced diversity identified in the Gambia River. In contrast, however, we found relatively high levels of SNP diversity in the Senegal River population, which may indicate this region is supporting larger, more stable populations compared to the nearby Gambia River. This is corroborated by differences in Ne, which in the absence of gene flow would lead to differing rates of diversity loss through genetic drift. Alternatively, the Senegal River may have had more frequent, intermittent historical connectivity to the Niger Basin40, possibly modulating the impacts of genetic drift on diversity loss. Two populations at Lake Sélingué in the Upper Niger River, and Kongoussi in the Upper Red Volta River also show a substantially lower percentage of polymorphic loci than others. This may be a result of their geographic location in the upper reaches of their respective river systems causing relative isolation. Consequently, limited gene flow between other populations or increasing exposure to seasonal fluctuations and intermittence in water flow could explain a likely increase the rate of genetic diversity loss caused by genetic drift.
Rapid population expansions as a result of the new water bodies created by dams can potentially have pronounced effect on the genetic properties of a population41. Lake Volta, created after the completion of the Akosombo Dam in 1965, is the largest man-made lake in the world by surface area and spans 8,502 km2. Despite such vast areas and the likely enormous expansion of Nile tilapia populations throughout the lake after its formation we detect no evidence for departures from Hardy-Weinberg Equilibrium (HWE) and little genetic differentiation amongst any Lake Volta populations, with some over 400 km apart. This could mean that there has either been a limited founder effect on Nile tilapia populations as a result of the formation of Lake Volta and that genetic structuring of these populations has not changed. Alternatively, any genetic differentiation prior to or post Akosombo Dam construction has been homogenized by sufficient gene flow across the lake and equilibrium of allele frequencies has been reached. Across all sites, only the population from the Afife Reservoir exhibited notable HWE departures. This site is located adjacent to the mouth of the Volta River, in the Kloukpa River basin, and is not thought to part of the natural distribution of Nile tilapia1. A departure from HWE coupled with moderate genetic differentiation and relatively low effective population size seen in the Afife Reservoir is a possible indication of founder effects due to being recently established, likely from nearby Volta River populations.
Previous population genetic studies of Nile tilapia have identified major breaks across its natural distribution, delineating three macro-geographic groups: (i) Sudano-Sahelian populations, covering West Africa, (ii) Ethiopian Rift valley populations and (iii) Nile drainage and Kenyan Rift Valley populations4,5. Our study identifies a greater overall FST amongst populations within the Sudano-Sahelian region than previous estimates based on microsatellite markers (RST = 0.09)5. This could be due to a greater sampling coverage, particularly the inclusion of populations from the Gambia River, which are highly differentiated from all other populations sampled. This, however, may also be due to differences in expected heterozygosity derived from highly polymorphic microsatellite markers compared to dimorphic SNP markers, which can influence the upper bounds of possible FST values that can be obtained42. Our study presents the first estimate of population diversity based on SNP markers in West Africa, providing an important baseline as SNP increasingly become the marker of choice for population studies and for commercial aquaculture breeding programs.
Climatic fluctuations in the Sahel region can be dramatic throughout the year, especially compared to the tropical regions of southern Ghana, which may exert greater selective pressure for commercially significant traits such temperature and salinity tolerance. Temperature ranges from 12 to 47 °C throughout the year in Burkina Faso43, contrasting to temperatures that rarely extend beyond 21 to 34 °C in southern Ghana44, which highlights the vastly different thermal amplitudes within the natural range of O. niloticus. Although hierarchical AMOVA based on grouping of populations according to their climatic zone did not explain as much variation as groupings based on river basins, the presence of pockets of genetically isolated populations throughout the Sahel region identified in this study, such as Lake Sélingué and Kongoussi, are of particular interest in this context. Notably, the population at Kongoussi may likely experience fluctuations in environmental conditions substantially greater than the populations in the main rivers due to low water exchange, extended periods of zero rainfall and its shallowness across that region. Genetically unique populations that are potentially valuable genetic resources have been recently discovered in other regions of Nile tilapia’s natural distribution, notably in East Africa7. Results such as this provide a basis for more in-depth investigation into whether genetically distinct populations represent potential genetic resources for future utilization in commercial breeding programs. Future genetic studies would be well placed to target greater sampling intensities and finer-scale geographic coverage around areas where genetically distinct populations are present. Complementary to this, research focusing on understanding whether such differentiation confers to any physiological differences at the population level would be especially valuable.
In addressing questions of genetic diversity and genetic structuring of West African Nile tilapia populations, the potential implications on the growing tilapia aquaculture industry throughout the region must be considered. We show there are high levels of genetic diversity amongst populations located across most major water basins, and identify some populations that are highly differentiated from others, indicating the presence of genetically unique populations. Our results suggest there is ample genetic diversity throughout wild Nile tilapia populations in West Africa to support the establishment and long-term development of selective breeding programs for aquaculture in the region. The consequence, however, of a large aquaculture industry based on selectively bred, domesticated populations, is that widespread use and distribution of these animals is inevitable. This may have potential impacts on natural populations when farmed animals unintentionally escape, and is a growing issue in other parts of Africa17,45,46,47,48. Knowledge of local population genetics developed through studies such as this (possibly utilising more targeted SNP genotyping tools) have an important role to detect, analyse and help manage the potential the impacts of likely escapes. Recent work identifying species-specific SNP markers among closely related tilapia species would also be of value where geographical overlaps occur, and there is potential for hybridization49.
Conclusions
This study identifies clear genetic structuring and differentiation amongst West African Nile tilapia populations, which concur with broad patterns found in previous studies4,5,8. In addition, this study provides new evidence for fine-scale genetic structuring within the Volta Basin and for further genetically differentiated populations in Gambia. Spatial genetic patterns and individual clustering methods indicate genetic structure can be largely explained by major rivers and sub-basins throughout the region. The 192 SNP marker suite used in this study is an effective tool for differentiating tilapia populations and we recommend incorporating this marker suite into future population screening of O.niloticus, potentially in conjunction with other species-specific SNPs. Our results form the basis of a solid platform for future research of wild tilapia genetic resources in West Africa, and the identification of potentially valuable germplasm for use in aquaculture breeding programs.
Data availability
The full dataset and metadata from this publication is available from the Dryad Digital Repository (https://doi.org/10.5061/dryad.bk3j9kd6m).
References
Trewevas, E. Tilapiine Fishes of the Genera Sarotherodon, Oreochromis and Danakilia. 583 (British Museum (Natural History), 1983).
Rognon, X., Andriamanga, M., McAndrew, B. & Guyomard, R. Allozyme variation in natural and cultured populations in two tilapia species: Oreochromis niloticus and Tilapia zillii. Heredity 76, 640–650 (1996).
Rognon, X. & Guyomard, R. Mitochondrial DNA differentiation among East and West African Nile tilapia populations. Journal of Fish Biology 51, 204–207 (1997).
Agnese, J.-F., Adepo-Gourene, B., Abban, E. K. & Fermon, Y. Genetic differentiation among natural populations of the Nile tilapia Oreochromis niloticus (Teleostei, Cichlidae). Heredity 79, 88–96 (1997).
Bezault, E. et al. Spatial and temporal variation in population genetic structure of wild Nile tilapia (Oreochromis niloticus) across Africa. BMC Genetics 12, 102 (2011).
Eknath, A. E. & Hulata, G. Use and exchange of genetic resources of Nile tilapia (Oreochromis niloticus). Reviews in Aquaculture 1, 197–213 (2009).
Nyingi, D., De Vos, L., Aman, R. & Agnèse, J.-F. Genetic characterization of an unknown and endangered native population of the Nile tilapia Oreochromis niloticus (Linnaeus, 1758) (Cichlidae; Teleostei) in the Loboi Swamp (Kenya). Aquaculture 297, 57–63 (2009).
Rognon, X. & Guyomard, R. Large extent of mitochondrial DNA transfer from Oreochromis aureus to O. niloticus in West Africa. Molecular Ecology 12, 435–445 (2003).
Nyingi, D. W. & Agnese, J.-F. Recent introgressive hybridization revealed by exclusive mtDNA transfer from Oreochromis leucostictus (Trewavas, 1933) to Oreochromis niloticus (Linnaeus, 1758) in Lake Baringo, Kenya. Journal of Fish Biology 70, 148–154 (2007).
FAO. FAO Yearbook - Fishery and Aquaculture Statistics 2017. (Food and Agriculture Organization of the United Nations, 2019).
Eknath, A. E. et al. Genetic improvement of farmed tilapias: the growth performance of eight strains of Oreochromis niloticus tested in different farm environments. Aquaculture 111, 171–188 (1993).
Dey, M. M. The impact of genetically improved farmed Nile tilapia in Asia. Aquaculture Economics & Management 4, 107–124, https://doi.org/10.1080/13657300009380263 (2000).
Asian Development Bank. An Impact Evaluation of the Development of Genetically Improved Farmed Tilapia and Their Dissemination in Selected Countries. (Asian Development Bank, 2005).
Attipoe, F. Y. K. Breeding and selection for faster growth strains of the Nile tilapia, Oreochromis niloticus, in Ghana, PhD Thesis, University of Cape Coast, Ghana (2006).
Attipoe, F. Y. K. & Abban, E. K. In Biodiversity, Management and Utilization of West African Fishes (eds Eddie K. Abban, Christine Marie V. Casal, Patrick Dugan, & Thomas M. Falk) 16–17 (WorldFish Center Conference Proceedings, 2004).
Ponzoni, R. W. & Brummett, R. E. In Pioneering Fish Genetic Resource Management and Seed Dissemination Programmes for Africa, CIFAA Occasional Paper No. 29 (eds Devin M. Bartley et al.) 32–41 (FAO, 2008).
Lind, C. E., Brummett, R. E. & Ponzoni, R. W. Exploitation and conservation of fish genetic resources in Africa: Issues and priorities for aquaculture development and research. Reviews in Aquaculture 4, 125–141 (2012).
Van Bers, N. E. M., Crooijmans, R. P. M. A., Groenen, M. A. M., Dibbits, B. W. & Komen, J. SNP marker detection and genotyping in tilapia. Molecular Ecology Resources 12, 932–941, https://doi.org/10.1111/j.1755-0998.2012.03144.x (2012).
Joshi, R. et al. Development and Validation of 58K SNP-Array and High-Density Linkage Map in Nile Tilapia (O. niloticus). Frontiers in Genetics 9, https://doi.org/10.3389/fgene.2018.00472 (2018).
Conte, M. A., Gammerdinger, W. J., Bartie, K. L., Penman, D. J. & Kocher, T. D. A high quality assembly of the Nile Tilapia (Oreochromis niloticus) genome reveals the structure of two sex determination regions. BMC Genomics 18, 341, https://doi.org/10.1186/s12864-017-3723-5 (2017).
Guyon, R. et al. A high-resolution map of the Nile tilapia genome: a resource for studying cichlids and other percomorphs. BMC Genomics 13, 222 (2012).
Jombart, T. adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24, 1403–1405, https://doi.org/10.1093/bioinformatics/btn129 (2008).
Goudet, J. hierfstat, a package for r to compute and test hierarchical F-statistics. Molecular Ecology Notes 5, 184–186, https://doi.org/10.1111/j.1471-8286.2004.00828.x (2005).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society (Series B) 57, 289–300 (1995).
R Core Team. (R Foundation for Statistical Computing, Vienna, Austria, 2013).
Waples, R. S. A bias correction for estimates of effective population size based on linkage disequilibrium at unlinked gene loci. Conservation Genetics 7, 167–184, https://doi.org/10.1007/s10592-005-9100-y (2006).
Do, C. et al. NeEstimator v2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Molecular Ecology Resources 14, 209–214, https://doi.org/10.1111/1755-0998.12157 (2014).
Excoffier, L. & Lischer, H. E. L. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Molecular Ecology Resources 10, 564–567https://doi.org/10.1111/j.1755-0998.2010.02847.x.
Vavrek, M. J. fossil: Palaeoecological and palaeogeographical analysis tools. Palaeontologia Electronica, 14.11.11T (2011).
Jombart, T., Devillard, S., Dufour, A. B. & Pontier, D. Revealing cryptic spatial patterns in genetic variability by a new multivariate method. Heredity 101, 92–103 (2008).
Pritchard, J. K., Stephens, M. & Donnelly, P. Inference of Population Structure Using Multilocus Genotype Data. Genetics 155, 945–959 (2000).
Falush, D., Stephens, M. & Pritchard, J. K. Inference of Population Structure Using Multilocus Genotype Data: Linked Loci and Correlated Allele Frequencies. Genetics 164, 1567–1587 (2003).
Evanno, G., Regnaut, S. & Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology 14, 2611–2620, https://doi.org/10.1111/j.1365-294X.2005.02553.x (2005).
Earl, D. A. & vonHoldt, B. M. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources 4, 359–361, https://doi.org/10.1007/s12686-011-9548-7 (2012).
Jakobsson, M. & Rosenberg, N. A. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23, 1801–1806, https://doi.org/10.1093/bioinformatics/btm233 (2007).
Rosenberg, N. A. distruct: a program for the graphical display of population structure. Molecular Ecology Notes 4, 137–138, https://doi.org/10.1046/j.1471-8286.2003.00566.x (2004).
Jombart, T., Devillard, S. & Balloux, F. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genetics 11, 94 (2010).
Léveque, C. Biodiversity dynamics and conservation: the freshwater fish of tropical Africa. 402 (Cambridge University Press, 1997).
Lind, C. E., Kilian, A. & Benzie, J. A. H. Development of Diversity Arrays Technology markers as a tool for rapid genomic assessment in Nile tilapia, Oreochromis niloticus. Animal Genetics 48, 362–364, https://doi.org/10.1111/age.12536 (2017).
Thieme, M. L. et al. Freshwater ecoregions of Africa and Madagascar: a conservation assessment. (Island Press, Washington, DC, 2005).
Neraas, L. P. & Spruell, P. Fragmentation of riverine systems: the genetic effects of dams on bull trout (Salvelinus confluentus) in the Clark Fork River system. Molecular Ecology 10, 1153–1164, https://doi.org/10.1046/j.1365-294X.2001.01269.x (2001).
Jakobsson, M., Edge, M. D. & Rosenberg, N. A. The Relationship Between FST and the Frequency of the Most Frequent Allele. Genetics 193, 515–528, https://doi.org/10.1534/genetics.112.144758.
Les Editions J. A. Jeune Afrique Atlases: Burkina Faso Atlas. 3rd edn, (Les Editions J. A., 1998).
WeatherSpark. Average Weather For Accra, Ghana, https://weatherspark.com/averages/28541/Accra-Greater-Accra-Ghana (2014).
Ansah, Y. B., Frimpong, E. A. & Hallerman, E. M. Genetically-Improved Tilapia Strains in Africa: Potential Benefits and Negative Impacts. Sustainability 6, 3697–3721 (2014).
Deines, A. M., Bbole, I., Katongo, C., Feder, J. L. & Lodge, D. M. Hybridisation between native Oreochromis species and introduced Nile tilapia O. niloticus in the Kafue River, Zambia. African. Journal of Aquatic Science 39, 23–34, https://doi.org/10.2989/16085914.2013.864965 (2014).
Firmat, C., Alibert, P., Losseau, M., Baroiller, J.-F. & Schliewen, U. K. Successive Invasion-Mediated Interspecific Hybridizations and Population Structure in the Endangered Cichlid Oreochromis mossambicus. PLOS ONE 8, e63880, https://doi.org/10.1371/journal.pone.0063880 (2013).
Shechonge, A. et al. Widespread colonisation of Tanzanian catchments by introduced Oreochromis tilapia fishes: the legacy from decades of deliberate introduction. Hydrobiologia, https://doi.org/10.1007/s10750-018-3597-9 (2018).
Syaifudin, M. et al. Species-Specific Marker Discovery in Tilapia. Scientific Reports 9, 13001, https://doi.org/10.1038/s41598-019-48339-2 (2019).
Acknowledgements
The authors would like to thank and acknowledge the following people for their valuable contributions towards sample collection: from Benin: Arsène d’Almeida, Togo: M. Agbokoussé; Burkina Faso: Bema Ouattara; Mali: M. Sanogo; Ghana: Olga Assemian, Lily Konadu; Côte d’Ivoire: Dr Ernest Kouakou, Olga Assemian; Senegal: M. Manel; Gambia: Moustapha Kébé, Musasaihou Mbenga, Njie Mariatou, Fatou Drammeh. We also thank John Benzie and Devin Bartley for useful comments on an earlier version of the manuscript. This study was supported by funding from Agencia Española de Cooperation Internacional/FAO TIVO Project GCP/RAF/417/SPA, from the European Commission (Grant No. 2000000466) as a contribution to the CGIAR Research Program on Livestock and Fish, and from the FAO sub-regional office for West Africa (SFW).
Author information
Authors and Affiliations
Contributions
C.L. led the design of the study, conducted the data analyses and wrote the manuscript draft; A.T. participated in the study design, sample collection and oversaw the DNA extraction; S.A. and F.A. participated in the study design and sample collection; C.N. participated in the study design, coordinated and participated in sample collection; R.C. designed the SNP assay and oversaw genotyping activities. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lind, C.E., Agyakwah, S.K., Attipoe, F.Y. et al. Genetic diversity of Nile tilapia (Oreochromis niloticus) throughout West Africa. Sci Rep 9, 16767 (2019). https://doi.org/10.1038/s41598-019-53295-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-53295-y
This article is cited by
-
Morphometric and genetic characterization of cultured and wild populations of tilapia, Oreochromis niloticus in India
3 Biotech (2024)
-
Pangenomics of the cichlid species (Oreochromis niloticus) reveals genetic admixture ancestry with potential for aquaculture improvement in Kenya
The Journal of Basic and Applied Zoology (2023)
-
Genetic resources of Nile tilapia (Oreochromis niloticus Linnaeus, 1758) in its native range and aquaculture
Hydrobiologia (2023)
-
Prevalence and co-infections of pathogenic bacteria in Nile tilapia broodstock farms: implications for aquaculture management in Togo
Environmental Monitoring and Assessment (2023)
-
An Investigation on Freshwater Fish Oreochromis niloticus (Linnaeus, 1758): Assessing Hemotoxic Effects of Different Copper Compounds Used as Nanomaterial or Pesticide
Bulletin of Environmental Contamination and Toxicology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.