Abstract
Banana (Musa spp.) is one of the world’s most important staple and cash crops. Despite accumulating genetic and transcriptomic data, low transformation efficiency in agronomically important Musa spp. render translational researches in banana difficult by using conventional knockout approaches. To develop tools for translational research in bananas, we developed a virus induced-gene silencing (VIGS) system based on a banana-infecting cucumber mosaic virus (CMV) isolate, CMV 20. CMV 20 genomic RNA 1, 2, and 3, were separately cloned in Agrobacterium pJL89 binary vectors, and a cloning site was introduced on RNA 2 immediately after the 2a open reading frame to insert the gene targeted for silencing. An efficient Agrobacterium inoculation method was developed for banana, which enabled the CMV 20 VIGS vector infection rate to reach 95% in our experiments. CMV 20-based silencing of Musa acuminata cv. Cavendish (AAA group) glutamate 1-semialdehyde aminotransferase (MaGSA) produced a typical chlorotic phenotype and silencing of M. acuminata phytoene desaturase (MaPDS) produced a photobleachnig phenotype. We show this approach efficiently reduced GSA and PDS transcripts to 10% and 18% of the control, respectively. The high infection rate and extended silencing of this VIGS system will provide an invaluable tool to accelerate functional genomic studies in banana.
Similar content being viewed by others
Introduction
Bananas belong to the family Musaceae and represent one of the world’s most important cash and staple crops1. This perennial crop is cultivated throughout the tropics and sub-tropics, and over 400 million people in developing regions are dependent on bananas as a vital food nutrient. In 2017, worldwide production of bananas was estimated to be over 113 million tons2. The vast majority of cultivated bananas are triploid, and are derived from inter- and intra-specific crosses between two diploid wild species, Musa acuminata (A genome) and M. balbisiana (B genome)3. Currently, commercialized banana is dominated by the Cavendish (AAA) cultivar group4.
The genome of M. acuminata ssp. malaccensis, a progenitor of the current cultivated banana, was the first Musa genome sequenced5, followed by a draft release of the genome of another banana progenitor, M. balbisiana6, and a close relative, M. itinerans7. In addition, a number of gene expression profiles of different banana cultivars, tissues, and conditions have also been investigated8,9,10,11,12,13,14. Despite the success of banana genomic and transcriptomic sequencing projects, reverse genetic approaches for identifying and characterizing gene functions in banana remain a challenge.
To date, characterizing target gene(s) of interest in banana has mainly relied on transgenic banana plants overexpressing the exogenous target gene15,16,17. More recently, a novel transgenic approach based on clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) are being reported for plants, and the use of CRISPR-Cas9 has been reported in banana cv. Rasthali18. However, low transformation efficiency and the time-exhaustive regeneration processes involved in this transgenic approach are disadvantageous in functional characterization of one or several candidate genes using CRISPR-Cas9 in banana. Moreover, polypoidy especially in the triploid Cavendish banana presents additional challenges for genetic knockout analysis19, as knockout of the target gene in all sets of chromosomes would be needed for loss-of-function analysis.
The availability of genomic and transcriptomic data have provided the basis for reverse genetic studies5,6,7,8,9,13,20,21,22. Virus-induced gene silencing (VIGS) has been used extensively as a tool for reverse genetic study in a wide range of plant species to characterize gene functions or for high throughput phenotypic screening of genes23,24,25,26,27,28,29,30,31,32,33,34. Instead of complete knockout of a gene, the VIGS system knocks down a gene at the RNA level thereby overcoming polyploidy concerns30. Such a system offers an attractively rapid method for loss-of-function assay23,35 in banana by circumventing the laborious transformation method. The ease of the VIGS system has led to the development of several VIGS vectors being used for gene function analysis or for high throughput phenotypic screening of genes in dicotyledonous plants. However, less vectors are available for studies in monocotyledonous plants26,29,32,36,37,38,39.
Cucumber mosaic virus (CMV) is known to infect a wide host range of more than 1200 species in over 100 plant families, including banana37,38,39. CMV is the type member of the genus Cucumovirus and consists of three positive sense single-stranded genomic RNAs (RNA1, 2, and 3) that expresses five proteins (see Supplementary Fig. S1)40,41,42. RNA1 and RNA2 encode the 1a and 2a proteins, respectively, involved in viral genome replication43. Additionally, RNA2 contains the 2b gene, which encodes the suppressor of RNA silencing41,44. The 2b has been reported to suppress RNA silencing through binding to short-interfering RNAs or double-stranded RNAs45,46, and impairing Argonaute 1 and 4 protein activities by direct interaction47,48,49,50,51,52. Previous studies indicate that without functional 2b gene, CMV systemically infects plants with mild symptoms53,54. RNA3 encodes the 3a movement protein and coat protein (CP). The CMV CP is a multifunctional factor that has roles in viral packaging as well as in systemic movement42. CMV has been successfully deployed as a VIGS in dicotyledonous plants such as Nicotiana benthamiana55, Glycine max56, Antirrhinum majus57, Solanum lycopersicum, and Capsicum annuum58, as well as monocotyledonous maize59; whether CMV can induce silencing in the monocotyledonous banana remains unknown.
It has been demonstrated that the successful development of VIGS vector largely depends on the virus strain26,55,59. Previously, we reported isolation of number of CMV isolates from bananas grown in the field60. Subsequently, by selecting a naturally banana-infecting CMV isolate and improving plant infection rates we successfully established a robust VIGS-based system in bananas. The developed tool will greatly expedite gene validation and selection prior to use in transgenic crops.
Results
Development of CMV VIGS vector
Initially, to analyze the suitability of the cucumber mosaic virus 20 (CMV 20)60 previously isolated from banana to induce gene silencing, we constructed cDNA infectious clones of CMV 20 RNAs 1, 2, and 3 (see Supplementary Fig. S2). The infectious clones contained T3 promoter immediately upstream of the full-length cDNA of CMV 20 RNAs 1, 2, and 3 (see Supplementary Fig. S2). We designated the CMV 20 RNA1 as pCMV20-R1, CMV 20 RNA2 as pCMV20-R2, and CMV 20 RNA3 as pCMV20-R3 (see Supplementary Fig. S2). No modification to full-length cDNA of CMV 20 RNAs 1 and 3 were made, whereas an AfeI restriction enzyme site was introduced in a position similar to that previously described55, which is immediately downstream of 2a and in the 2b ORF of CMV to generate pCMV20-R2E (see Supplementary Fig. S2). The AfeI site served as a cloning site for our target gene, and disrupts CMV 20 when a target gene fragment is inserted. VIGS vector construction is detailed in the Materials and Methods section.
CMV 20 induces gene silencing in N. benthamiana by agroinfection
As we were unable to inoculate in vitro transcripts of CMV 20 vector on banana, we sought alternative methods using agroinfection. We cloned CMV 20 genomic RNA 1, 2 and 3 from pCMV20-R1, -R2E, and -R3 to binary vector pJL8961 to generate pJLCMV20-R1, pJLCMV20-R2E, and pJLCMV20-R3 (Fig. 1a). We inserted a DNA fragment of CaMV 35S promoter (35S) in pJLCMV20-R2E AfeI cloning site, which disrupted the CMV 20 2b gene, and designated it as pJLCMV20-35S (Fig. 1a). 35S promoter is a foreign DNA, which is not endogenously found in N. benthamiana or banana. Thus, pJLCMV20-35S was subsequently used as a control vector for pJLCMV20-R2E carrying host endogenous target gene in VIGS assay. DNA fragment of NbGSA and N. benthamiana phytoene desaturase gene (NbPDS) were inserted to pJLCMV20-R2E to generate pJLCMV20-NbGSA and pJLCMV20-NbPDS, respectively (Fig. 1a).
Schematic representation of CMV 20 VIGS vectors. pJLCMV20-R1, pJLCMV20-R2E, and pJLCMV20-R3 were generated by cloning CMV 20 RNA1, RNA2E (RNA2 containing AfeI restriction enzyme site), and RNA3 to binary vector pJL89, respectively. T-DNA left border (LB) and right border (RB) of the binary vector (pJL89) are indicated in light grey. Open rectangles represent open reading frames (ORF) encoded by CMV genomic RNAs or of the inserted DNA fragments (35S, NbGSA, NbPDS, MaGSA, and MaPDS). 35S promoter (35S) is indicated with black arrow immediately adjacent to the most 5′ end of CMV 20 ORF, and hepatitis delta virus ribozyme (Rz) to the 3′ end. Schematic depiction of AfeI cloning site is indicated with a grey line and triangle.
CMV20-R2E agroinfection (a mixture of Agrobacterium carrying pJLCMV20-R1, pJLCMV20-R3, and pJLCMV20-R2E), CMV20-35S agroinfection (a mixture of Agrobacterium carrying pJLCMV20-R1, pJLCMV20-R3, and pJLCMV20-35S), CMV20-NbGSA agroinfection (a mixture of Agrobacterium carrying pJLCMV20-R1, pJLCMV20-R3, and pJLCMV20-NbGSA), and CMV20-NbPDS agroinfection (a mixture of Agrobacterium carrying pJLCMV20-R1, pJLCMV20-R3, and pJLCMV20-NbPDS) were infiltrated with a needleless syringe onto N. benthamiana leaves and assayed for infectivity and silencing efficacy. Phenotypically, CMV20-R2E infected N. benthamiana plants scored an average disease severity level of 3, whereas CMV20-35S agro-inoculated plants exhibited mild (disease severity level of 1) to asymptomatic phenotype (Fig. 2a, see Supplementary Fig. S3). In addition, visible chlorotic and photobleaching phenotype was observed in all plants agroinfected with CMV20-NbGSA or of CMV20-NbPDS at 6–8 dpi, respectively (Fig. 2a).
Agro-inoculation of Nicotiana benthamiana and banana with CMV 20 VIGS vectors. (a) Phenotypes of N. benthamiana inoculated with Agrobacterium containing pJL89 (mock), or Agrobacterium mixture carrying pJLCMV20-R1, pJLCMV20-R3 plus pJLCMV20-R2E (CMV20-R2E), pJLCMV20-35S (CMV20-35S), pJLCMV20-NbGSA (CMV20-NbGSA), or pJLCMV20-NbPDS (CMV20-NbPDS) at 8 dpi. Bar = 5 cm. (b) Quantification of the mRNA levels of CMV 20 coat protein in N. benthamiana inoculated with mock, CMV20-R2E, CMV20-35S, CMV20-NbGSA, and CMV20-NbPDS. Data represent mean ± SD, n = 5 biological replicates. Different letters indicate statistically significant differences analyzed by one-way analysis of variance (ANOVA) Tukey’s test (P < 0.05). (c) Quantification of the mRNA levels of NbPDS, and NbGSA of N. benthamiana analyzed by quantitative RT-PCR, normalized relative to the level of actin at 8 dpi. Data represent mean ± SD; n = 5 biological replicates; ND, not detectable. ****P < 0.001, Student’s t test compared to CMV20-35S agro-inoculum inoculated plants. (d) Schematic representation of Agrobacterium-assisted CMV 20 inoculation method by injecting Agrobacterium mixture carrying CMV 20 into bananas at the pseudostem-rhizome junction. (e) Phenotypes of healthy, mock inoculated, or CMV20-R2E agro-infected banana plants. Scale bar = 10 cm.
Total RNA extracted from mock, CMV20-R2E, CMV20-35S, CMV20-NbGSA, and CMV20-NbPDS agro-infiltrated plants at 8 dpi were used for detection of CMV by real- time qRT-PCR. The infection of CMV was confirmed in all CMV20-R2E, CMV20-35S, CMV20-NbGSA, and CMV20-NbPDS agro-infiltrated plants in our three independent experiments (Fig. 2b). In addition, significant reduction of GSA and PDS mRNA was observed in plants with CMV20-NbGSA and CMV20-NbPDS agroinfection, respectively, compared to CMV20-35S agroinfected plants (Fig. 2c).
Development of efficient CMV 20 vector inoculation methods on banana by pseudostem-rhizome junction injection of Agrobacterium
Despite successfully agro-infiltrating CMV 20 vectors in N. benthamiana, we were not able to transmit CMV 20 vectors in banana using a regular agro-infiltration method. Therefore, we optimized an agroinfection method for banana. We used inoculation buffer that has been reported to enable high inoculation efficiency62,63, and used needled injection instead of needleless infiltration of Agrobacterium-mixture to minimize the wounding as we previously demonstrated on orchids32,64. In addition, we tested agro-injection or CMV 20 vector on different banana tissues (Fig. 2d). In our repeated sets of experiments, 95% (41 of 43 plants) of bananas inoculated with CMV20-R2E at the pseudostem-rhizome junction showed severe viral symptoms, similar to the slender leaves previously described60, starting at 25 dpi to 36 dpi (Fig. 2e). In addition to symptom observation, qRT-PCR was used to detect CMV in ten randomly selected plants that had been inoculated with CMV20-R2E, which confirmed consistent CMV infection.
Silencing of the banana GSA and PDS genes by CMV 20 vectors
Building on the success of our inoculation system, we next tested whether CMV 20 vector could be used to induce gene silencing in bananas. A 205-bp Musa GSA (MaGSA) and a 204-bp Musa PDS (MaPDS) DNA fragment were cloned into pJLCMV20-R2E to generate pJLCMV20-MaGSA and pJLCMV20-MaPDS, respectively (Fig. 1). CMV20-R2E, CMV20-35S, CMV20-MaGSA (mixture of Agrobacterium carrying pJLCMV20-R1, pJLCMV20-R3, and pJLCMV20-MaGSA), or CMV20-MaPDS (mixture of Agrobacterium carrying pJLCMV20-R1, pJLCMV20-R3, and pJLCMV20-MaPDS) were agroinfected into banana plants.
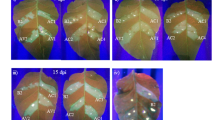
In our experiments, banana inoculated at 25 °C for 2 weeks and then shifted to 28 °C showed GSA silencing phenotype that could be observed on CMV20-MaGSA agroinfected banana starting at 30 dpi (Fig. 3a, Table 1). In three repeated sets of experiments, the chlorotic phenotype was observed in at least 70% and up to 80% of CMV20-MaGSA agroinfected bananas (Table 1), and the photobleaching phenotype was observed in at least 60% and up to 85% of CMV20-MaPDS agroinfected bananas (Experiments 2 and 3, Table 1). The majority of the CMV20-35S, CMV20-MaGSA, or CMV20-MaPDS agroinfected bananas were asymptomatic or mildly symptomatic, and severe symptoms were rarely observed (1 out of 28 CMV20-35S agroinfected bananas, 1 out of 28 CMV20-MaGSA agroinfected bananas, and 1 out of 23 CMV20-MaPDS agroinfected bananas) (Experiments 1–3, Table 1). In addition to CMV20-R2E, real-time qRT-PCR also revealed that the infection rate reached 92%, 100%, and 100% for bananas with CMV20-35S agroinfection, CMV20-MaGSA agroinfection, and CMV20-MaPDS agroinfection, respectively (Experiment 3, Table 1).
CMV20-based VIGS of glutamate 1-semialdehyde aminotransferease (MaGSA) or phytoene desaturase (MaPDS) in banana. (a) Phenotype of banana inoculated with Agrobacterium containing pJL89 (mock), or Agrobacterium mixture carrying pJLCMV20-R1, pJLCMV20-R3 plus pJLCMV20-R2E (CMV20-R2E), pJLCMV20-35S (CMV20-35S), or pJLCMV20-MaGSA (CMV20-MaGSA) at 30 dpi. Bar = 5 cm. Quantification of the mRNA levels of CMV 20 coat protein and MaGSA was analyzed by quantitative RT-PCR, normalized relative to the level of actin. (b) Phenotypes of banana inoculated with Agrobacterium containing pJL89 (mock), CMV20-R2E, CMV20-35S, or pJLCMV20-MaPDS (CMV20-MaPDS) at 30 dpi. Bar = 5 cm. Quantification of the mRNA levels of CMV20 coat protein and MaPDS was analyzed by quantitative RT-PCR, normalized relative to the level of actin. Quantitative RT-PCR data represent mean ± SD; 3 representative technical repeats; ND, not detectable. Different letters indicate statistically significant differences analyzed by one-way analysis of variance (ANOVA) Tukey’s test (P < 0.05).). ****P < 0.001, Student’s t test compared to CMV20-35S agro-inoculum inoculated plants.
The silencing phenotypes that could be observed starting around 30 dpi (Fig. 3a,b) could also be observed in newly developed leaves up to 63 dpi. In addition, the results of qRT-PCR revealed that the RNA level of MaGSA in CMV20-MaGSA agroinfected bananas and MaPDS in CMV20-MaPDS agroinfected bananas was significantly reduced by 90% and 82% of the CMV20-35S agroinfected bananas (Fig. 3a,b), respectively, and corresponded to the phenotype observed. Taken together, these data showed that the currently developed CMV 20-based VIGS system could be effectively used to silence genes of interest in banana plants.
CMV 20-vectors induce generation of siRNA
To further confirm that reduced expression of GSA and PDS RNA was due to RNA interference caused by VIGS, small RNA-seq was conducted using mock, CMV20-35S, CMV20-MaGSA, and CMV20-MaPDS agro-inoculum inoculated plants. Short sense and antisense reads of 21–24 nt were mapped to the cDNA sequences of MaGSA and MaPDS (Fig. 4). Although short sense and antisense reads that mapped to either MaGSA or MaPDS can be identified in all plant groups (mock, CMV20-35S, CMV20-MaGSA, and CMV20-MaPDS agro-inoculum inoculated plants), the relative abundance of sense and antisense reads of MaGSA and MaPDS was significantly higher in CMV20-MaGSA and CMV20-MaPDS bananas compared to control plants (mock and CMV20-35S plants), respectively. In CMV20-MaGSA agroinfected bananas, the small RNA (21–24 nt) abundance mapped to MaGSA was 1052 fold higher than the mock and 446 fold higher than the CMV20-35S agroinfected bananas (see Supplementary Table S1). In CMV20-MaPDS agroinfected bananas, the small RNA (21–24 nt) abundance mapped to MaPDS was 1195 fold higher compared to mock and 451 fold higher compared to CMV20-35S agroinfected bananas (see Supplementary Table S1). These results indicate the target specificity of the established VIGS using CMV 20 as viral vector.
Discussion
Here, we report the development of a robust and efficient CMV-VIGS system for bananas and demonstrated that this approach enabled efficient silencing of genes using PDS and GSA. The developed CMV 20 vector allows silencing of targeted genes of interest with high infection rate and mild disease symptoms. The success of our VIGS system in banana may be partly attributed to using a naturally banana infecting CMV isolate for the construction of banana VIGS system, as well as overcoming the difficulties of CMV 20 inoculation on banana using a pseudostem-rhizome junction needled agroinjection system (Fig. 2d).
In our study, we tested a number of methods to inoculate CMV 20 empty vector or CMV 20 vector with a targeted silence gene insert. While agro-inoculation systems have been widely adopted for virus delivery into plants65,66,67, inoculation of plant viruses with an agrobacterium-assisted system on Musa plants has not been successfully established prior to this study. Compared to inoculating CMV 20 on N. benthamiana, the thicker banana leaves, latex, and other inhibitory substances present in bananas add to the challenge of establishing an efficient viral inoculation system68,69. Of several inoculation methods, we were able to obtain a desirable infection rate (Table 1) by replacing regular needleless agro-infiltration methods with a syringe and fine needle for inoculum injection. Such an approach may allow minimal wounding on banana, and injecting an Agrobacterium mixture carrying CMV 20 to the center of banana pseudostem-rhizome juncture, likely allowed virus infection to take place on the meristem or yet-to-emerge leaves before the true banana leaf is developed. Interestingly, it has been previously reported that 2b protein-defective mutant is unable to invade the shoot apical meristem70. This may partly explain why CMV 20 accumulation is significantly higher in CMV20-R2E agroinfected plants compared to CMV20-GSA or CMV20-PDS.
The GSA and PDS genes are commonly used as phenotypic markers for VIGS vector development in many plant species59. Using CMV 20-based VIGS vector, silencing MaGSA and MaPDS in banana resulted in a chlorotic or photobleaching phenotype, respectively, that was observed starting 30 dpi and persisted until 63 dpi. We speculate that the meristem tissue is vulnerable to transformation of agrobacterium, and serves as the initial infection site for CMV. As no vascular system in the meristem is available for virus movement, virus infection is confined to the meristem, and VIGS phenotype is only visualized with the development of the leaves. Moreover, the visual phenotype was observed in 77% or 85% of the infected bananas inoculated with CMV20-MaGSA agro-inoculum or CMV20-MaPDS agro-inoculum, respectively (Experiment 3, Table 1). We found initial temperature set at 25 °C for two weeks followed by constant 28 °C provided the best VIGS results compared to constant temperature maintained at 28 °C. It has been reported that lower temperature during inoculation may increase the transformation efficiency of Agrobacterium71. The temperature control allowed us to achieve a balance for plant growth and VIGS efficacy.
Small RNA sequencing from mock, CMV20-35S agro-inoculum, CMV20-MaGSA agro-inoculum, and CMV20-MaPDS agro-inoculum inoculated banana revealed that higher levels of small RNAs were mapped to the targeted gene and corresponded to the reduction in the mRNA transcript level of the targeted gene, and this higher level of MaGSA and MaPDS small RNAs are found in both the sense and anti-sense orientation indicating that double stranded small RNA is formed and induced post-transcriptional silencing response in this VIGS system.
In this study, the CMV 20-based VIGS vector developed effectively silenced endogenous genes in both N. benthamiana and banana, demonstrating that CMV 20 is able to induce silencing in both dicots and monocots, respectively. We demonstrated that the combination of the developed CMV 20 vector and inoculation strategy enabled silencing of the targeted gene of interest in banana with high infection rate and silencing efficacy. Although VIGS may be limited by the duration of silencing or inherently having to observe plant phenotype under virus infection, the availability of the system developed here may enable previously unavailable rapid functional analysis of genes in banana and expedite a more complete understanding of different biological questions in this important staple and cash crop.
Materials and Methods
Plant material and growth conditions
Nicotiana benthamiana was grown in pots at 25 °C, in insect-proof growth chamber under 16 h light/8 h dark cycle with 60 μmol m−2 s−1 illumination. ‘Tai-Chiao No. 7’ (Cavendish, AAA subgroup) plantlets were purchased from Taiwan Banana Research Institute (Pingtung, Taiwan). Unless otherwise indicated, banana plants in jars were first placed in growth chamber for seven days for acclimation prior to potting in sterilized soil containing 6 parts peat, 1 part perlite, and 1 part vermiculite, and grown in insect-proof growth chamber at 28 °C under 16 h light/8 h dark cycle with 60 μmol m−2 s−1 illumination. For VIGS experiments, unless otherwise specified, inoculated banana plantlets were placed at 25 °C under 16 h light/8 h dark cycle with 60 μmol m−2 s−1 illumination for 2 weeks and transferred to 28 °C of the same illumination.
Construction of infectious CMV20-derived vectors
All primers used are listed in Supplementary Table S2. The full-length cucumber mosaic virus (CMV20) cDNA of RNAs 1, 2, and 3 (NCBI accession number: MH709142-MH709144) were first cloned into pGEM-T vectors (Promega, Madison, WI, USA) and designated pCMV20-R1, pCMV20-R2, and pCMV20-R3. Total RNA extracted from CMV20 infected N. benthamiana using TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions, was used as template for synthesis of first strand cDNA of CMV 20 RNA1 by use of CMV 3′ primer and M-MLV Reverse Transcriptase (Promega). Two overlapping CMV20 RNA 1 fragments were PCR amplified using primer pairs R125 (with T3 promoter)/YR1 1646RE and YR1 1584FR/CMV3′. The two amplified fragments were cloned into pGEM-T (Promega). The two plasmids were digested with EcoRV and NotI and separated on 1% agarose gel. The 4.6 and 1.7 kb fragments were gel purified and ligated to generate pCMV20-R1.
The construction of pCMV20-R2 is similar to construction of pCMV20-R1 except primer pairs R125 (with T3 promoter)/YR2 1720RE and YR2 1536FR/CMV3′ were used instead to amplify two overlapping CMV fragments for cloning to pGEM-T (Promega), and both cloned plasmids were digested with XhoI and SpeI then, separated on 1% agarose gel. The 4.5 kb and 1.5 kb fragments were gel purified and ligated to generate pCMV20-R2.
For construction of pCMV20-R3, primer pair RNA35′20-2 (with T3 promoter)/CMV 3′ were used to amplify full-length CMV 20 RNA3. The amplified fragment was cloned into pGEM-T easy (Promega) to generate pCMV20-R3.
An AfeI restriction enzyme site was added on the 2b ORF to generate a gene insertion site similar to the method previously described55. Two fragments were amplified by PCR with pCMV20-R2 used as template and primer pairs CMV2-2225F/CMV2-2663AfeIR and CMV2-2663AfeIF/CMV2-294R. The amplified fragments were gel extracted and mixed as templates for overlapping extension PCR reaction by use of primer pairs CMV2-2225F/CMV2-2949R. The amplified fragments were gel extracted and digested with SfoI and AvrII. The digested fragments were ligated into pCMV20-R2 that had been digested with SfoI and AvrII to generate pCMV20-R2E.
To generate pJLCMV20-R1, pCMV20-R1 was used as template and was amplified by PCR with primer pairs CMV20-R1R2-F/CMV20-R1-R. The amplified fragment was ligated to pJL8961 digested with StuI and SmaI. The construction of pJLCMV20-R2E and pJLCMV20-R3 followed a similar method to the construction of pJLCMV20-R1 except pCMV20-R2E and pCMV20-R3 were used as a template, and primer pairs CMV20-R1R2-F/CMV20-R2-R and CMV20-R3-F/CMV20-R3-R were used to amplify the CMV20-R2E and CMV20-R3 fragments in the PCR reaction and ligated to pJL8961.
Construction of CMV20 gene-silencing vectors
Total RNA extracted from N. benthamiana using TRIzol reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions, was used to synthesize cDNA with PrimeScript RT-PCR Kit (TaKaRa Bio Inc., Otsu, Shiga, Japan). The gene fragment of NbPDS (NCBI accession number: EU165355.1) was amplified by using the synthesized cDNA and primer pair NbPDS-F/NbPDS-R. The amplified fragment was cloned into AfeI-digested pJLCMV20-R2E to generate pJLCMV20-NbPDS. The PCR amplification of NbGSA (Sol Genomic Network sequence ID: Niben101Scf06382g00006.1) fragment is similar to amplification of NbPDS fragment except NbGSA-F/NbGSA-R primer pair was used instead. The amplified NbGSA was cloned into AfeI digested pJLCMV20-R2E to generate pJLCMV20-NbGSA. 35S DNA fragment was PCR amplified with template pCymMV-Gateway vector (Lu et al., 2012) and primer pair 35S-F/35S-R, and cloned into AfeI-digested pJLCMV20-R2E to generate pJLCMV20-35S.
The construction of pJLCMV20-MaPDS and pJLCMV20-MaGSA was similar to that described above for pJLCMV20-NbPDS and pJLCMV20-NbGSA except total RNA was extracted from banana using PureLink Plant RNA Reagent (Thermo Fisher Scientific) and MaPDS (NCBI accession number: XM_018821595.1); and MaGSA (NCBI accession number: XM_009405501.2) fragments were amplified by primers MusaPDS-F/R or MusaGSA-F/R and cloned into the AfeI site of the pJLCMV20-R2E, which yielded plasmids pJLCMV20-MaPDS and pJLCMV20-MaGSA, respectively.
Agroinfection of N. benthamiana and banana
All T-DNA clones were separately introduced into A. tumefaciens EHA105 by electroporation using a Gene Pulser (Bio-Rad Laboratories, Hercules, CA, USA). For N. benthamiana, the vectors pJLCMV20-R1, pJLCMV20-R2E, pJLCMV20-R3, pJLCMV20-35S, pJLCMV20-NbPDS, pJLCMV20-NbGSA were transformed individually into A. tumefaciens strain EHA105. Preparation of Agrobacterium cultures harboring pJLCMV20-R1, pJLCMV20-R2E, pJLCMV20-R3, or pJLCMV20-R2E derivatives carrying the specified gene inserts were grown and prepared as described previously62. Selective medium kanamycin of 50 mg in 1 L of Yeast Extract Peptone (YEP) liquid medium was used. The cell mixtures with 1:1:1 ratio between pJLCMV20-R1, pJLCMV20-R2E, pJLCMV20-R3, or pJLCMV20-R2E derivatives carrying the specified gene inserts (i.e. 35S, NbPDS, NbGSA) were infiltrated into true leaves of N. benthamiana plants. Mock plants were infiltrated with infiltration solution containing A. tumefaciens strain EHA105. Systemic leaves were collected at 8 days post-inoculation (dpi) for further analysis.
For bananas, preparation of the inoculum was similar to that previously described. For inoculation of banana, aliquots of 400 μl of Agrobacterium mixture was injected into pseudostem-rhizome junction of banana plants (1-month after transplanting tissue culture plantlets to soil pots) using a 0.5-ml sterile syringe.
RT-PCR and qRT-PCR analysis
For N. benthamiana, total RNAs were extracted using TRIzol Reagent (Thermo Fisher Scientific) from 1 mg of leaf sample, and treated with Turbo DNAse (Thermo Fisher Scientific) to remove genomic DNA. cDNA for qRT-PCR was synthesized from 1 μg of DNA-free RNA and oligo (dT) using PrimeScript RT Reagent Kit (Takara Bio), following the manufacturer’s instructions. For banana samples, total RNAs were extracted using Purelink Plant Reagent (Thermo Fisher Scientific), and the synthesis of cDNA followed the same instructions as N. benthamiana.
For detection and quantification of CMV20 in both N. benthamiana and banana, primers pairs CMV20CP-F/R that targeted CMV20 coat protein were used. All qRT-PCR was carried out using SYBR Premix EX Taq II Kit (Takara Bio) with ABI Prism 7500 sequence detection system (Applied Biosystems). Actin of N. benthamiana or banana was used as a control to calculate the relative CMV20 level.
For qRT-PCR assay of silencing experiments, gene-specific primers were designed to prime outside the region targeted for silencing. For qRT-PCR of N. benthamiana PDS or GSA, primer pairs NbPDS-qF/qR or NbGSA-qF/qR were used, respectively. N. benthamiana actin (NCBI accession number: JQ256516.1) was used as an internal control using primer pairs NbActin-qF/qR. For banana PDS or GSA, primer pairs MusaPDS2-qF/qR or MusaGSA-qF/qR were used, respectively. Banana actin (NCBI accession number: XM_009412781.2) was used as in internal control using primer pairs MusaActin-qF/qR. RNA derived from mock- or 35S control vector-inoculated plants, as specified in the figures, was set as 1 for relative quantitative analysis.
Small RNA sequencing and small RNA-Seq raw read processing
Total RNA of mock, CMV20-35S, CMV20-MaGSA, and CMV20-MaPDS were extracted from banana using PureLink Plant Reagent (Thermo Fisher Scientific) and sent underwent small RNA sequencing conducted by Biotools (Taipei, Taiwan). In brief, the small RNA was size excised from agarose gel and used as input material for a small RNA library construction using NEBNext Multiplex Small RNA Library Prep Set for Illumina (New England Biolabs, Ipswich, MA, USA) according to the manufacturer’s protocol. The libraries were pooled and sequenced on NextSeq 500 (Illumina, San Diego, CA, USA), and the sequenced data were uploaded into the NCBI Sequence Read Archive (Accession number: SRP159135). For raw read processing, the FastX-Toolkit v.0.0.14 was used to trim the adapter and for quality control. The trimmed reads of 18–28 nt were quality-filtered with patterns [-q 30 -p 90]. Next, the trimmed reads were mapped to the banana genome and cDNA sequences retrieved from the banana genome hub (Musa acuminata DH-Pahang version 2; banana-genome-hub.southgreen.fr) by BOWTIE 1.2.1.1 with 1 mismatch. The reads with >20 genomic hits or mapped to the mitochondria genome, chloroplast genome from GenBank (HF677508.1), non-coding RNA from Rfam 13.0, such as ribosomal RNAs, transfer RNAs, small nuclear RNAs and small nucleolar RNAs, were discarded. Reads derived from the transcript of MaPDS and MaGSA sequences were identified using BOWTIE 1.2.1.1 with 1 mismatch.
References
De Langhe, C., Vrydaghs, L., De Maret, P., Perrier, X. & Denham, T. Why bananas matter: an introduction to the history of banana domestication. Ethnobotany Research and Applications 7, 165–177 (2009).
FAOSTAT. http://faostat.fao.org (2019).
Heslop-Harrison, J. S. & Schwarzacher, T. Domestication, genomics and the future for banana. Ann Bot 100, 1073–1084, https://doi.org/10.1093/aob/mcm191 (2007).
Arias, P. The World Banana Economy, 1985–2002. (Food & Agriculture Org., 2003).
D’Hont, A. et al. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 488, 213–217, https://doi.org/10.1038/nature11241 (2012).
Davey, M. W. et al. A draft Musa balbisiana genome sequence for molecular genetics in polyploid, inter- and intra-specific Musa hybrids. BMC Genomics 14, 683, https://doi.org/10.1186/1471-2164-14-683 (2013).
Wu, W. et al. Whole genome sequencing of a banana wild relative Musa itinerans provides insights into lineage-specific diversification of the Musa genus. Scientific Reports 6, https://doi.org/10.1038/srep31586 (2016).
Li, C. et al. Transcriptome profiling of resistant and susceptible Cavendish banana roots following inoculation with Fusarium oxysporum f. sp. cubense tropical race 4. BMC Genomics 13, 374, https://doi.org/10.1186/1471-2164-13-374 (2012).
Wang, Z. et al. De Novo characterization of the banana root transcriptome and analysis of gene expression under Fusarium oxysporum f. sp. Cubense tropical race 4 infection. BMC Genomics 13, 650, https://doi.org/10.1186/1471-2164-13-650 (2012).
Bai, T. T. et al. Transcriptome and Expression Profile Analysis of Highly Resistant and Susceptible Banana Roots Challenged with Fusarium oxysporum f. sp. cubense Tropical Race 4. PLoS ONE 8, https://doi.org/10.1371/journal.pone.0073945 (2013).
Guo, L. et al. Genome and Transcriptome Analysis of the Fungal Pathogen Fusarium oxysporum f. sp. cubense Causing Banana Vascular Wilt Disease. PLoS ONE 9, https://doi.org/10.1371/journal.pone.0095543 (2014).
Backiyarani, S. et al. Genome-wide analysis and differential expression of chitinases in banana against root lesion nematode (Pratylenchus coffeae) and eumusa leaf spot (Mycosphaerella eumusae) pathogens. Applied biochemistry and biotechnology 175, 3585–3598, https://doi.org/10.1007/s12010-015-1528-z (2015).
Hu, W. et al. The core regulatory network of the abscisic acid pathway in banana: genome-wide identification and expression analyses during development, ripening, and abiotic stress. BMC plant biology 17, 145, https://doi.org/10.1186/s12870-017-1093-4 (2017).
Wang, Y. et al. Differential gene expression in banana roots in response to Fusarium wilt. Canadian Journal of Plant Pathology 39, 163–175, https://doi.org/10.1080/07060661.2017.1342693 (2017).
May, G. D. et al. Generation of transgenic banana (Musa acuminata) plants via Agrobacterium-mediated transformation. Nature biotechnology 13, 486 (1995).
Yadav, K., Patel, P., Srivastava, A. K. & Ganapathi, T. Overexpression of native ferritin gene MusaFer1 enhances iron content and oxidative stress tolerance in transgenic banana plants. PLoS ONE 12, e0188933 (2017).
Kovács, G. et al. Expression of a rice chitinase gene in transgenic banana (‘Gros Michel’, AAA genome group) confers resistance to black leaf streak disease. Transgenic research 22, 117–130 (2013).
Kaur, N. et al. CRISPR/Cas9-mediated efficient editing in phytoene desaturase (PDS) demonstrates precise manipulation in banana cv. Rasthali genome. Functional & integrative genomics 18, 89–99 (2018).
Liu, J. et al. Efficient regeneration and genetic transformation platform applicable to five Musa varieties. Electronic Journal of Biotechnology 25, 33–38 (2017).
Dash, P. K. & Rai, R. Translating the “Banana genome” to delineate stress resistance, dwarfing, parthenocarpy and mechanisms of fruit ripening. Frontiers in plant science 7, 1543 (2016).
Dash, P. K., Rai, R., Mahato, A. K., Gaikwad, K. & Singh, N. K. Transcriptome landscape at different developmental stages of a drought tolerant cultivar of flax (Linum usitatissimum). Frontiers in chemistry 5, 82 (2017).
Shivaraj, S. et al. Genome-wide identification, characterization, and expression profile of aquaporin gene family in flax (Linum usitatissimum). Scientific Reports 7, 46137 (2017).
Baulcombe, D. C. Fast forward genetics based on virus-induced gene silencing. Current opinion in plant biology 2, 109–113 (1999).
Turnage, M. A., Muangsan, N., Peele, C. G. & Robertson, D. Geminivirus‐based vectors for gene silencing in Arabidopsis. The Plant Journal 30, 107–114 (2002).
Burch‐Smith, T. M., Anderson, J. C., Martin, G. B. & Dinesh‐Kumar, S. P. Applications and advantages of virus‐induced gene silencing for gene function studies in plants. The Plant Journal 39, 734–746 (2004).
Lu, H.-C. et al. Strategies for functional validation of genes involved in reproductive stages of orchids. Plant Physiology 143, 558–569 (2007).
Padmanabhan, M. & Dinesh-Kumar, S. P. Virus-induced gene silencing as a tool for delivery of dsRNA into plants. Cold Spring Harbor Protocols 2009, pdb. prot5139 (2009).
Tang, Y. et al. Virus-based microRNA expression for gene functional analysis in plants. Plant physiology, pp. 110.155796 (2010).
Zhang, C., Bradshaw, J. D., Whitham, S. A. & Hill, J. H. The development of an efficient multipurpose bean pod mottle virus viral vector set for foreign gene expression and RNA silencing. Plant physiology 153, 52–65 (2010).
Senthil-Kumar, M. & Mysore, K. S. New dimensions for VIGS in plant functional genomics. Trends in plant science 16, 656–665 (2011).
Huang, C., Qian, Y., Li, Z. & Zhou, X. Virus-induced gene silencing and its application in plant functional genomics. Science China Life Sciences 55, 99–108 (2012).
Lu, H.-C. et al. A high-throughput virus-induced gene-silencing vector for screening transcription factors in virus-induced plant defense response in orchid. Molecular plant-microbe interactions 25, 738–746 (2012).
Zhang, J. et al. Vacuum and co-cultivation agroinfiltration of (germinated) seeds results in tobacco rattle virus (TRV) mediated whole-plant virus-induced gene silencing (VIGS) in wheat and maize. Frontiers in plant science 8, 393 (2017).
de Oliveira, M. V. et al. Specific control of Arabidopsis BAK1/SERK4-regulated cell death by protein glycosylation. Nature plants 2, 15218 (2016).
Ratcliff, F., Martin‐Hernandez, A. M. & Baulcombe, D. C. Technical advance: tobacco rattle virus as a vector for analysis of gene function by silencing. The Plant Journal 25, 237–245 (2001).
Scofield, S. R., Huang, L., Brandt, A. S. & Gill, B. S. Development of a virus-induced gene-silencing system for hexaploid wheat and its use in functional analysis of the Lr21-mediated leaf rust resistance pathway. Plant Physiology 138, 2165–2173 (2005).
Douine, L., Quiot, J., Marchoux, G. & Archange, P. Index of plants susceptible to cucumber mosaic virus (CMV). Bibliographical study. Annales de Phytopathologie (France) (1979).
King, A. M., Lefkowitz, E., Adams, M. J. & Carstens, E. B. Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses. (Elsevier, 2011).
Scholthof, K. B. G. et al. Top 10 plant viruses in molecular plant pathology. Molecular plant pathology 12, 938–954 (2011).
Palukaitis, P., Roossinck, M. J., Dietzgen, R. G. & Francki, R. I. In Advances in virus research Vol. 41 281–348 (Elsevier, 1992).
Ding, S.-W., Anderson, B. J., Haase, H. R. & Symons, R. H. New overlapping gene encoded by the cucumber mosaic virus genome. Virology 198, 593–601 (1994).
Palukaitis, P. & García-Arenal, F. Cucumoviruses. Advances in virus research 62, 241–323 (2003).
Hayes, R. J. & Buck, K. W. Complete replication of a eukaryotic virus RNA in vitro by a purified RNA-dependent RNA polymerase. Cell 63, 363–368 (1990).
Guo, H. S. & Ding, S. W. A viral protein inhibits the long range signaling activity of the gene silencing signal. The EMBO journal 21, 398–407 (2002).
Goto, K., Kobori, T., Kosaka, Y., Natsuaki, T. & Masuta, C. Characterization of silencing suppressor 2b of cucumber mosaic virus based on examination of its small RNA-binding abilities. Plant and cell physiology 48, 1050–1060 (2007).
González, I. et al. RNA binding is more critical to the suppression of silencing function of Cucumber mosaic virus 2b protein than nuclear localization. RNA 18, 771–782 (2012).
Zhang, X. et al. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes & development 20, 3255–3268 (2006).
Duan, C.-G. et al. Suppression of Arabidopsis ARGONAUTE1-mediated slicing, transgene-induced RNA silencing, and DNA methylation by distinct domains of the Cucumber mosaic virus 2b protein. The Plant Cell 24, 259–274 (2012).
Feng, L., Duan, C. G. & Guo, H. S. Inhibition of in vivo S licer activity of A rgonaute protein 1 by the viral 2b protein independent of its dsRNA‐binding function. Molecular plant pathology 14, 617–622 (2013).
González, I. et al. Cucumber mosaic virus 2b protein subcellular targets and interactions: their significance to RNA silencing suppressor activity. Molecular plant-microbe interactions 23, 294–303 (2010).
Hamera, S., Song, X., Su, L., Chen, X. & Fang, R. Cucumber mosaic virus suppressor 2b binds to AGO4‐related small RNAs and impairs AGO4 activities. The Plant Journal 69, 104–115 (2012).
Fang, Y.-Y. et al. CMV2b-AGO interaction is required for the suppression of RDR-dependent antiviral silencing in Arabidopsis. Frontiers in microbiology 7, 1329 (2016).
Lewsey, M. et al. The role of the Cucumber mosaic virus 2b protein in viral movement and symptom induction. Molecular plant-microbe interactions 22, 642–654 (2009).
Lewsey, M. G. et al. Symptom induction and RNA silencing suppression by the cucumber mosaic virus 2b protein. Plant signaling & behavior 5, 705–708 (2010).
Otagaki, S. et al. Rapid induction of transcriptional and post-transcriptional gene silencing using a novel Cucumber mosaic virus vector. Plant biotechnology 23, 259–265 (2006).
Nagamatsu, A. et al. Functional analysis of soybean genes involved in flavonoid biosynthesis by virus‐induced gene silencing. Plant biotechnology journal 5, 778–790 (2007).
Kim, B. M., Inaba, J.-I. & Masuta, C. Virus induced gene silencing in Antirrhinum majus using the Cucumber Mosaic Virus vector: functional analysis of the AINTEGUMENTA (Am-ANT) gene of A. majus. Horticulture, Environment, and Biotechnology 52, 176 (2011).
Hong, J.-S. et al. Application of a reassortant Cucumber mosaic virus vector for gene silencing in tomato and chili pepper plants. The Plant Pathology Journal 28, 81–86 (2012).
Wang, R. et al. An efficient virus‐induced gene silencing vector for maize functional genomics research. The Plant Journal 86, 102–115 (2016).
Chou, C. N., Chen, C. E., Wu, M. L., Su, H. J. & Yeh, H. H. Biological and molecular characterization of Taiwanese isolates of Cucumber mosaic virus associated with banana mosaic disease. Journal of Phytopathology 157, 85–93 (2009).
Lindbo, J. A. TRBO: a high-efficiency tobacco mosaic virus RNA-based overexpression vector. Plant physiology 145, 1232–1240 (2007).
Wu, H.-Y. et al. AGROBEST: an efficient Agrobacterium-mediated transient expression method for versatile gene function analyses in Arabidopsis seedlings. Plant methods 10, 19 (2014).
Wang, Y.-C., Yu, M., Shih, P.-Y., Wu, H.-Y. & Lai, E.-M. Stable pH Suppresses Defense Signaling and is the Key to Enhance Agrobacterium-Mediated Transient Expression in Arabidopsis Seedlings. Scientific Reports 8, 17071 (2018).
Chang, L. et al. Plant A20/AN1 protein serves as the important hub to mediate antiviral immunity. PLoS pathogens 14, e1007288 (2018).
Kant, R., Sharma, S. & Dasgupta, I. In Plant Gene Silencing 201–217 (Springer, 2015).
Lu, R. et al. High throughput virus‐induced gene silencing implicates heat shock protein 90 in plant disease resistance. The EMBO journal 22, 5690–5699 (2003).
Vaghchhipawala, Z., Rojas, C. M., Senthil-Kumar, M. & Mysore, K. S. In Plant Reverse Genetics 65–76 (Springer, 2011).
Lockhart, B. Purification and serology of a bacilliform virus associated with banana streak disease. Phytopathology 76, 995–999 (1986).
Lockhart, B. Banana streak badnavirus infection in Musa: epidemiology, diagnosis and control. (ASPAC Food & Fertilizer Technology Center, 1995).
Sunpapao, A., Nakai, T., Dong, F., Mochizuki, T. & Ohki, S. T. The 2b protein of cucumber mosaic virus is essential for viral infection of the shoot apical meristem and for efficient invasion of leaf primordia in infected tobacco plants. Journal of general virology 90, 3015–3021 (2009).
Dillen, W. et al. The effect of temperature on Agrobacterium tumefaciens‐mediated gene transfer to plants. The Plant Journal 12, 1459–1463 (1997).
Acknowledgements
This work was supported by an Innovative Translational Agricultural Research grant (AS-KPQ-107-ITAR-TD02) and by Academia Sinica, Taipei, Taiwan. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. We thank Miranda Loney of the Agricultural Biotechnology Research Center, Academia Sinica for English editing.
Author information
Authors and Affiliations
Contributions
H.-H.Y. conceived and coordinated the study. M.-C.L., H.-H.J., Y.-S.C., T.-C.T., C.-N.C. and Y.T. performed experiments and data analysis. H.-M.C. and B.-.H.H. supervised and conducted small RNA-seq analysis. H.-H.Y. and Y.T. wrote the manuscript. H.-H.Y. agrees to serve as the author responsible for contact and ensures communication.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tzean, Y., Lee, MC., Jan, HH. et al. Cucumber mosaic virus-induced gene silencing in banana. Sci Rep 9, 11553 (2019). https://doi.org/10.1038/s41598-019-47962-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-47962-3
This article is cited by
-
Cucumber mosaic virus-induced gene and microRNA silencing in water dropwort (Oenanthe javanica (Blume) DC)
Plant Methods (2024)
-
Modern plant biotechnology as a strategy in addressing climate change and attaining food security
Agriculture & Food Security (2022)
-
Effect of mutations in the 2b protein of tomato aspermy virus on RNA silencing suppressor activity, virulence, and virus-induced gene silencing
Archives of Virology (2022)
-
First report of viruses infecting Vigna marina and Vigna riukiuensis in Japan
Journal of General Plant Pathology (2022)
-
Simultaneous silencing of two different Arabidopsis genes with a novel virus-induced gene silencing vector
Plant Methods (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.