Abstract
Top-Down Proteomics (TDP) is an emerging proteomics protocol that involves identification, characterization, and quantitation of intact proteins using high-resolution mass spectrometry. TDP has an edge over other proteomics protocols in that it allows for: (i) accurate measurement of intact protein mass, (ii) high sequence coverage, and (iii) enhanced identification of post-translational modifications (PTMs). However, the complexity of TDP spectra poses a significant impediment to protein search and PTM characterization. Furthermore, limited software support is currently available in the form of search algorithms and pipelines. To address this need, we propose ‘SPECTRUM’, an open-architecture and open-source toolbox for TDP data analysis. Its salient features include: (i) MS2-based intact protein mass tuning, (ii) de novo peptide sequence tag analysis, (iii) propensity-driven PTM characterization, (iv) blind PTM search, (v) spectral comparison, (vi) identification of truncated proteins, (vii) multifactorial coefficient-weighted scoring, and (viii) intuitive graphical user interfaces to access the aforementioned functionalities and visualization of results. We have validated SPECTRUM using published datasets and benchmarked it against salient TDP tools. SPECTRUM provides significantly enhanced protein identification rates (91% to 177%) over its contemporaries. SPECTRUM has been implemented in MATLAB, and is freely available along with its source code and documentation at https://github.com/BIRL/SPECTRUM/.
Similar content being viewed by others
Introduction
Mass spectrometry-based proteomics is a well-established technique for protein identification, characterization, and quantitation1,2,3. The conventional Bottom-Up Proteomics (BUP)4 protocol involves mass spectrometry (MS) analysis of peptides obtained from enzymatic digestion of whole proteins4,5. Several software tools such as SEQUEST6, Mascot7 and ExPASy tools8 (FindPept9 and EasyProt10) have been reported for BUP data analysis. However, BUP spectra and its analysis have limited power in: (i) identification of post-translational modifications (PTMs)2, (ii) sequence coverage11,12, and (iii) characterization of very small proteins13. Recent advancements in proteomics protocols and instrumentation have enabled precise mass measurements of large proteins by employing soft ionization techniques14 coupled with high-resolution mass analyzers15. This has led to the emergence of Top-Down Proteomics16 (TDP) protocol which is becoming increasingly popular for analyzing intact proteins17,18. TDP offers an enhanced sequence coverage19 as compared to BUP4 along with an improved identification of proteoforms (proteins and its variants)20,21. However, the complexity of high-resolution TDP spectral data poses a significant challenge for analysis tools. Current tools for TDP include ProSight PTM12, ProSight PTM 2.022, MS-Align+23, pTop24, TopPIC25, and MSPathFinder26 amongst others. ProSight PTM, the first tool reported for TDP data analysis, employed shotgun annotation27 for protein identification and PTM localization. ProSight PTM 2.0 enhanced ProSight PTM by providing an improved database annotation along with a capability to search variable, fixed as well as terminal modifications. However, the tool’s protein identification search space was limited to organism-specific protein sequence variations. Also, the shotgun annotation led to a significant increase in the size of search database. In 2012, MS-Align+ addressed this issue by using spectral alignment methodology28 to elicit unknown PTMs and truncated proteins. The tool, however, had a command line interface (CLI) rendering it difficult to use. In 2016, TopPIC and pTop were reported. TopPIC provided an improved implementation of MS-Align+ and facilitated high-throughput novel proteoforms discovery by including primary structure alterations. However, the tool was limited in its capability to identify proteins with multiple variable modifications. pTop, on the other hand, employed de novo sequencing to shortlist proteins and search combinations of user-provided variable modifications. This approach was particularly effective for searching multiple PTMs but was unable to cater for unknown modifications and truncated proteins. Recently reported MSPathFinder, a high-throughput tool employing parametric dynamic programming for spectral alignment, uses sequence graphs for efficient filtering of combinatorial proteoforms. However, it also lacks support for searching unknown modifications and its CLI makes it difficult to use. Taken together, TDP data analysis tools continue to suffer from limitations in: (i) identification of truncated proteins, (ii) identification, characterization and localization of unknown and multiple PTMs, (iii) identification of truncated proteoforms having PTMs, and (iv) an intuitive visualization of results. Moreover, the lack of open-architecture software practice impedes the development and benchmarking of TDP algorithms to address these shortcomings.
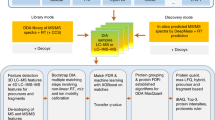
In this work, we propose “SPECTRUM”, an open source and open architecture top-down proteoform identification toolbox for MATLAB. Several algorithms have been systematically integrated to form the core of SPECTRUM search pipeline (Fig. 1). These algorithms include a novel intact protein mass tuner to augment MS1 measurements for scoring and filtering protein databases. De novo sequencing has been employed for extracting and scoring peptide sequence tags (PSTs)29,30. A novel PTM prediction strategy employs dbPTM31,32 for evaluating the shortlisted candidate proteins for known PTM binding sites besides supporting a blind PTM search. SPECTRUM also provides search support for single-side truncated proteins. Lastly, the canonical spectral comparison between theoretical and experimental spectra33,34,35 has also been employed for refining candidate protein list. To develop an overall ranking of candidate proteins, a composite scoring scheme has been implemented wherein users can tune weights for individual component scores to obtain the final score. For data interoperability36, SPECTRUM currently supports plain text files (columns of mass to charge ratios (m/z) and relative intensities), eXtensible Markup Language (XML) files with m/z and relative abundances (mzXML)37, Mass Spectrometry Markup Language (mzML)38,39 and Mascot Generic Format (MGF)7 data formats in both single and batch file processing modes. Users can access the toolbox by a set of intuitive graphical user interfaces (GUIs) for setting up search parameters as well as viewing results. Each GUI has been developed using MATLAB GUI development environment (GUIDE)40 and can, therefore, be readily customized or refactored.
We have validated and benchmarked SPECTRUM toolbox by undertaking case studies on two published datasets. Case study I was performed to evaluate protein identification accuracy and blind PTM characterization using an experimental dataset41 with known target protein (HeLa Histone H4). Results obtained from SPECTRUM were compared with those from ProSightPC42 (a commercial version of ProSight PTM 2.0), TopPIC, and pTop. SPECTRUM correctly identified the target protein which was reported by ProSightPC and TopPIC (see Case Study I – Results Section). For evaluating SPECTRUM’s ability to identify unknown proteins, a second case study was carried out using an Escherichia coli dataset25. SPECTRUM results reported up to 47% more spectral matches and over 91% more proteins in comparison with other tools (see Case Study II – Results Section).
In conclusion, SPECTRUM is a state-of-the-art tool for protein identification and characterization and is available in the form of a conveniently customizable MATLAB toolbox. This open-architecture toolbox stands to impart impetus to the advancement of TDP by assisting in design, implementation and benchmarking of novel TDP algorithms leading to an improved proteoform identification.
Results
In this work, we have reported SPECTRUM, a next-generation open-source MATLAB40 toolbox for top-down proteomics. The toolbox is available as a GitHub repository. Documentation (see Supplementary Information –E. Availability) and video tutorials have also been made available (see Supplementary Information –F. Video Tutorials).
The toolbox provides a comprehensive graphical user interface (GUI) framework (Fig. 2). The main GUI window (Fig. 2a) acts as the entry-point for setting up spectral data, protein databases, and search parameters. Elaborate GUIs have been provided for each step in the search process (Fig. 2b–f) and the summary of search results can be visualized as a ranked protein list (Fig. 2g). Using the “Detailed Protein View” (Fig. 2h), users can also view details of candidate proteins including information on predicted modifications, peptide sequence tags (PSTs) and theoretical fragments (Fig. 2i,k).
Overview of SPECTRUM GUIs. The set of graphical user interfaces (GUIs) in SPECTRUM created using MATLAB GUIDE to undertake the search process and visualize results. (a) Main SPECTRUM GUI to provide general search parameters, (b) GUI to tune intact protein mass, (c) GUI to specify special fragmentation ions and mass mode in the search process, (d) GUI to provide peptide sequence tag (PST) search parameters, and (e) GUI to specify instrument-based chemical modification(s) along with terminal modifications. (f) GUI to adjust weights in the scoring scheme, (g–h) GUIs to provide users with brief as well as detailed results, (i) GUI to describe spectral matching details, (j) GUI providing a legend for use in detailed result view, and (k) GUI for mass spectrum visualization.
Salient search features and algorithms of SPECTRUM
SPECTRUM’s top-down protein search pipeline comprises of three major components, i.e. (a) intact protein mass tuner and filter, (b) de novo sequencing and PST filter, and (c) in silico spectral comparator. SPECTRUM provides search support for chemical, terminal, fixed and variable modifications along with terminally truncated proteoforms. A blind post-translational modification (PTM) search module has also been included to search for unknown PTMs without requiring prior information. Data file format support for MGF7, mzXML36,43, mzML38,39 and flat text peak list file has been provided (see Supplementary Information – H. Feature Comparison). Alongside, SPECTRUM supports search in single as well as batch modes. Single-mode permits the users to search the four file formats while batch-mode allows for an automated search of multiple flat text files. Lastly, a multifactorial and customizable scoring scheme has been designed to tune the search process by weighing each component of protein search pipeline towards calculating the final scores.
Case Study I – Evaluation of SPECTRUM search with known target protein
To validate the protein identification accuracy of SPECTRUM, we searched a HeLa spectral dataset41 with known target protein (Histone H4). The dataset consisted of ten files containing monoisotopic data (see Supplementary Data S1). The search results obtained from SPECTRUM were compared with pTop24, TopPIC25 and ProSightPC22,42 (see Supplementary Data S2). Target protein’s rank in the candidate protein list and search runtime were then obtained and compared. The first comparison was performed between SPECTRUM and pTop wherein de novo sequencing was employed (search parameters in Supplementary Table S1). pTop took 13 seconds to perform protein search, however, it failed to identify any protein from the dataset. SPECTRUM on the other hand, completed the search in 28 seconds and reported Histone H4 as the top-ranked protein in eight out of ten experiments. SPECTRUM did not report any protein for remaining two files (summary and complete results in Supplementary Tables S2 and S3, respectively). Next, we compared SPECTRUM with TopPIC, a spectral alignment tool (search parameters in Supplementary Table S4). TopPIC took 2350 seconds and reported Histone H4 for seven data files; one file reported a false positive and two did not report any protein. SPECTRUM took 21 seconds to search the complete dataset and correctly identified the true protein from eight data files while false positives were reported for the remaining two files (summary and complete results in Supplementary Tables S5 and S6, respectively). We then compared spectral comparison capability of SPECTRUM with ProSightPC (search parameters in Supplementary Table S7). For this purpose, PST-based filtering was disabled, and the weight of intact protein mass score was set to zero. ProSightPC completed the search in 24 seconds and reported Histone H4 as top-ranked protein for eight data files while false-positives were reported for the remaining two. SPECTRUM executed the search in 19 seconds and reported eight true-positives besides two false-positive entries (summary and complete results in Supplementary Table S8 and S9, respectively). An overall comparison of the search results obtained from each tool has been provided in Supplementary Table S10.
Having validated protein identification, we then evaluated SPECTRUM’s blind PTM search feature for identifying unknown PTMs without prior information from the user. TopPIC reported unknown mass shifts for seven correct identifications but could not translate them into PTMs. SPECTRUM not only captured these mass shifts but also successfully characterized PTMs from three data files (see Supplementary Table S11 and Supplementary Information – B. Supplementary Results).
To evaluate the sensitivity of the search process to various parameters, a sensitivity analysis was performed on intact mass, PST and in silico comparison components. The parameter variations used for intact protein mass tolerance were 250, 500, 1000 and 2000 Da, PST lengths between 4 to 6 and 3 to 6, and in silico spectral comparison tolerances of 15 and 25 ppm, respectively. By increasing PST length range, an improvement in protein identification was observed. However, variations in protein mass tolerance had a minimal impact. The results from parameter sensitivity have been tabulated in Supplementary Table S12 (also see Supplementary Information – B. Supplementary Results: Case Study I).
Case Study II – Evaluation of SPECTRUM search with unknown target protein
After validating SPECTRUM search accuracy with known target proteins, we employed the toolbox to search a dataset with unknown target protein(s). Published Escherichia coli dataset25 obtained using alternating CID and ETD fragmentation modes (see Supplementary Data S3) was employed for the search. The search parameters have been provided in Supplementary Tables S13 and S14 for search with and without PSTs, respectively. The results were compared with those from MSPathFinder26, TopPIC25, and pTop24 at 1% false discovery rate and E-value of 1E-10 (summary of overall results in Supplementary Table S15).
The first comparison in this case study was performed between SPECTRUM and MSPathFinder. Peptide sequence tag (PST) filter was enabled for both the tools. SPECTRUM identified 245 proteins as compared to MSPathFinder which identified 128 proteins, indicating a 91% improvement. SPECTRUM also demonstrated an enhancement in number of PrSMs (1739) in comparison with MSPathFinder (1458). Next, the PST filter was turned off and the search was performed again. SPECTRUM reported 305 proteins and 1911 PrSMs in comparison to MSPathFinder’s 110 proteins and 1319 PrSMs, an improvement of 177% and 44% in proteins and PrSMs, respectively. We then compared SPECTRUM with TopPIC. Since TopPIC does not support tag-based search, SPECTRUM’s PST filter was disabled. SPECTRUM identified 305 proteins as compared to TopPIC which identified 128 proteins, indicating a 138% improvement. In comparison with 1911 PrSMs reported by SPECTRUM, TopPIC reported 1262 PrSMs. Lastly, we compared SPECTRUM toolbox with pTop. Since pTop’s search employs PSTs, we enabled SPECTRUM’s PST filter to search the dataset. SPECTRUM reported 245 proteins while pTop reported 128 proteins, marking a 91% improvement. Moreover, SPECTRUM reported 1739 PrSMs as compared to 1181 PrSMs from pTop, a 47% improvement.
Taken together, SPECTRUM identified a significantly larger number of proteins as compared to MSPathFinder, TopPIC, and pTop from Escherichia coli dataset (Fig. 3). A summary of search results has been provided in Fig. 3 and Supplementary Table S15. The complete results for both target and decoy databases search for each fragmentation mode (CID and ETD) have been provided in Supplementary Tables S16–S23. A summary table listing the result files has been provided in Supplementary Information – B. Supplementary Results: Case Study II.
Discussion
High-resolution top-down proteomics (TDP) is increasingly being employed for understanding mechanisms underpinning disease towards biomarker discovery21,44,45,46. Specifically, information-rich top-down mass spectra have a significant potential towards an enhanced proteoform identification47. For an optimal searching of TDP data, continuous advancement in top-down search algorithms and software is required. Contemporary tools for TDP have achieved remarkable protein identification rates, however, these tools provide partial search pipelines, are closed source or only available commercially. Besides, there is still a significant room for improvement in protein identification and characterization.
Towards addressing this need, we have proposed SPECTRUM, an open-source and open-architecture MATLAB toolbox for proteoform identification in top-down proteomics. SPECTRUM algorithmic pipeline advances the state-of-the-art by significantly enhancing proteoform identification and characterization as compared to the contemporary TDP tools (see Supplementary Table S24). To demonstrate the search capabilities of SPECTRUM, two case studies were conducted using published data25,41. In the first study, SPECTRUM successfully identified the known target protein, Hela - Histone H4, as was reported by pTop, ProSightPC and TopPIC. In the second study on Escherichia coli dataset with unknown target proteins, SPECTRUM reported up to 177% more proteins over other tools. Computational runtimes for the toolbox were also profiled and compared with MSPathFinder, pTop and TopPIC, for each case study. SPECTRUM runtimes were comparable with other tools for the HeLa dataset which comprised of 10 files41. However, for the larger Escherichia coli dataset, SPECTRUM runtime lagged behind other tools which can be attributed to the MATLAB interpreter. This can, however, be overcome by parallelizing the toolbox or by using MATLAB GPU computing routines. The blind PTM search module of SPECTRUM also improves upon TopPIC25 (see Supplementary Information – B. Supplementary Results) with an enhanced mass-shift identification and characterization. In terms of parameter sensitivity, three core modules including intact mass filter, peptide sequence tags (PST) generator and in silico spectral comparator influence the search to varying degrees (see Supplementary Information – B. Supplementary Results). Specifically, results were improved by increasing the range of PST length while no significant effect was observed for intact protein mass and spectral comparison. Prospectively, SPECTRUM can provide a significantly enhanced proteoform identification to its users. The batch-mode search also adds a high-throughput capability. Fixed, variable and blind modifications can be characterized besides reporting unexplained mass shifts. SPECTRUM pipeline also caters for truncated protein search. Users can customize the scoring scheme towards sensitizing the search process to their experimental setups. The graphical user interface (GUI) can be conveniently modified or enhanced using MATLAB GUIDE.
As with other spectral analysis tools, search results from SPECTRUM are dependent on the quality of MS data. Hence, the accuracy of search results may vary with mass spectrometer resolution. In terms of limitations, since SPECTRUM has been implemented in MATLAB, it requires a MATLAB license, thereby impeding the non-MATLAB users to run SPECTRUM. This need has been met with provision of the toolbox in form of an executable file (see Supplementary Information – E. Availability). SPECTRUM currently offers one-sided truncation and does not accommodate for double-sided truncations and amino acid substitutions. SPECTRUM’s blind-PTM module only characterizes those PTMs which are supported by spectral data. A natural extension will be incorporation of a probabilistic model in blind-PTM module for enhanced PTM characterization. Proteoform identification can be further enhanced by using combined spectral data obtained from alternating fragmentation mode of mass spectrometers. A useful extension of the toolbox can also come in the form of relative and absolute protein quantitation.
In conclusion, SPECTRUM is a state-of-the-art MATLAB-based top-down proteomics (TDP) toolbox that has been developed with an aim to assist in next-generation mass spectrometry data analysis. The toolbox is capable of identifying a significantly larger number of proteins as compared to its contemporaries besides characterizing post-translational modifications without requiring any prior knowledge. The proposed toolbox has been developed to facilitate biomedical research along with assisting in proteomics education by providing a versatile training platform for proteoform identification.
Material and Methods
Methodology and flow of SPECTRUM search pipeline
MATLAB 2017a40, a popular scientific computing platform, was used to develop SPECTRUM. A set of interactive GUIs were constructed using MATLAB graphical user interface (GUI) development environment (GUIDE)40 for taking user parameters and displaying search results. Figure 4 represents the overall methodology employed by SPECTRUM to search TDP data. Details on SPECTRUM search methodology, scoring scheme, validation, and data conversion have been provided below.
SPECTRUM data processing flowchart. User-selected protein database is filtered on intact protein mass followed by scoring of shortlisted proteins. De novo sequencing is performed to obtain peptide sequence tags (PSTs). Each candidate protein from the database is evaluated and scored for these sequence tags. Experimental and theoretical spectra of each candidate protein are compared to obtain in silico component score. Intact protein mass, PST and in silico scores are then used to determine final protein rank.
SPECTRUM search methodology and scoring algorithms
Intact protein mass tuner
MS2 data comprising of mass to charge ratios of intact protein’s fragments and relative abundances, was used to tune the intact protein mass, MS1. Fragment-pairs were generated for each element in MS2 data and a tuned precursor whole protein mass (MS1) was computed from a sum of each pair. The fragment-pair sums within the user-defined tolerance were selected (FPSmz). The average of abundances for each shortlisted constituent element in FPSmz were also computed. A window of size equal to the mass of a proton was used to scan the sorted fragment-pair sums to obtain the tuned mass. The window was progressively shifted by a user-defined step size and the number of fragment-pair sums falling within each window, at each shift, were counted. The window with the highest number of fragment-pair sums was selected, and tuned mass was computed as the intensity weighted average of fragment-pair sums within this window. A conceptual outline of the methodology has been shown in Fig. 5 and the complete set of mathematical equations have been provided in Supplementary Methods A1 - Intact Protein Mass Tuner.
Intact protein mass tuner workflow. (a) Fragment-pair sums of MS2 data are computed and sorted in an ascending order. Sliding window of a size equal to the mass of a proton is used to determine the window with maximum number of peaks. Finally, tuned mass is obtained by calculating the intensity weighted average of tuple sums from selected window. (b) Contextual explanation of intact protein mass tuner, Step 1: Obtain experimental spectrum, Step 2: Compute fragment-pair sums of MS2 data, Step 3: Sliding window of size equal to mass of a proton is used to determine the number of peaks in each window, Step 4: Obtain window with maximum peak count, and Step 5: Tuned mass is obtained by calculating the intensity weighted average of tuple sums from selected window.
Scoring proteins by intact protein mass
The absolute differences between theoretical masses (details in Supplementary Methods A2 - Computing Theoretical Mass of a Protein) of candidate proteins and the experimental mass (tuned mass or MS1) were calculated towards computing the protein score using intact protein mass. The proteins with mass difference within the user-defined tolerance were shortlisted and scored (equations (1, 2)).
where,
Massdiff is absolute difference between theoretically calculated mass of protein and experimental mass, Massexperimental is experimental mass of sample protein (tuned mass or MS1), and Masstheoretical is theoretical protein mass calculated using protein sequence.
where,
Scoremass is the mass score of shortlisted protein, and Thr is user-defined intact protein mass tolerance.
Methodology for extracting peptide sequence tags
De novo sequencing was used to construct peptide sequence tag (PST) ladders. Incorporation of PSTs in the database search provided for tandem scoring of the candidate proteins. PST extractor was designed to take mass differences between successive experimental peaks within a user-specified tolerance. The mass difference corresponding to mass of any of the twenty amino acid residues constituted an amino acid tag. User-provided tolerance was used to determine the matching stringency for hops that mismatch the monoisotopic molecular weights of amino acids. The hops, with the starting peaks, ending peaks, the mass difference between these peaks, matching amino acid names and their molecular weights were stored. Hops having equal starting peak and ending peak values were joined together to form PST ladders. User-provided range of PST lengths was used to filter out anomalous (i.e. very short or very long) PST ladders to avoid biasing of the protein search process. The methodology is outlined in Fig. 6 and complete details have been provided in Supplementary Methods A3 - Extraction of Peptide Sequence Tags.
Workflow of peptide sequence tags (PSTs) extraction. (a) De novo sequencing of experimental data is performed to obtain peptide sequence tags. Each candidate protein from database is evaluated and scored for these PSTs. (b) Contextual explanation, Step 1: Obtain experimental spectrum, Step 2: Compute fragment-pair difference of MS2 data, Step 3: Obtain amino acids corresponding to fragment-pair differences, and Step 4: Tags having the same starting and ending peaks are joined together.
Scoring proteins using peptide sequence tags
PST scoring utilizes cumulative root mean squared error, peak intensities, PST occurrence count and PST length. RMSE over the entire PST length was computed and employed for shortlisting PSTs by user-defined tolerance. For each filtered tag, intensity of the constituent amino acids was determined by taking the average intensity of representative experimental peaks. Cumulative intensity of tag was then computed using average intensities for scoring. The influence of PSTs towards protein filtering and scoring was implemented to increase exponentially with length. The PST-based score for shortlisted proteins was computed using the frequency score, accumulative tag error score and occurrence of PST tags that reported these proteins. The scoring process has been defined in equations (3–10).
where,
ErrorAA is the difference between Massexperimental and Massmonoisotopic, Massexperimental is the experimental mass of a residue present in an extracted PST, and Massmonoisotopic is the monoisotopic mass of a standard amino acid residue in the PST.
where,
RMSE is cumulative root mean squared error calculated over the entire PST length, \(Erro{r}_{i}^{AA}\) is the difference between experimental and theoretical mass of ith residue in the PST, and N is length of peptide sequence tag.
where,
Errorscore is the cumulative score of PST error computed using RMSE.
where,
intPST is the average intensity of constituent amino acids of PST; inthome and inthop are the intensities of the peaks in the PST ladder.
where,
IntensityPST is the cumulative intensity of all the amino acids in the PST.
where,
Lenscore is the score for length of a tag.
where,
Freqscore is the PST component score computed using IntensityPST and Lenscore.
where,
ScorePST is the PST score of shortlisted proteins, Occurence is the frequency of occurrence of a PST tag in a protein sequence, and M is the total number of tags.
Spectral generation and comparisons
A total of nine fragmentation techniques including collision-induced dissociation (CID), electron-capture dissociation (ECD), electron-transfer dissociation (ETD) and electron-detachment dissociation (EDD) etc. have been employed in SPECTRUM search pipeline. Additionally, single-sided truncations have also been incorporated. The mass of N-terminus ion was computed by summing up the masses of its constituent amino acids while for C-terminus ion, the mass was obtained by calculating the mass difference between the N-terminus ion and protein molecular weight. Also, during fragmentation, a hydroxyl group and a proton were added to N-terminus ion and C-terminus ion, respectively (see Supplementary Methods A4 - Spectral Generation and Comparison). User-specified neutral ion loss parameters were used to cater for fragments which have gained or lost functional groups.
For a given experimental dataset, its intensity values were normalized between 0 and 1 followed by their scaling (NormalizedIntensity) using a step function described in equation (11). Note that the threshold of 9.2 × 10−5 was set after performing a sensitivity analysis on several available spectral datasets. Towards scoring the proteins using the in silico spectrum, N-terminus ions and C-terminus ions were compared with the experimental data within a certain user-specified tolerance. For every match, the candidate protein was awarded a score, based on the number of consecutive fragment matches in experimental spectrum (ConsecutivePeakCounter), as shown in equation (12). Next, the final score was computed for each protein using equation (13). The process has been outlined in Fig. 7.
where,
Spectral generation and comparisons workflow and contextual explanation. (a) After retrieving protein sequences from user-selected protein database, theoretical fragments of each protein are generated. Experimental and theoretical spectra are then compared to get in silico component score. (b) Step 1: Obtain experimental spectrum, Step 2: Generate theoretical fragments of candidate protein, Step 3: Experimental and theoretical spectra are compared to get number of matches, and Step 4: In silico component score is computed.
NormalizedIntensity is the scaled intensity value of experimental spectrum, and Intensity is the intensity of experimental spectrum normalized to 1.
where,
MatchScorei is the score of fragment match corresponding to ith experimental peak, NormlizedIntensityi is the sigmoid weighted intensity value of ith experimental peak, and ConsecutivePeakCounter is the number of consecutive experimental peak matches.
where,
MatchScoreiis the score of ith fragment match, Fragexperimental is the total number of experimental fragments, and n is the number of spectral matches.
Composite scoring scheme
The candidate protein list was ranked using (i) intact protein mass filtering (Scoremass), (ii) PST filtering (Scorepst) and (iii) spectral matching (Scoreinsilico). The weight of each scoring component can be adjusted towards sensitizing the scoring to their experimental settings using equation (14).
where,
Scorefinal is the final score for each candidate protein shortlisted from the database, W1 is the weight set by the user for intact protein mass score, W2 is the weight set by the user for PSTs score, and W3 is the weight set by the user for in silico score. Note that the default weight (‘1’) elicits maximal sensitivity from each scoring sub-system in SPECTRUM.
Methodology for predicting post-translational modifications
SPECTRUM provides support for searching fixed, variable and blind post-translational modifications (PTMs) (Fig. 8). For fixed modifications, each instance of the implicated amino acid site was modified. For variable modifications32, the product of amino acid occurrence propensities within a certain enzyme binding site was obtained. An enzyme binding site could be a single or multi-residue substrate site containing the amino acid to be modified. Binding sites scoring above a user-specified threshold were selected for onward modifications (see equation (15)). In case multiple sites were shortlisted, all combinations of modified protein were created.
where,
PTM_Score is the product of amino acid occurrence propensities within the binding site, and PTM_Thr is the user-specified threshold selected for modifications.
Datasets used for validating SPECTRUM’s search pipeline
SPECTRUM validation was performed using datasets from two published top-down proteomics experiments including a HeLa41 and an Escherichia coli25 dataset. The HeLa dataset, which was used in case study 1, comprised of 10 MS spectra of Hela Histone H4 protein obtained using a Q-FTICR hybrid mass spectrometer. The spectra were calibrated externally using an electron-capture dissociation (ECD) bovine ubiquitin spectrum. Case study II employed Escherichia coli K-12 MG1655 dataset, which was acquired using an LTQ Orbitrap Velos mass spectrometer in an alternating fragmentation setting. The resulting data comprised of two sets of spectra, each containing 2027 scans from collision-induced dissociation (CID) and electron-transfer dissociation (ETD), respectively. SPECTRUM was employed to search the two datasets and the results were compared with those obtained from ProSightPC42 (a commercial version of ProSight PTM 2.022), TopPIC25, pTop24 and MSPathFinder26.
Validating SPECTRUM results
Target-decoy approach48,49 was employed to estimate the false discovery rate (FDR). The decoy database was generated by shuffling the protein sequences followed by the incorporation of three random amino acid mutations26,50. To further enhance the stability of FDR estimate, three decoy proteins were assembled for each protein entry in the target database. FDR was computed using equation48 (16). To estimate the statistical significance of each candidate protein, E-values were computed using an adaptation of generating function method51. For that, the probability of each amino acid is computed in the database. These amino acid probabilities are then used to calculate the probability of each protein sequence in the database. Using the number of spectral matches, the spectral probability51 of each sequence is then computed using equation (17). This is followed by an adjustment51 for truncation and computation of E-value using equation (18).
Data conversion to supported file formats
SPECTRUM requires experimental data in standardized input file formats. These formats include Mascot Generic Format (MGF)7, eXtensible Markup Language (XML) file containing mass to charge ratios (mz) and relative abundances (mzXML)36,43, and Mass Spectrometry Markup Language (mzML)38,39. Raw data files such as Thermo Xcalibur ‘.raw’, ABI/Sciex ‘.WIFF’ and Bruker ‘.YEP’, therefore, need to be converted into the aforementioned formats. For that, file format conversion and deconvolution tools such as MS-Convert52 and MS-Deconv53 can be employed. mzXML and mzML files with centroided and peak-picked data, obtained using MS-Convert52, can be imported into SPECTRUM. SPECTRUM then relies on MS-Deconv53 and OpenMS54 to extract monoisotopic peak lists. Deconvolved MGF files containing monoisotopic peaks are automatically converted into searchable flat text files, using a custom file reader that has been implemented in SPECTRUM.
References
Wasinger, V. C. et al. Progress with gene‐product mapping of the Mollicutes: Mycoplasma genitalium. Electrophoresis 16, 1090–1094 (1995).
Han, X., Aslanian, A. & Yates, J. R. Mass spectrometry for proteomics. Curr. Opin. Chem. Biol. 12, 483–490 (2008).
Smith, L. M. et al. Proteoform: a single term describing protein complexity. Nat. Methods 10, 186 (2013).
Zhang, Y., Fonslow, B. R., Shan, B., Baek, M.-C. & Yates, J. R. III. Protein analysis by shotgun/bottom-up proteomics. Chem. Rev. 113, 2343–2394 (2013).
Gundry, R. L. et al. Preparation of Proteins and Peptides for Mass Spectrometry Analysis in a Bottom‐Up Proteomics Workflow. Curr. Protoc. Mol. Biol. 10.25. 1–10.25. 23 (2009).
Qian, W.-J. et al. Probability-based evaluation of peptide and protein identifications from tandem mass spectrometry and SEQUEST analysis: the human proteome. J. Proteome Res. 4, 53–62 (2005).
Perkins, D. N., Pappin, D. J. C., Creasy, D. M. & Cottrell, J. S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 (1999).
Gasteiger, E. et al. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788 (2003).
Gattiker, A., Bienvenut, W. V., Bairoch, A. & Gasteiger, E. FindPept, a tool to identify unmatched masses in peptide mass fingerprinting protein identification. Proteomics 2, 1435–1444 (2002).
Gluck, F. et al. EasyProt—an easy-to-use graphical platform for proteomics data analysis. J. Proteomics 79, 146–160 (2013).
Tran, J. C. et al. Mapping intact protein isoforms in discovery mode using top-down proteomics. Nature 480, 254–258 (2011).
LeDuc, R. D. et al. ProSight PTM: an integrated environment for protein identification and characterization by top-down mass spectrometry. Nucleic Acids Res. 32, W340–W345 (2004).
Wu, S. et al. Top-down characterization of the post-translationally modified intact periplasmic proteome from the bacterium Novosphingobium aromaticivorans. Int. J. Proteomics 2013 (2013).
El-Aneed, A., Cohen, A. & Banoub, J. Mass spectrometry, review of the basics: electrospray, MALDI, and commonly used mass analyzers. Appl. Spectrosc. Rev. 44, 210–230 (2009).
Monge, M. E., Harris, G. A., Dwivedi, P. & Fernández, F. M. Mass spectrometry: recent advances in direct open air surface sampling/ionization. Chem. Rev. 113, 2269–2308 (2013).
Yates, J. R. & Kelleher, N. L. Top down proteomics. Anal Chem 85, 6151 (2013).
Armirotti, A. & Damonte, G. Achievements and perspectives of top-down proteomics. Proteomics 10, 3566–3576 (2010).
Zhou, M. & Veenstra, T. Mass spectrometry: m/z 1983-2008. Biotechniques 44, 667–668,670 (2008).
Fornelli, L. et al. Top-down proteomics: Where we are, where we are going? J. Proteomics (2017).
Cai, W., Tucholski, T. M., Gregorich, Z. R. & Ge, Y. Top-down proteomics: technology advancements and applications to heart diseases. Expert Rev. Proteomics 13, 717–730 (2016).
Gregorich, Z. R. & Ge, Y. Top‐down proteomics in health and disease: Challenges and opportunities. Proteomics 14, 1195–1210 (2014).
Zamdborg, L. et al. ProSight PTM 2.0: improved protein identification and characterization for top down mass spectrometry. Nucleic Acids Res. 35, W701–W706 (2007).
Liu, X. et al. Protein identification using top-down spectra. Mol. Cell. Proteomics 11(M111), 008524 (2012).
Sun, R.-X. et al. pTop 1.0: a high-accuracy and high-efficiency search engine for intact protein identification. Anal. Chem. 88, 3082–3090 (2016).
Kou, Q., Xun, L. & Liu, X. TopPIC: a software tool for top-down mass spectrometry-based proteoform identification and characterization. Bioinformatics 32, 3495–3497 (2016).
Park, J. et al. Informed-Proteomics: open-source software package for top-down proteomics. Nat. Methods 14, 909 (2017).
Pesavento, J. J., Kim, Y.-B., Taylor, G. K. & Kelleher, N. L. Shotgun annotation of histone modifications: a new approach for streamlined characterization of proteins by top down mass spectrometry. J. Am. Chem. Soc. 126, 3386–3387 (2004).
Tsur, D., Tanner, S., Zandi, E., Bafna, V. & Pevzner, P. A. Identification of post-translational modifications via blind search of mass-spectra. In Computational Systems Bioinformatics Conference, 2005. Proceedings. 2005 IEEE 157–166 (IEEE, 2005).
Tanner, S. et al. InsPecT: identification of posttranslationally modified peptides from tandem mass spectra. Anal. Chem. 77, 4626–4639 (2005).
Mann, M. & Wilm, M. Error-tolerant identification of peptides in sequence databases by peptide sequence tags. Anal. Chem. 66, 4390–4399 (1994).
Eisenhaber, B. & Eisenhaber, F. Prediction of posttranslational modification of proteins from their amino acid sequence. Data Min. Tech. Life Sci. 365–384 (2010).
Lu, C.-T. et al. DbPTM 3.0: an informative resource for investigating substrate site specificity and functional association of protein post-translational modifications. Nucleic Acids Res. gks1229 (2012).
Eng, J. K., McCormack, A. L. & Yates, J. R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 (1994).
Cottrell, J. S. & London, U. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20, 3551–3567 (1999).
Baumgardner, L. A., Shanmugam, A. K., Lam, H., Eng, J. K. & Martin, D. B. Fast parallel tandem mass spectral library searching using GPU hardware acceleration. J. Proteome Res. 10, 2882–2888 (2011).
Deutsch, E. W. File formats commonly used in mass spectrometry proteomics. Mol. Cell. Proteomics 11, 1612–1621 (2012).
Pedrioli, P. G. A. et al. A common open representation of mass spectrometry data and its application to proteomics research. Nat. Biotechnol. 22, 1459–1466 (2004).
Turewicz, M. & Deutsch, E. W. In Data mining in proteomics 179–203 (Springer, 2011).
Martens, L. et al. mzML—a community standard for mass spectrometry data. Mol. Cell. Proteomics 10(R110), 000133 (2011).
MathWorks. MATLAB. Available at: https://www.mathworks.com (1994).
Frank, A. M., Pesavento, J. J., Mizzen, C. A., Kelleher, N. L. & Pevzner, P. A. Interpreting top-down mass spectra using spectral alignment. Anal. Chem. 80, 2499–2505 (2008).
Inc., T. F. S. ProSightPC 4.0. Available at: http://proteinaceous.net/product/prosightpc-4-0/(2013).
Lin, S. M., Zhu, L., Winter, A. Q., Sasinowski, M. & Kibbe, W. A. What is mzXML good for? Expert Rev. Proteomics 2, 839–845 (2005).
Peng, Y. et al. Top-down targeted proteomics for deep sequencing of tropomyosin isoforms. J. Proteome Res. 12, 187–198 (2012).
Calligaris, D., Villard, C. & Lafitte, D. Advances in top-down proteomics for disease biomarker discovery. J. Proteomics 74, 920–934 (2011).
Siuti, N. & Kelleher, N. L. Decoding protein modifications using top-down mass spectrometry. Nat. Methods 4, 817 (2007).
Savaryn, J. P., Catherman, A. D., Thomas, P. M., Abecassis, M. M. & Kelleher, N. L. The emergence of top-down proteomics in clinical research. Genome Med. 5, 53 (2013).
Aggarwal, S. & Yadav, A. K. In Statistical Analysis in Proteomics 119–128 (Springer, 2016).
Navarro, P. & Vázquez, J. A refined method to calculate false discovery rates for peptide identification using decoy databases. J. Proteome Res. 8, 1792–1796 (2009).
Park, J. K. et al. Informed-Proteomics: Open Source Software Package for Top-Down Proteomics. (Pacific Northwest National Laboratory (PNNL), Richland, WA (US), Environmental Molecular Sciences Laboratory (EMSL), 2017).
Liu, X., Segar, M. W., Li, S. C. & Kim, S. Spectral probabilities of top-down tandem mass spectra. In BMC genomics 15, S9 (BioMed Central, 2014).
Chambers, M. C. et al. A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol. 30, 918 (2012).
Liu, X. et al. Deconvolution and database search of complex tandem mass spectra of intact proteins a combinatorial approach. Mol. Cell. Proteomics 9, 2772–2782 (2010).
Röst, H. L. et al. OpenMS: a flexible open-source software platform for mass spectrometry data analysis. Nat. Methods 13, 741 (2016).
Acknowledgements
We acknowledge the support provided by Osama Shiraz Shah for the fruitful discussions and suggestions during the development of the toolbox. This work was supported by HEC (21-320SRGP/R&D/HEC/2014, 20-2269/NRPU/R&D/ HEC/12/4792 and 20-3629/NRPU/R&D/HEC/14/585), Ignite (SRG-209), TWAS (RG 14-319 RG/ITC/AS_C) and LUMS (STG-BIO-1008, FIF-BIO-2052 and FIF-BIO-0255) grants.
Author information
Authors and Affiliations
Contributions
C.S.U. designed the project and supervised the research; C.S.U., A.R.B., K.I., Z.A., M.F.K., R.H., H.G.K., A.S., M.H., H.A.H., M.M. and M.A.S. carried out the toolbox development; C. S. U., A.R.B. and K.I. carried out the case study and analyses; C.S.U., A.R.B., K.I., M.F.K., M.T., Z.U., S.Z., S.A., E.U., S.H., and F.A. wrote the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Basharat, A.R., Iman, K., Khalid, M.F. et al. SPECTRUM – A MATLAB Toolbox for Proteoform Identification from Top-Down Proteomics Data. Sci Rep 9, 11267 (2019). https://doi.org/10.1038/s41598-019-47724-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-47724-1
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.