Abstract
Tuberculosis (TB) is the ninth leading cause of death worldwide, ranking above human immunodeficiency virus. Latency is the major obstacle in the eradication of this disease. How the physiology of the pathogen changes in transition to the latent stage needs to be understood. The latent bacteria extracted from animal hosts exist in a nonculturable (NC) phase, whereas bacteria extracted from most in vitro models are culture-positive. In the present study, we observed that nitrite, up to a concentration of 5 mM, shows the growth of Mycobacterium tuberculosis (MTB) in liquid media, but this effect starts reversing at higher concentrations. At a concentration of 10 mM, nitrite induces rapid nonculturability of MTB at the aerobic stage. This noncultivable dormancy was confirmed by analyzing the characteristics of NC bacteria. Further differential gene expression analyses clearly supported the formation of a dormancy phenotype. This study will be helpful for the use of this bacillus as a dormancy model in future studies on TB latency.
Similar content being viewed by others
Introduction
Tuberculosis (TB) is one of the most ancient infectious diseases of mankind and is one of the top 10 causes of death worldwide, affecting a third of the global population1. Mycobacterium tuberculosis (MTB) is the causative agent of TB, which can persist within humans for long periods of time without causing any symptoms2,3. Latent bacteria in the nonculturable (NC) stage cannot form colonies on solid media, which is why current diagnostic techniques are unable to detect more than 40% of TB recurrence cases. In addition, recent evidence suggests an association between nonreplicating bacilli and disease manifestations4,5. Recently, some reports showed that the development of drug resistance is not only because of the inheritable genetic resistance mechanism, but rather it is governed by changes in the physiological state of MTB6. However, more studies are needed in order to understand the metabolic processes that are critical for the pathogen to shift into dormancy and survive under stress conditions.
The factors that probably limit bacterial growth in granulomas include oxygen and nutrient deprivation, exposure to acidic pH, and the presence of nitric oxide or carbon monoxide7,8,9. On the basis of these factors, different dormancy models have been proposed. Dormant bacteria that were isolated from most in vitro models, such as the Wayne model and multiple stress model, were nonreplicating but remained cultivable10,11. However, bacteria obtained from in vivo models of latency, such as the Cornell model and noncultivable model developed by Shleeva et al. (2004), remained noncultivable12,13. It was observed that persistent and viable but noncultivable (VBNC) stages of MTB coexist and remain as part of the dormancy continuum. They could survive together as mixed population but differs in their physiological positions in the dormancy range4. VBNC cells have a higher toxin-to-antitoxin ratio as compared to persisters, so the VBNC stage could be considered a deeper stage of dormancy than persisters4,14. In all of these models, the external environment of the bacilli is modified. However, the internal targets whose modification plays a significant role in the development of noncultivability are still not fully explored.
However, in this study, we demonstrated that exposure of the MTB bacilli to nitrite induces dormancy under in vitro aerobic conditions. This dormancy is characteristically similar to the noncultivable type of dormancy, which was earlier reported and observed in isolates from in vivo animal models and humans. Thus, our study provides a new insight into the development of noncultivable dormancy in MTB.
Results
Effect of nitrite on the growth of MTB
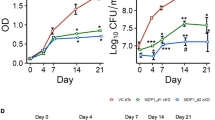
Sodium nitrite has been reported to inhibit the growth of a wide variety of bacteria, such as Clostridia and Staphylococcus aureus15. Thus, in order to assess the effect of nitrite on MTB, we measured the OD620 and colony forming unit (CFU) count of MTB bacilli at different time points. This study was carried out on MTB strain H37Ra, which has a >99.9% genome-wide similarity, and it was expected that the results will be applicable to pathogenic cells too. The results clearly indicated that nitrite supports the growth of MTB at concentrations below 5 mM, whereas at higher concentrations, nitrite significantly inhibits cell growth (Fig. 1A).
Growth kinetics of MTB in the presence of nitrite. The effect of nitrite on the growth of cells was determined by measuring (A) the OD at 620 nm and (B) the CFU count, at different time points with different nitrite concentrations. Log-phase cells treated with vehicle as a control (▪) and 5 mM (⚫), 10 mM (▴) 15 mM (▾), and 20 mM (♦) nitrite, respectively. More details are provided in the Materials and Methods section. The results are shown as the mean of three identical experiments ± standard deviation (SD). This experiment was repeated three times. The error bar represents the SD.
Further, a simultaneous assessment of the CFU count was performed at nitrite concentrations of 10, 15, and 20 mM at different time points. The CFU on an agar medium was also found to decrease by 3.23 log10 CFU/mL within 72 h of 10 mM nitrite treatment, with no CFU on day 6 (Fig. 1B). As compared to 7.37 ± 0.38 log10 CFU/mL on day 1, the CFU count of the control was found to be 8.03 ± 0.33 log10 CFU/mL on day 6. The reduction in the CFU count raises the possibility of death or acquired nonreplicative feature of the bacilli.
Effect of nitrite on the viability of MTB bacilli
In order to further elucidate the reason behind the observed decrease in the CFU, we performed live–dead staining of these bacilli. Bacilli treated with nitrite (10 mM) remained 100% viable for more than 10 days without forming any colonies on agar (Figs 1B and 2A,B). However, treatment with higher concentrations of nitrite also did not reduce the cell viability of MTB cells in any significant manner.
Relative viability analysis of MTB cells. (A) Bacterial cells were exposed to 10 mM (⚫), 15 mM (▴), and 20 mM (▾) nitrite, and percent viability was analyzed up to 10 days of treatment with respect to the untreated control (▪) as 100% viable. The experiment was performed in triplicate. Data are shown as the mean ± SD. (B) Fluorescence microscopic images of cells treated with nitrite (10 mM) were captured at different time intervals using an EVOS microscope as mentioned in the Materials and Methods section. The data shown is representative of three independent experiments (scale bar: 50 µm).
Effect of nitrite on the morphological properties of MTB
It is well known that the shape of the bacilli changes to more rounded with a concomitant reduction in their size when actively growing cells shift to the NC state11,16,17. The morphological characteristics of noncultivable bacilli developed after treatment with nitrite were analyzed using scanning electron microscopy (SEM). It was observed that nitrite-treated cells decreased in size compared to untreated control cells. Interestingly, on day 6, the size of the cells treated with nitrite (10 mM) was reduced to a level between 0.8 and 1.2 µm, as compared to actively growing controls (size: 3.0–6.0 µm) (Fig. 3, Table 1).
Electron microscopic measurement of nitrite-treated and nontreated MTB cells. (A) The log-phase culture was treated with nitrite (10 mM), and SEM images were taken on (B) day 3 and (C) day 6 of treatment. Scale bar: 1 µm. More details on the experiment were described in the Materials and Methods section.
After analyzing the cell size distribution (Table 1), it was found that the size of 98% of the cells was between 0.6 and 1.5 µm on day 6 of nitrite treatment. However, in the controls, the size of 41% of the cells was in the range of 3.5–4.5 µm, and only 2% of the cells were in the range of 0.6–1.5 µm. This observation strengthened the notion of the reduction of cell size in nitrite-treated MTB cells being a characteristic feature of the VBNC stage.
Loss of acid fastness in nonculturable cells
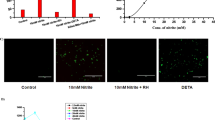
Loss of acid fastness is another major characteristic change that takes place when mycobacteria shift from the active state to a dormant state. Thus, in order to support our observation of shifting of bacilli to the noncultivable dormant state upon nitrite treatment, we assessed the acid-fast property using the fluorescent acid-fast staining dye Auramine O in combination with the neutral lipid staining Nile Red dye. The log-phase culture and NRP stage 2 culture of the Wayne model were used as two extreme controls (Fig. 4A,B).
Effect of nitrite exposure on the acid-fast property of MTB bacilli. MTB cells were stained with Auramine O and Nile Red fluorescence dyes at different time intervals. (A) Untreated cells were used as the vehicle control. (B) NRP stage 2 culture of the Wayne model was stained as a positive control. Cells treated with nitrite (10 mM) were stained and observed on day 3 (C) and day 6 (D) of treatment. More details are described in the Materials and Methods section. Fluorescence microscopic images were captured at 60× objective using an EVOS microscope (Life Technologies) (scale bar: one unit = 50 µm).
However, with respect to the controls (Fig. 4A), cells treated with nitrite (10 mM) showed red fluorescence after six days, indicative of the loss of the acid-fast property of the bacilli (Fig. 4D). Interestingly, cells stained after three days of treatment were observed to have yellow fluorescence due to the combined effect of Auramine O and Nile Red stains, indicative of the gradual shift of the acid-fast property from positive to negative (Fig. 4C).
Effect of nitrite on the antibiotic sensitivity of MTB
Resistance to antibiotics is a hallmark of latency in mycobacteria7,18,19,20. Standard antimycobacterial drugs like rifampin (RIF) and isoniazid (INH) were used to assess the sensitivity of nitrite-treated cells. Nitrite-treated MTB cells showed total resistance toward standard antitubercular drugs like RIF and INH, even at concentrations higher than IC90 (Fig. 5A). These results were in accordance with most of the dormancy models, including few that demonstrated significant susceptibility toward RIF (Fig. 5B). RIF inhibits RNA synthesis, which is essential for dormant cells at a low level6. On the other hand, INH impacts cell wall biosynthesis, a process that is inactive in dormant cells15. Thus, cells in all dormancy models were expected to be characterized by their resistance to INH.
Determination of the sensitivity of nitrite-treated bacilli to antitubercular drugs. (A) Actively growing and nitrite-treated MTB cells were treated with RIF and INH at different concentrations (0.5, 1, 5, and 10 µg/mL) for seven days, and viable bacilli were enumerated by the live–dead staining method. (B) A table depicting a comparison between the percent viable cells that remained with other dormancy models after independent treatment with 5 µg/mL INH and RIF each. The details of the protocol followed in both experiments were described in the Materials and Methods section. #Percent resistance values taken from Antimicrob Agents Chemother. 2014.
Transcriptional response of MTB bacilli to nitrite stress
The effect of nitrite on gene regulation of MTB was analyzed using microarray analysis. The gene expression pattern on days 1, 3, and 6 of nitrite treatment was compared with the transcriptome data obtained from log-phase cells without nitrite treatment as a control. On day 6, in nitrite-treated cells, 30 genes were upregulated and 110 genes were downregulated, as compared to untreated cells. However, on days 1 and 3 of nitrite treatment, 107 and 98 genes were upregulated, whereas 8 and 50 genes were downregulated, respectively. With the increase of the duration of nitrite treatment, the number of upregulated genes decreased, with a simultaneous increase in the number of downregulated genes (Tables S1–S6).
On day 1, as compared to the control, more genes were induced in nitrite-treated cells, possibly to cope up with the stress condition. However, on day 6, the number of downregulated genes was high, which might be due to the low metabolic state of bacteria.
Overlap between differentially expressed genes of MTB
The number of overlapping genes between the three data points (1, 3, and 6 day nitrite treated) is depicted as a Venn diagram (Fig. S1). Among the differentially expressed genes, ppe65, Rv1815, and mce1c were repressed (Table S6), whereas the rrl, rnpB, and cysK2 genes were induced (Fig. S1B), at all three time points after nitrite treatment. The cysK2 gene is involved in the cysteine biosynthetic pathway. Sulfur metabolism and de novo cysteine biosynthesis are important for redox homeostasis in persistent MTB, and these pathways could provide promising targets for novel antibiotics for the treatment of the latent form of the disease21.
Comparative profile of overlapping genes between nitrite Treatment and other dormancy models
The differentially regulated genes identified after nitrite treatment were compared with other dormancy models. The gene expression profile in nitrite-treated cells exhibited a significant difference between overlapping genes with other dormancy models (Table 2). From the analysis, it was observed that the expression pattern obtained from most of the dormancy models as well as on days 1 and 3 of nitrite treatment has indicated similarity from the higher number of overlapping genes compared to day 6. The NC cells obtained in a potassium deficiency model and on day 6 of nitrite treatment showed a high number of overlapping genes, as compared to days 1 and 3 of nitrite treatment.
Tracing of pathways involving Up- and downregulated genes
The differentially expressed genes were classified into various functional categories, such as lipid metabolism, transcription, translation, and carbohydrate metabolism. Some of the functional classes that showed changes after nitrite treatment are discussed below.
Chaperones and heat shock proteins
Genes under the devR regulon are upregulated under stress conditions such as hypoxia, oxidative stress, pH stress, and bacteria infecting macrophages22,23,24,25,26. In the presence of nitrite, the dosR regulon gene hsp (heat shock protein) was upregulated on days 1 and 6, whereas the hspX gene was downregulated on days 3 and 6. The hsp gene gets induced generally under all stress conditions and 24 h after infection of macrophages23.
The HspX protein showed low abundance in potassium-deficient NC cells, also downregulated under acidic conditions27. 21 dosR-regulon-related genes in potassium-deficient cells and 27 genes in nitrate-treated cells were downregulated10,28. The expression dosR regulon is not found to change in response to H2O2, in the persisting cells during starvation, as well as the gradual depletion of the carbon source16,24. In recent literature, it has been hypothesized that the dosR regulon is induced when oxygen tension is reduced in aerobically respiring bacteria. In this study, the cells did not experience oxygen stress, which could be preventing the induction of dosR regulon as observed in other dormancy models.
Detoxification-related genes
ahpC is an alkyl hydroperoxide reductase, which is important for resistance against oxidative stress and is induced in different dormancy models23,24,29,30,31,32. The ahpC gene was found to be upregulated in MTB bacilli on day 1 of exposure to nitrite.
Sulfur metabolism genes like cysA2, cysK1, and cysH were found to be upregulated on day 1, whereas the cysK2 gene was found to be upregulated at all three time points. The sulfur assimilation genes cysD and cysN are induced during iron deficiency, in oxidative stress, and in the presence of a vitamin-C-induced dormancy condition33, whereas the cysH gene is also induced in the iron deficiency condition29. Bacteria regulate sulfur assimilation in response to toxic oxidants, and the cell protects itself from reactive species by the induction of these genes. It was reported that sulfur metabolism is involved in the antioxidant defense mechanism of MTB34.
Lipid and mycolic acid metabolism
Genes related to the fatty acid degradation pathway, such as fadD9 and fadD31, coding for fatty acid CoA ligase, and genes adhD, alkB, and adhC were upregulated on day 1, whereas genes fadA2 and Rv0914C were downregulated on day 3. Like other dormancy models, mycobacteria alter their own metabolism to use fatty acid as a carbon source after nitrite treatment10.
Genes involving fatty acid biosynthesis were downregulated on days 3 and 6. As fatty-acid-synthesis-related genes were downregulated from day 3 onwards, the bacilli may have reduced their energy consumption required for fatty acid synthesis. The acpM gene, which is involved in mycolic acid biosynthesis, and the wag31 gene (cell wall synthesis protein) were also downregulated on day 635,36. wag31 regulates the cell morphology of bacteria and is involved in the oxidative stress response of MTB36. The ald gene, which catalyzes alanine hydrolysis, an important constituent of the peptidoglycan layer, is upregulated on days 1 and 6, which can be correlated with the morphological changes that occurred in MTB after nitrite treatment.
Many dehydrogenases genes like ald (alanine dehydrogenase), icd2 (isocitrate dehydrogenase), fadB2 (3-hydroxybutyryl-CoA dehydrogenase), adhD (alcohol dehydrogenase), pdhB (putative 2-oxoisovalerate dehydrogenase), and lldD2 (lactate dehydrogenase) were downregulated on day 6. This might be needed to maintain the NADH pool under dormant conditions.
Replication, transcription, and translation
In consensus with the earlier reports on NC cells10, a large number of genes involved in replication, transcription, and translation were downregulated in the presence of nitrite37,38. In this study, the DNA replication initiation protein dnaA was downregulated on day 6. A differential expression of transcriptional factors was observed in nitrite-treated cells. The genes sigB, sigH, and sigE were upregulated on day 1, whereas Rv0516c was downregulated on day 3 and anti-sigma E factor RseA encoding Rv1222 was upregulated on day 6. sigH may be involved in the response of MTB toward reactive oxygen species (ROS) and reactive nitrogen intermediates (RNIs). During the transition to the NC state, some marker genes of stress conditions were induced, such as hsp, the chaperones Rv0440 and Rv3417с, and sigma-factors like sigG and sigE. sigG regulates the genes necessary for survival inside macrophages, whereas sigB controls the stationary phase regulon and general resistance to stress16.
recC (exodeoxyribonuclease V) and Rv3828c genes involved in recombination were upregulated on day 3, whereas Rv3750C was downregulated on day 6. The DNA repair mechanism may become activated after nitrite treatment to protect the DNA under stress conditions.
Translation-related genes like IF-3, IF-1, tuf, and fusA1 were downregulated on day 6. 16 proteins of 30 S and nine proteins of 50 S ribosomal encoding genes were downregulated on days 6 and 3, respectively. Overall, DNA replication, transcription, ribosome assembly, translation, and purine and pyrimidine metabolism were downregulated in nitrite-treated MTB bacilli.
Gluconeogenesis, tricarboxylic acid (TCA) cycle, and glyoxylate metabolism
In the persistent phase, metabolism is shifted from glucose to lipids, glycolysis is downregulated, and the glyoxylate shunt is upregulated, allowing anaplerotic maintenance of TCA cycle39,40,41.
In the presence of nitrite, gluconeogenesis and TCA cycle genes were upregulated on day 1. The genes involved in glyoxylate metabolism and pyruvate metabolism were also upregulated on day 1, and then downregulation was observed. It can be concluded that, from the first day of nitrite treatment, the cells shifted from glucose to lipids as a source of carbohydrate.
The ndh gene encoding NADH dehydrogenase and nirA protein involved in sulfate reduction were upregulated on day 1. During aerobic and nitrate respiration, NADH dehydrogenase I (NDH-2) enzyme is used42. It was reported that MTB also utilizes substrates like nitrate for respiration at the dormant state33,43,44,45. It has been shown that, during the NC stage, bacteria respire anaerobically, even though oxygen is available, with uncoupled non-proton-pumping NADH dehydrogenase II playing an important role.
Regulators
Transcriptional regulators like Rv0485, Rv2621C, and Rv3249c (TetR family transcriptional regulators); Rv2499c (oxidase regulator); and whiB3 (redox-responsive transcriptional regulator) were upregulated on day 1. hrcA (heat-inducible transcriptional repressor) and ESX-1 (transcriptional regulator EspR) were upregulated on day 3 and the ArsR family transcriptional regulator was upregulated on day 6, whereas HTH and Rv2166 gene encoding the transcriptional regulator MraZ were downregulated on day 6.
The furA gene product negatively regulates the iron scavenging response, is induced under oxidative stress and hypoxia conditions, and was upregulated on day 610,23,26. The same gene was upregulated in the presence of vitamin C, which suggested that an iron-deficient environment may aid in the repair/replacement of iron-containing proteins that may be damaged during stresses caused by ROIs and RNIs33.
The rpfC gene encoding resuscitation promoting factor was downregulated on days 3 and 6. Other dormancy models even show repression of the resuscitation promoting factor24.
ESAT-6 secretion system
The ESAT-6 secretion system 1 (ESX-1) is associated with the virulence and pathogenesis of MTB46,47. 10 ESAT-protein-encoding genes were upregulated on day 1, whereas three and seven genes were downregulated on days 3 and 6, respectively. It was reported that, at a dormant state, ESAT protein abundance is low, and a similar result was found in cells treated with nitrite for six days27,29,47.
PE and PPE family protein
A variable gene expression pattern was observed in nitrite-treated cells. Most of the genes were upregulated on day 1, whereas similar genes were downregulated on days 3 and 6. These proteins show a variable expression in MTB-infected macrophages and the mouse model, which could provide a dynamic antigenic profile during changing microenvironments within the host48,49,50.
Transmembrane proteins
Six genes encoding integral or transmembrane proteins were upregulated on day 1, and the ftsK gene a transmembrane protein encoding for the cell division was upregulated on day 3. Similar types of transmembrane proteins were also found to be upregulated under low-iron conditions, 24 h after infection to the macrophage, hypoxia at 2% O2 for 2 h20,23,29. Similarly, four membrane-protein-coding genes were upregulated on day 6. Novel pathways involving membrane proteins play an important role in the transition of growing cells to the NC stage10. After nitrite treatment, most of the transmembrane proteins are upregulated and may be involved in the transition of bacilli from the active to the NC state.
Toxin–antitoxin system
The gene rv2063 encoding antitoxin MazE7 was upregulated on day 3. Antitoxin VapB14 encoding genes Rv0968 and Rv1955 coding for toxin HigB were upregulated on day 6, and Rv3408 gene coding for ribonuclease VapC47, a toxin, was downregulated on day 6. The available literature shows that the toxin–antitoxin system plays a role in the stress physiology of bacteria, and it could be part of the specific mechanism involving different enzymes and regulators. Activation of this system results in the shutdown of protein synthesis, until favorable conditions occur, and this system is also involved in the transition of bacteria from the active to a dormant and NC phenotype14,51,52,53,54,55.
Based on the results of the transcription analysis of the noncultivable cells obtained in our study (treated with nitrite for six days) and those obtained in other models of persisters, some genes were found to be common, which are listed in Table 3. These genes may play an important role in the noncultivable state of MTB.
Validation of microarray data by quantitative polymerase chain reaction (qPCR)
In order to validate the microarray data, the fold change of selected genes was determined using qPCR. Even though the fold change measured by the qPCR was higher than that measured by microarray, a similar pattern of gene expression was observed by both methods (Fig. 6).
Differential gene expression level measurement by microarray and qPCR. The gene expression levels of nitrite-treated cells after (A) one, (B) three, and (C) six days were measured using microarray and qPCR. The expression level of the genes was compared with aerobically grown log-phase cells (control). The blue-colored column shows the fold change obtained by microarray, whereas the red-colored column shows the fold change obtained by qPCR for each respective gene. Each experiment was performed three times, and the SD is shown on the graph.
Discussion
It is well known that bacilli can survive the latency stage within the host for many years. Therefore, it can be assumed that the endogenous inducer should be chemically and metabolically very stable. Among the known factors like oxygen depletion, nutrient starvation, acidic pH, CO, and NO, cannot be sourced for years within the host milieu to maintain the non-replicating status of the bacilli. Nitrate and nitrite are the most predominantly available RNIs observed in the lung cavities. However, the possibility of nitrite or nitrate playing a major role is not yet understood. Nitrite has been known to interfere with energy conservation by inhibiting oxygen uptake, oxidative phosphorylation, and proton-dependent active transport, where it acts as an uncoupler, causing the collapse of the proton gradient. Nitrite has also been known to inhibit intracellular thiol groups and certain metabolic enzymes like glycolytic enzymes2. Endogenously generated nitrite may be beneficial for host cells to survive in hostile environments28,56.
In the present study, nitrite was added to an MTB culture under aerobic conditions. As reported in the literature, the effect of nitrite was observed on MTB strain H37Ra28. The concentration of nitrite and strain used in the study was different in our experiments than reported earlier. We observed that the bacteriostatic effect of nitrite appears on MTB with the sharp reduction in the CFU count, without any adverse effect on the viability of cells (Figs 1A,B and 2A,B). Moreover, when these nitrite-treated bacilli were inoculated into nitrite-free media, there was no growth on both the Dubos broth and agar plate. Thus, it can be concluded that nitrite restricts the growth of MTB bacilli, probably shifting it to a noncultivable state. Owing to the similarity of nitrite-treated MTB cells to the bacilli isolated from patients with TB57 and infected animals, a comprehensive study of these forms will be important to elucidate the molecular mechanisms that underlie the development and maintenance of nonreplicating persistence of mycobacteria.
Furthermore, development of antibiotic resistance is associated with the loss of acid fastness. For example, INH treatment impacts cell wall biosynthesis, a process that is inactive in dormant cells58. As a result, cells in all dormancy models are expected to be characterized by their resistance to INH. Moreover, most of the dormancy models demonstrated significant susceptibility to RIF. In our study, under nitrite stress, nearly all of the MTB cells with lipid droplets failed to show acid-fast staining and developed phenotypic antibiotic resistance during the development of dormancy (Figs 4 and 5).
Global changes in the gene expression level occurred in nitrite-induced dormant MTB. As reported earlier, we found a similar shift in metabolism from the available set of genes and proteins when compared between the active and dormant states6,10,28,33 (Table 2, Fig. 7).
Effect of nitrite on the metabolic pathways of MTB. Nitrite affects the metabolic pathways of MTB. Nitrite treatment given for one ( ), three (
), three ( ), and six (
), and six ( ) days alters the metabolic pathways, which is shown in the figure. The figure shows the upregulation and downregulation of genes involved in the marked pathway.
) days alters the metabolic pathways, which is shown in the figure. The figure shows the upregulation and downregulation of genes involved in the marked pathway.
From the first day of nitrite treatment, genes involved in fatty acid degradation were upregulated and fatty-acid-synthesis-related genes were downregulated. This indicates that cells shifted from glucose to lipids for an energy source and confirms the previous evidence59. Genes encoding proteins such as the ABC transporter and ATP binding protein were downregulated on day 6. Four genes encoding dehydrogenases were upregulated, which are required to maintain the NADH pool under dormant conditions60.
The 34 genes involved in DNA replication, transcription, ribosome assembly, translation, and purine and pyrimidine metabolism were downregulated after day 3 of nitrite exposure. Thus, it is very clear that the cell was investing minimum energy for cell division/replication33.
During the transition to the NC state, some genes like hsp, Rv0440, Rv3417с, sigG, and sigE, used as markers of stress conditions, were induced in the presence of nitrite. dosR-regulon gets induced under dormant conditions, but recently it was reported that it acts as a regulator of oxygen tension and gets induced under hypoxia conditions16,30.
The wag31 gene, member of the nrdHIEF operon genes, involved in the oxidative stress response of MTB, also protects cells from ROS and regulates the cell morphology of bacteria. It was observed that transmembrane proteins were upregulated after nitrite treatment, which may be involved in the change in morphology of bacteria during the transition from the active to the dormant state33,61.
Transcriptional regulators (nine genes) and members of a two-component system (six genes) were differentially expressed in response to nitrite exposure. These proteins are involved in many cellular functions, such as the control of multidrug efflux pumps; pathways for the biosynthesis of glyoxylate shunt; and responses to osmotic stress, environmental stimuli, oxidative stress, and toxic chemicals, and are involved in the transition of cells from the active to the dormant state. Thus, these factors could lead us to understand the signaling processes that are necessary for survival under stress conditions.
Redox-sensitive proteins, namely, ferredoxin, thiosulfate sulfur transferase, and methyl transferase, which maintain cellular redox homeostasis, were found to be downregulated62. All of these results showed that the bacilli possess a well-equipped machinery to survive under stress conditions. From the transcriptional study of our model and other models, some common genes were pinpointed in Table 3, which may play an important role in the survival of NC bacteria. These genes can be further studied as good drug targets against latent MTB.
In summary, exposure of cells to nitrite-induced noncultivable dormancy. Moreover, noncultivability was well achieved in our nitrite-induced dormant culture. The rapid noncultivability and dormancy phenotype in nitrite-treated MTB bacilli could characteristically make it one of the most favored dormancy models because of its very similar characteristics to those of cells obtained from clinical samples. This finding could be useful in designing new in vitro and in vivo models for studying latency and screening of potent and specific therapeutic agents against TB. The major advantage of using a nitrite-induced dormant culture is that a large amount of noncultivable cells can be obtained within a short period of time using this robust protocol. In addition, this report mainly helps understand the metabolic shift that occurs during the dormancy of tubercular bacilli. Thus, the presence of nitrite in the medium could act as an extracellular robust inducer of noncultivable dormancy in MTB.
Materials and Methods
Organism and media used
MTB strain H37Ra was a gift from AstraZeneca, Karnataka, India. All chemicals were purchased from Sigma (USA), unless otherwise mentioned. MTB was cultured in a Mycobacterium phlei medium containing 0.5 g of potassium dihydrogen orthophosphate, 0.25 g of sodium citrate, 60 mg of magnesium chloride, 0.5 g of asparagine, and 2% (v/v) glycerol in 100 mL of distilled water at pH 6.6 ± 0.2. The stock culture was maintained at −70 °C and subcultured once in a liquid medium before inoculation into an experimental culture.
Cultivation of aerobic bacilli
For aerobic cultivation, bacterial cultures were grown in a 30 mL defined medium in a 100 mL flask under aerobic conditions in a shaker incubator (Model 481; Thermo Electron Corporation, Waltham, MA, USA) maintained at 150 rpm and 37 °C until the logarithmic phase (OD620 ~ 1.0) was reached.
Growth kinetics of MTB in the presence of nitrite
Different concentrations (5, 10, 15, and 20 mM) of nitrite were added when the cell OD620 reached 0.3, and its effect on growth was monitored by reading the absorbance of the culture at 620 nm as well as by counting the CFU.
Estimation of the cell viability assay
Viability was measured using a live–dead (BacLight) staining kit (Molecular Probes, Eugene, OR, USA) following the manufacturer’s instructions. The assay utilizes a mixture of SYTO 9 green fluorescent nucleic acid stain and red fluorescent nucleic acid stain, propidium iodide (PI). SYTO 9 stains both live and dead cells having an intact or damaged membrane, whereas PI stains dead cells or dying cells. Briefly, log-phase cells were grown in a M. phlei medium up to an optical density of 0.2 (620 nm) and treated with nitrite at different concentrations (10, 15, and 20 mM). The samples (200 µL) were incubated with SYTO 9 (5.1 μM) and PI (80 μM) for 15 min. Fluorescence was scanned, keeping excitation fixed at 488 nm, with emission spanning from 490 to 700 nm. In addition to this, cells treated with nitrite (10 mM) were further used for imaging analysis. The cells were first washed with phosphate-buffered saline and suspended in a fresh medium. A smear was then prepared with 10 µL of cell suspension on a grease-free slide, and a fluorescence image was taken using an EVOS microscope (Life Technologies, Darmstadt, Germany). Standardization of both the dyes and the exposure time was performed with respect to 100% live and 100% dead MTB cells. (Fig. S2). Representative data for INH killing is provided in the Supplementary Data (Fig. S3).
Sample preparation for SEM
SEM of MTB cells was carried out with some modifications in the established protocol63. Microscopy was performed with a Zeiss Supra 55VP microscope (Carl Zeiss, Oberkochen, Germany). Secondary electron microscopic images were taken at low electron energy at 15 kV.
Acid-Fast staining
The Auramine O stain was used in combination with Nile Red (9-diethylamino-5H-benzo[α]phenoxazine-5 -one) to detect the acid-fast property of cells by following an earlier established protocol6.
Extraction of RNA from MTB for microarray analysis
RNA isolation from nitrite-treated and nontreated bacilli was performed at different time points using the TRIzol method64. Genes with >2 fold change and \(p\)-value ≤ 0.05 (i.e., 95% data confidence interval) were considered to be significantly up- or downregulated.
Microarray data processing
In order to study the gene expression profile of nitrite-treated MTB, we availed the services of Genotypic India, Pvt., Ltd. RNA was used as a starting material, and samples (nitrite-treated and nontreated) were processed independently in duplicate. Total RNA was isolated using Qiagen’s RNeasy Minikit (Cat. #74104). From the total RNA (1 μg) extracted, 50 ng/μL RNA was further processed for microarray analysis. Single color labeling was performed, and an 8 × 15k array chip was used. Briefly, the RNA isolated from the samples was converted into cDNA and then into c-RNA with a Cy3-labeled probe using Agilent Quick Amp labeling kit. The random hexamer method of labeling was performed, followed by T7 promoter based linear amplification to generate labeled c-RNA (one-color microarray-based gene expression analysis). 600 ng of c-RNA was used for one array. For hybridization, Agilent in situ hybridization kit (part number 5188–5242) was used. RNA quality was checked using a bioanalyzer, which is a chip-based capillary electrophoresis machine. Probes were spotted on glass slides (eight arrays on one glass slide; each array has 60k probes). After hybridization, the slides were scanned using an Agilent microarray scanner and raw data were extracted using the Agilent Feature Extraction Software. Raw data were then used in GeneSpring GX 12.0 software for normalization by the percentile shift normalization method. Data were filtered on the basis of fold change in nitrite-treated cells as compared to untreated cells. Genes with > 1.5 fold change and \(p\)-value ≤ 0.05 (i.e., 95% data confidence interval) were considered to be significantly up- or downregulated. Data obtained from the series of microarray experiments were analyzed using mainly three bioinformatics tools: Database for Annotation, Visualization and Integrated Discovery (DAVID), KEGG pathway and Cytoscape, and TB Database (http://www.tbdb.org/). Using DAVID and KEGG, a functional analysis of differentially expressed genes (GO analysis) along with the analysis of signaling pathways was performed. These differentially expressed genes were compared with different dormancy models of H37Rv strain.
Real-Time PCR/qPCR Study
For qPCR, 1 µg of total RNA was reverse-transcribed to cDNA using a single-strand synthesis kit (Sigma) as per the manufacturer’s instructions. qPCR was carried out using a QuantiTect SYBR Green PCR kit as per the manufacturer’s instructions. Gene-specific primers were used as mentioned in Table S7, where their respective amplification conditions are also mentioned. The transcript levels between various RNA samples were normalized using sigA. The experiments were carried out in triplicate for each time point.
References
Global Tuberculosis report, W.H.O. (WHO) (2016)
Bloom, B. R. & Murray, C. J. L. Tuberculosis: commentary on a reemergent killer. Science 257, 1055–1064 (1999).
Rustad, T. R., Sherrid, A. M., Minch, K. J. & Sherman, D. R. Hypoxia: a window into Mycobacterium tuberculosis latency. Cellular Microbiology 11, 1151–1159 (2009).
Global Tuberculosis report, W.H.O. (WHO) (2017)
Ayrapetyan, M., Williams, T. C., Baxter, R. & Oliver, J. D. Viable but Nonculturable and Persister Cells Coexist Stochastically and Are Induced by Human Serum. Infect Immun. 83, 4194–4203 (2015).
Deb, C. et al. A Novel in Vitro Multiple-Stress Dormancy Model for Mycobacterium tuberculosis generates a Lipid-Loaded, Drug-Tolerant, Dormant Pathogen. Plos One. 4(6), e6077, https://doi.org/10.1371/journal.pone.0006077 (2009).
Betts, J., Lukey, P., Robb, L., McAdam, R. & Duncan, K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol. 43, 717–731 (2002).
Voskuil, M. Mycobacterium tuberculosis gene expression during environmental conditions associated with latency. Tuberculosis. 84, 138–143 (2004).
Wayne, L. & Hayes, L. An in vitro model for sequential study of shift down of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect Immun. 64, 2062–2069 (1996).
Salina, E. G. et al. Potassium availability triggers Mycobacterium tuberculosis transition to, and resuscitation from, non-culturable (dormant) states. Open Biology. 10, 1098 (2014).
Shleeva, M., Mukamolova, G., Young, M., Williams, H. & Kaprelyants, A. Formation of ‘non-culturable’ cells of Mycobacterium smegmatis in stationary phase in response to growth under suboptimal conditions and their Rpf-mediated resuscitation. Microbiology. 150, 1687–1697 (2004).
Dhillon, J., Lowri, E. D., Mitchson, D. Mycobacterium tuberculosis from chronic murine infections that grow in liquid but not on solid medium. BMC infect.dis. 4(51), https://doi.org/10.1186/1471-2334-4-51 (2004).
Shleeva, M. O. et al. Formation and resuscitation of ‘non-culturable’ cells of and Mycobacterium tuberculosis in prolonged stationary phase. Microbiology. 148, 1581–1591 (2002).
Demidenok, O., Kaprelyants, A. & Goncharenko, A. Toxin–antitoxin vapBC locus participates in formation of the dormant state in Mycobacterium smegmatis. FEMS Microbiol Lett. 352, 69–77 (2014).
Yarbrough,J., Rake, J. & Eagon, R. Bacterial Inhibitory Effects of Nitrite: Inhibition of Active Transport, But Not of Group Translocation, and of Intracellular Enzymes. Applied And Environmental Microbiology. doi: 0099-2240/80/04-0831/04$02.00/0 (1980).
Salina, E., Mollenkopf, H., Kaufmann, S. & Kaprelyants, A. M. Tuberculosis Gene Expression during Transition to the “Non-Culturable” State. Acta Naturae. 1, 73–77 (2009).
Medlab, E., Bernstein, S. & Steward, D. A bacteriologic study of resected tuberculous lesions. Am. Rev. Tuberc. 66, 36–43 (1952).
Gomez, J. & McKinney, J. Tuberculosis persistence, latency, and drug tolerance. Tuberculosis. 84, 29–44 (2004).
Haapanen, J., Kass, I., Gensini, G. & Middlebrook, G. Studies on the gaseous content of tuberculous cavities. Am Rev Respir Dis. 80, 1–5 (1959).
Wayne, L. & Sohaskey, C. Nonreplicating persistence of Mycobacterium tuberculosis. Annu Rev Microbiol. 55, 139–163 (2001).
Steinera, E. et al. CysK2 from Mycobacterium tuberculosis Is an O-Phospho-L-Serine-Dependent S-Sulfocysteine Synthase. J Bacteriol. 196, 3410–3420 (2014).
Schnappinger, D. et al. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J Exp Med. 198, 693–704 (2003).
Rohde, K., Abramovitch, R. & Russell, D. Mycobacterium tuberculosis Invasion of Macrophages: Linking Bacterial Gene Expression to Environmental Cues. Cell Host & Microbe. 2, 352–364 (2007).
Voskuil, M. I. et al. The response of Mycobacterium tuberculosis to reactive oxygen and nitrogen species. Front Microbiol, https://doi.org/10.3389/fmicb.2011.00105 (2011).
Rustad, T., Harrell, M., Liao, R., Sherman, D. The enduring hypoxic response of Mycobacterium tuberculosis. PLoS ONE, https://doi.org/10.1371/journal.pone.0001502 (2008).
Voskuil, M., Visconti, K. & Schoolnik, G. Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis. 84, 218–227 (2004).
Fisher, M., Plikaytis, B. & Shinnick, T. Microarray Analysis of the Mycobacterium tuberculosis Transcriptional Response to the Acidic Conditions Found in Phagosomes. J. Bacteriol. 184, 4025–4032 (2002).
Bussel, A., Zhang, T. & Carl, F. Nitrite produced by Mycobacterium tuberculosis in human macrophages in physiologic oxygen impacts bacterial ATP consumption and gene expression. Proc Natl Acad Sci. 110, E4256–E4265 (2013).
Rodriguez, G., Voskuil, M., Gold, B., Schoolnik, G. & Smith, I. ideR, an Essential Gene in Mycobacterium tuberculosis: Role of IdeR in Iron-Dependent Gene Expression, Iron Metabolism, and Oxidative stress response. Infection and Immunity. 70, 3371–3381 (2002).
Sherman, D. et al. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding α-crystallin. Proc Natl Acad Sci. 98, 7534–7539 (2001).
Springer, B. et al. Silencing of oxidative stress response in Mycobacterium tuberculosis: expression patterns of ahpC inactivation. Infect Immun. 69, 5967–5973 (2001).
Master, S. et al. Oxidative stress response genes in Mycobacterium tuberculosis: role of ahpC in resistance to peroxynitrite and stage-specific survival in macrophages. Microbiology 148, 3139–3144 (2002).
Sikri, K. et al. The pleiotropic transcriptional response of Mycobacterium tuberculosis to vitamin C is robust and overlaps with the bacterial response to multiple intracellular stresses. Microbiology 161, 739–753 (2015).
Pinto, R., Tang, Q., Britton, W., Leyh, T. & Triccas, J. The Mycobacterium tuberculosis cysD and cysNC genes form a stress-induced operon that encodes a tri-functional sulfate-activating complex. Microbiology 150, 1681–1686 (2004).
Zhang, M. et al. Expression and characterization of the carboxyl esterase Rv3487c from Mycobacterium tuberculosis. Protein Expr Purification. 42, 59–66 (2005).
Zimhony, O. et al. AcpM, the meromycolate extension acyl carrier protein of Mycobacterium tuberculosis, is activated by the 4′-phosphopantetheinyl transferase PptT, a potential target of the multistep mycolic acid biosynthesis. Biochemistry, https://doi.org/10.1021/bi501444e (2015)
Yamamoto, K., Muniruzzaman, S., Rajagopalan, M. & Madiraju, M. Modulation of Mycobacterium tuberculosis DnaA protein–adenine-nucleotide interactions by acidic phospholipids. Biochem J. 363, 305–311 (2002).
Andries, K. et al. A diarylquinoline drug active on the ATP synthesis of Mycobacterium tuberculosis. Science 307, 223–227 (2005).
McKinney, J. et al. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyas. Nature 406, 735–748 (2000).
Lorenz, G. & Fink, M. The glyoxylate cycle is required for fungal virulence. Nature 412, 83–96 (2001).
Kumar, R. & Sanyal, S. Mycobacterium tuberculosis: Dormancy, Persistence and Survival in the Light of Protein Synthesis. In: Cardona P, editor. Bacilli Understanding Tuberculosis - Deciphering the Secret Life of the. Infectious diseases, https://doi.org/10.5772/31098 (2012).
Gyan, S., Shiohira, Y., Sato, I., Takeuchi, M. & Sato, T. Regulatory Loop between Redox Sensing of the NADH/NAD Ratio by Rex (YdiH) and Oxidation of NADH by NADH Dehydrogenase Ndh in Bacillus subtilis. Journal of Bacteriology 188, 7062–7071 (2006).
Sohaskey, C. & Wayne, L. Role of narK2X and narGHJI in hypoxic upregulation of nitrate reduction by Mycobacterium tuberculosis. J Bacteriol. 185, 7247–7256 (2003).
Malm, S. et al. The roles of the nitrate reductase NarGHJI, the nitrite reductase NirBD and the response regulator GlnR in nitrate assimilation of Mycobacterium tuberculosis. Microbiology 155, 1332–1339 (2009).
Kumar, R. Glyoxylate shunt: Combating Mycobacterium at forefront. International Journal of Integrative Biology. 7, 69–72 (2009).
Samten, B., Wang, X. & Barnes, P. Mycobacterium tuberculosis ESX-1 system-secreted protein ESAT-6 but not CFP10 inhibits human T-cell immune responses. Tuberculosis 601, 574–586 (2009).
Tan, T., Lee, W., Alexander, D., Grinstein, S. & Liu, J. The ESAT-6/CFP-10 secretion system of Mycobacterium marinum modulates phagosome maturation. Cell Microbiol. 8, 1417–29 (2006).
McEvoy, C. et al. Comparative Analysis of Mycobacterium tuberculosis pe and ppe Genes Reveals High Sequence Variation and an Apparent Absence of Selective Constraints. PLoS ONE, https://doi.org/10.1371/journal.pone.0030593 (2012).
Tiwari, B., Kannan, N., Vemu, L., Raghunand, T. The Mycobacterium tuberculosis PE Proteins Rv0285 and Rv1386 Modulate Innate Immunity and Mediate Bacillary Survival in Macrophages. Plos One, https://doi.org/10.1371/journal.pone.0051686 (2012)
Sampson, S. Mycobacterial PE/PPE Proteins at the Host-Pathogen Interface. Journal of Immunology Research, https://doi.org/10.1155/2011/497203 (2011)
Albrethsen, J. et al. Proteomic Profiling of Mycobacterium tuberculosis Identifies Nutrient-starvation responsive Toxin–antitoxin Systems. The American Society Biochemistry and Molecular Biology, 1180–1191 (2013)
Fozo, E. et al. Abundance of type I toxin-antitoxin systems in bacteria: Searches for new candidates and discovery of novel families. Nucleic Acid Res. 38, 3743–3759 (2010).
Makarova, K., Wolf, Y. & Koonin, E. Comprehensive comparative-genomic analysis of type-2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biol.Direct, https://doi.org/10.1186/1745-6150-4-19 (2009).
Pandey, D. & Gerdes, K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acid Res. 33, 966–976 (2005).
Zhu, L. et al. Characterization of mRNA interferases from Mycobacterium tuberculosis. Biol Chem. 281, 18638–18643 (2006).
Wayne, L. & Hayse, L. Nitrate reduction as a marker for hypoxic shift down of Mycobacterium tuberculosis. Tuber Lung Dis. 2, 127–132 (1998).
Khomenko, A. The variability of Mycobacterium tuberculosis in patients with cavitary pulmonary tuberculosis in the course of chemotherapy. Tubercle. 68, 243–253 (1987).
Gandhi, N. R. et al. Extensively drug resistant tuberculosis as a cause of death in patients co-infected with tuberculosis and HIV in a rural area of South Africa. Lancet 368, 1575–1780 (2006).
Shi, L. et al. Carbon flux rerouting during Mycobacterium tuberculosis growth arrest. Mol Microbiol. 78, 1199–1215 (2010).
Hutter, T. & Dick, B. Increased alanine dehydrogenase activity during dormancy in Mycobacterium smegmatis. FEMS Microbiol Lett. 167, 7–11 (1998).
Monje-casas, F. et al. Expression analysis of the nrdHIEF Operon from Escherichia coli conditions that trigger the transcript level in vivo. The Journal of Biological Chemistry. 276, 18031–18037 (2001).
Akif, M., Khare, G., Tyagi, A., Mande, S. & Sardesai, A. Functional Studies of Multiple Thioredoxins from Mycobacterium tuberculosis. Journal of bacteriology. 190, 7087–7095 (2008).
Takayama, K., Wang, L. & Merkal, R. Scanning Electron Microscopy of the H37Ra Strain of Mycobacterium tuberculosis Exposed to Isoniazid. Am Soc Microbiol. 4, 62–65 (1973).
Akhtar, S., Sarkar, S., Mishra, A. & Sarkar, D. A method to extract intact and pure RNA from mycobacteria. Analytical Biochemistry. 2, 286–288 (2011).
Voskuil, M. et al. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J Exp Med. 198, 705–713 (2003).
Taneja, N. K., Dhingra, S., Mittal, A., Naresh, M. & Tyagi, J. S. Mycobacterium tuberculosis Transcriptional Adaptation, Growth Arrest and Dormancy Phenotype Development Is Triggered by Vitamin C. PLoS ONE, https://doi.org/10.1371/journal.pone.0010860 (2010).
Acknowledgements
The authors would like to thank AstraZeneca R&D Bangalore, India for providing the M. tuberculosis strain as a gift also thankful to Genotypic Technology for servicing Microarray data. SG and SA are thankful to CSIR and UGC for their research fellowships, respectively. This work was supported by Council of Scientific and Industrial Research (CSIR) India through grant aid projects BSC0103 and CSC0406.
Author information
Authors and Affiliations
Contributions
The study was carried out and written by S.G. and S.A. helped in experimental work, S.G. and S.A. analyzed the data as well as contributed in manuscript writing under the supervision of the D.S.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gample, S.P., Agrawal, S. & Sarkar, D. Evidence of nitrite acting as a stable and robust inducer of non-cultivability in Mycobacterium tuberculosis with physiological relevance. Sci Rep 9, 9261 (2019). https://doi.org/10.1038/s41598-019-45652-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-45652-8
This article is cited by
-
Viable but nonculturable bacteria and their resuscitation: implications for cultivating uncultured marine microorganisms
Marine Life Science & Technology (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.