Abstract
Toxic dinoflagellates belonging to the genus Dinophysis acquire plastids indirectly from cryptophytes through the consumption of the ciliate Mesodinium rubrum. Dinophysis acuminata harbours three genes encoding plastid-related proteins, which are thought to have originated from fucoxanthin dinoflagellates, haptophytes and cryptophytes via lateral gene transfer (LGT). Here, we investigate the origin of these plastid proteins via RNA sequencing of species related to D. fortii. We identified 58 gene products involved in porphyrin, chlorophyll, isoprenoid and carotenoid biosyntheses as well as in photosynthesis. Phylogenetic analysis revealed that the genes associated with chlorophyll and carotenoid biosyntheses and photosynthesis originated from fucoxanthin dinoflagellates, haptophytes, chlorarachniophytes, cyanobacteria and cryptophytes. Furthermore, nine genes were laterally transferred from fucoxanthin dinoflagellates, whose plastids were derived from haptophytes. Notably, transcription levels of different plastid protein isoforms varied significantly. Based on these findings, we put forth a novel hypothesis regarding the evolution of Dinophysis plastids that ancestral Dinophysis species acquired plastids from haptophytes or fucoxanthin dinoflagellates, whereas LGT from cryptophytes occurred more recently. Therefore, the evolutionary convergence of genes following LGT may be unlikely in most cases.
Similar content being viewed by others
Introduction
Plastids in photosynthetic dinoflagellates are classified into five types according to their origin1. Plastids in most photosynthetic dinoflagellates are bound by three membranes and contain peridinin as the major carotenoid. Although peridinin is considered to have originated from an endosymbiotic red alga1, this hypothesis remains controversial2. Some photosynthetic dinoflagellates possess plastids originated from other eukaryotic alga. Such occasional plastid replacements have reportedly occurred in several dinoflagellate lineages, resulting in these dinoflagellates possessing plastids originated from haptophytes3, stramenopiles4, chlorophytes5 or cryptophytes6. Dinoflagellates with plastids derived from haptophytes contain 19′-hexanoyloxy-fucoxanthin (fucoxanthin) as the major carotenoid3. Initial plastid acquisition possibly occurs through the predator–prey interactions, which result in the establishment of a permanent plastid likely through gene loss and lateral gene transfer (LGT) from the plastid to the host cell nucleus7,8. Moreover, plastids in some dinoflagellates (e.g. Dinophysis spp. and Nusuttodinium spp.) are derived from their photosynthetic prey through kleptoplasty9,10,11, which is believed to have been the driving force behind the evolutionary transition towards the establishment of a permanent plastid.
Dinophysis spp. can serve as potential models for studying plastid establishment since they acquire plastids from their ciliate prey Mesodinium rubrum12, which itself derives plastids from its cryptophyte prey9,13. Therefore, Dinophysis spp. acquire plastids of cryptophyte origin (kleptoplastids). Although Dinophysis spp. primarily depend on their prey for nutrition, they can survive for several months without feeding by relying on photosynthesis through kleptoplastids14,15. However, unlike permanent plastids, kleptoplastids in Dinophysis spp. are not maintained for a sufficiently long-term and eventually need to be replenished via the consumption of more prey. Expression of genes related to cryptophyte plastid function and maintenance is lower in D. acuminata than in completely phototrophic algae with permanent plastids16, suggesting that D. acuminata cannot establish permanent plastids. However, products of at least five genes are reportedly transported to kleptoplastids, three of which are acquired by LGT from fucoxanthin dinoflagellates, haptophytes and cryptophytes16. Furthermore, additional genes that likely function in plastids have been reported17, although their phylogenetic origins have not been analysed in detail. Studies conducted to date have focused on D. acuminata alone, and the extent of dominance of laterally transferred genes in the kleptoplastids of Dinophysis remains unknown.
In the present study, we sequenced D. fortii transcripts and identified proteins that are generally considered to functions in plastids. The origins of both newly identified and known D. acuminata proteins were analysed through phylogenetic studies. Two or more isoforms of the same D. fortii protein were examined, and their transcript levels were compared. The findings of this study shed light on the evolutionary transition towards plastid retention in Dinophysis.
Results
Genes expressed in kleptoplastid-retaining D. fortii
Sequencing of cDNA libraries of the cryptophyte alga Teleaulax amphioxeia, the ciliate M. rubrum and the dinoflagellate D. fortii using NextSeq 500 (Illumina Inc., San Diego, CA, USA) yielded a total of 44–110 million reads per species, which were deposited in the DNA Data Bank of Japan (DDBJ) Sequence Read Archive under accession numbers DRX131336–DRX131339, DRX131340–DRX131341 and DRX131342–DRX131343, respectively. Once the adapter and low-quality sequences were trimmed, the remaining reads from each pair of libraries were assembled by Trinity18 into 132,239, 144,278 and 217,120 contigs for T. amphioxeia, M. rubrum and D. fortii, respectively (Supplementary Table 1). Removal of the prey sequences from the assembled contigs for D. fortii yielded 185,121 contigs as D. fortii-derived sequences. Open reading frames (ORFs) of >300 bp were extracted from 122,676 of the assembled D. fortii contigs and translated into 423,018 amino acid sequences. Following the clustering of redundant amino acids with up to 95% homology, 372,783 distinct amino acid sequences were obtained (Supplementary Table 1), which were used for sequence homology searches. Overall, 59,907 (16.1%) and 61,878 (16.6%) amino acid sequences showed significant similarities (e-value < 1e–3) to protein sequences in the non-redundant proteins (nr) and UniRef90 databases, respectively. Moreover, 3,365 gene ontology (GO) numbers were assigned to 39,850 amino acid sequences (10.7%), and 711 enzyme commission (EC) numbers were assigned to 10,328 amino acid sequences (2.8%) (Supplementary Table 1).
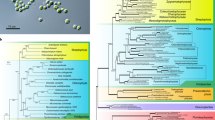
Based on the assigned EC numbers and annotated descriptions, 58 of the amino acid sequences were found to be related to isoprenoid, carotenoid, porphyrin and chlorophyll biosyntheses as well as to photosynthesis; all sequences were registered with DDBJ as transcriptome shotgun assembly (TSA) sequences (Supplementary Table 2). High-resolution phylogenetic trees revealed that 12 D. fortii enzymes originated from other organisms, while another 13 originated from peridinin dinoflagellates. Of note, in phylogenetic trees, almost all proteins identified protein in D. fortii branched with those in D. acuminata with high statistical support and genes of both species shared almost the same evolutionary backgrounds.
Porphyrin and chlorophyll biosynthesis genes
The phylogenetic trees indicated that the following six porphyrin biosynthetic enzymes originated from peridinin dinoflagellate: glutamate-tRNA ligase, glutamyl-tRNA reductase (HemA), delta-aminolevulinate dehydratase (HemB), uroporphyrinogen decarboxylase (HemE), coproporphyrinogen oxidase (HemF) and protoporphyrinogen oxidase (HemY) (Supplementary Fig. 1). Moreover, ferrochelatase (HemH) was clustered with peridinin dinoflagellates, although as a part of the delta-proteobacteria clade. There was insufficient support to resolve the phylogenetic relationships of glutamate-1-semialdehyde 2,1-aminomutase (HemL), hydroxymethylbilane synthase (HemC) and uroporphyrinogen-III synthase (HemD).
Regarding chlorophyll biosynthetic enzymes, magnesium-protoporphyrin IX chelatase comprised three subunits (H, D and I). Subunit H (ChlH) of Dinophysis was clustered with that of the fucoxanthin dinoflagellate Karenia mikimotoi, with moderate statistical support [bootstrap probability (BP) = 85%, Bayesian posterior probability (BPP) = 1.00; Fig. 1a]. Moreover, both subunits were clustered with those of Karlodinium micrum and haptophytes, and their monophyly was completely supported (Fig. 1a). Meanwhile, subunit D (ChlD) of Dinophysis was clustered with that of cyanobacteria (BP = 100%, BPP = 1.00), and its trans-spliced leader sequence was attached to that of the 5′ end of D. fortii mRNA, albeit with six mismatches (Fig. 1b). Magnesium-protoporphyrin IX methyltransferase (ChlM) of Dinophysis formed a clade with that of fucoxanthin dinoflagellates and haptophytes, with moderate statistical support (BP = 87%, BPP = 1.00; Fig. 1c). Two distinct isoforms of protochlorophyllide oxidoreductase (POR) were identified in Dinophysis, both of which were clustered with those of fucoxanthin dinoflagellates (BP = 99%, BPP = 1.00 in clade I, BP = 95%, BPP = 1.00 in clade II; Fig. 1d). Moreover, one isoform was clustered with haptophyte isoforms (BP = 97%, BPP = 1.00; clade I), while the other with E. huxleyi and T. amphioxeia isoforms, with moderate statistical support (BP = 83%, BPP = 1.00; clades II). The expression of one POR isoform of D. fortii in clade I was significantly higher than that of the other isoforms (one-way analysis of variance [ANOVA], P < 0.05; Fig. 1d). Finally, chlorophyll synthase (ChlG) of Dinophysis branched with that of the chlorarachniophyte Bigelowiella natans, with moderate statistical support (BP = 81%, BPP = 1.00; Fig. 1e).
Maximum-likelihood trees of (a) ChlH, (b) ChlD, (c) ChlM, (d) POR and (e) ChlG. Bootstrap values ≥50% are shown. Nodes supported by Bayesian posterior probabilities ≥0.95 are highlighted with bold lines. Where two or more isoforms were identified, transcription levels are shown in the inset bar chart in the top left corner and the isoform with the highest transcription level is highlighted with a red bar in the bar chart. Transcriptome shotgun assembly (TSA) accession numbers are shown in parentheses. The alignment of the dinoflagellate splice leader sequence and 5′-end of the chlD mRNA sequence is shown in (b). Coloured circles indicate proteins of peridinin dinoflagellates (orange), fucoxanthin dinoflagellates (yellow), Haptophyta (blue), Chlorarachniophyta (magenta), Cryptophyta (light blue), Chromerida (purple), Rhodophyta (red), Stramenopiles (brown), Viridiplantae (green) and Bacteria (grey). Proteins of Dinophysis fortii are presented on black background with white font.
Isoprenoid and carotenoid biosynthesis genes
Two isoforms each of the enzymes 4-hydroxy-3-methylbut-2-en-1-yl diphosphate reductase (IspH) and phytoene synthase, which are related to isoprenoid and carotenoid biosyntheses (Fig. 2a,b), and one isoform of phytoene synthase was identified in D. acuminata. One isoform of Dinophysis IspH in clade I was tightly clustered with that of fucoxanthin dinoflagellates (BP = 94%, BPP = 1.00) and with that of haptophytes (BP = 100%, BPP = 1.00; clade I). The other isoforms of Dinophysis IspH was tightly clustered with that of peridinin dinoflagellates (BP = 100%, BPP = 1.00; clade II). The expression of D. fortii isoform in clade II was significantly higher than that of the isoform in clade I (t-test, P < 0.05; Fig. 2a). Similarly, the Dinophysis phytoene synthase isoform in clade I was clustered with that of fucoxanthin dinoflagellate isoform, with moderate statistical support (BP = 80%, BPP = 0.99; clade I), whereas the other isoform in clade II was clustered with peridinin dinoflagellates (BP = 100%, BPP = 1.00; clade II). The expression of D. fortii isoform in clade I was significantly higher than that of D. fortii isoform in clade II (t-test, P < 0.05; Fig. 2b). Geranylgeranyl reductase (ChlP) of Dinophysis, which produces phytyl diphosphate for chlorophyll biosynthesis, was clustered with that of haptophytes, with weak statistical support (BP = 67%, BPP = 1.00), but was distinct from that of peridinin dinoflagellates (Fig. 2c). Since the fucoxanthin dinoflagellate sequences were not available, the relationship among those could not be elucidated.
Maximum-likelihood trees of (a) IspH, (b) phytoene synthase and (c) ChlP. Bootstrap values ≥50% are shown. Nodes supported by Bayesian posterior probabilities ≥0.95 are highlighted with bold lines. Where two or more isoforms were identified, transcription levels are shown in the inset bar chart in the top left corner, and the isoform with the highest transcription level is highlighted with a red bar in the bar chart. Transcriptome shotgun assembly (TSA) accession numbers are shown in parentheses. Coloured circles indicate proteins from the same organisms as described in Fig. 1.
Other enzymes related to isoprenoid biosynthesis in Dinophysis, including 1-deoxy-D-xylulose-5-phosphate synthase (DXS), 1-deoxy-D-xylulose-5-phosphate reductoisomerase (IspC), 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase (IspD), 4-(cytidine 5′-diphospho)-2-C-methyl-D-erythritol kinase (IspE), 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase (IspF) and 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase (IspG) were estimated to have originated from peridinin dinoflagellates (Supplementary Fig. 2). There was insufficient support to resolve the phylogenetic relationships of the two isoforms of farnesyl-diphosphate synthase (FDSP), which is related to carotenoid biosynthesis.
Photosynthesis genes
Seven and three isoforms of ascorbate peroxidase were identified in D. fortii, and D. acuminata, respectively. Five and two of the D. fortii and D. acuminata isoforms, respectively, were clustered in clade I, with moderate statistical support (BP = 90%, BPP = 1.00), which also comprised another D. fortii isoform as well as isoforms of cryptophytes Guillardia theta and T. amphioxeia (BP = 89%, BPP = 1.00). The remaining isoforms of Dinophysis were clustered with isoforms of fucoxanthin and peridinin dinoflagellates, although the monophyly was not well supported (BP = 43%, BPP = 0.55; clade II in Fig. 3a). The expression of D. fortii isoform in clade II (IADC01000026) was the highest among the seven isoforms, and differences were statistically significant (one-way ANOVA, P < 0.01; Fig. 3a). Three isoforms of cytochrome b6/f complex iron-sulfur subunit (PetC) were identified in D. fortii, but none in D. acuminata. These isoforms branched together (BP = 100%, BPP = 1.00) and were tightly clustered with cryptophyte isoforms (BP = 93%, BPP = 1.00; Fig. 3b). However, there were no significant differences in the expression levels of these isoforms (one-way ANOVA, P > 0.05; Fig. 3b). Two ferredoxin-NADP( + ) reductase (PetH) isoforms were identified in Dinophysis. One isoform was clustered with fucoxanthin dinoflagellate isoforms, with moderate statistical support (BP = 80%, BPP = 1.00; clade I in Fig. 3c), while the remaining isoform was clustered with peridinin dinoflagellate isoforms, with strong statistical support (BP = 94%, BPP = 1.00; clade II in Fig. 3c). The expression of D. fortii isoform in clade I was significantly higher than that of isoform in clade II (t-test, P < 0.01; Fig. 3c). Finally, the oxygen-evolving enhancer protein (PsbO) of Dinophysis was tightly clustered with that of fucoxanthin dinoflagellates and haptophytes (BP = 96%, BPP = 1.00; Fig. 3d).
Maximum-likelihood trees of (a) ascorbate peroxidase, (b) PetC, (c) PetH and (d) PsbO. Bootstrap values ≥50% are shown. Nodes supported by Bayesian posterior probabilities ≥0.95 are highlighted with bold lines. Where two or more isoforms were identified, transcription levels are shown in the inset bar chart in the top left corner, and the isoform with the highest transcription level is highlighted with a red bar in the bar chart. Transcriptome shotgun assembly (TSA) accession numbers are provided in parentheses. Coloured circles indicate proteins from the same organisms as described in Fig. 1.
Discussion
In this study, we examined the origins of genes encoding D. fortii proteins, which are involved in the biosyntheses of porphyrins, chlorophylls and isoprenoids as well as in photosynthesis. We identified 58 proteins involved in these processes, 30 of which originated from peridinin dinoflagellates, 21 from other species via LGT and the origin of the remaining 7 could not be identified. Our findings indicate that the mosaic origin of plastid genes may be a common characteristic of Dinophysis spp. and that LGT occurred in common ancestral species of D. fortii and D. acuminata. Moreover, gene replacement may have occurred, followed by LGT, which is rather rare in some pathways. All proteins involved in porphyrin and isoprenoid biosyntheses appear to have originated from peridinin dinoflagellates, although the phylogenies of three of these proteins (HemL, C and D) could not be resolved in the present study (Supplementary Fig. 1). In addition, HemH originated from peridinin dinoflagellates, although it formed a cluster with the HemH of red algae and delta-proteobacteria (Supplementary Fig. 1j). According to a previous phylogenetic analysis, HemH originated from proteobacteria19. Thus, the ancestral species of the peridinin dinoflagellates likely obtained HemH from red algae.
Genes involved in porphyrin and isoprenoid biosyntheses are essential because they produce the chlorophyll backbone as well as haem, which acts as the prosthetic group of cytochromes, catalases and peroxidases during porphyrin biosynthesis20 and as a backbone for steroids, sterol and carotenoids in isoprenoid biosynthesis. Thus, the genes involved in essential pathways may be highly conserved and unlikely to be replaced by genes of other origins via LGT. Therefore, Dinophysis may possess conserved proteins derived from peridinin dinoflagellate, which are involved in essential biosynthetic pathways. However, ChlH, ChlM and POR, which are involved in chlorophyll biosynthesis following porphyrin biosynthesis, originated from fucoxanthin dinoflagellates, whereas ChlG and ChlD originated from chlorarachniophytes and cyanobacteria, respectively. The phylogenetic tree indicated that the gene encoding ChlD, a subunit of magnesium-protoporphyrin IX chelatase, was transcribed with a trans-spliced leader sequence in D. fortii, whereas the ChlD of Dinophysis was derived from cyanobacteria via LGT. (Fig. 1b). Thus, genes involved in chlorophyll biosynthesis appear to have originated from organisms different from which genes involved in porphyrin and isoprenoid biosyntheses were derived from. Dinophysis spp. contain 59–221 times higher volumes of chlorophyll a (Chl a) per cell than T. amphioxeia; thus, Chl a may be synthesised even in Dinophysis cells21. However, because ChlE/ChlA and DVR involved in chlorophyll biosynthesis were not identified in the present study, the assumption of additional Chl a biosynthesis in Dinophysis cells is not supported by our data. If ChlE/ChlA and DVR are not transcribed, Mg-protoporphyrin IX 13-methyl ester, which is produced by ChlM from Mg-protoporphyrin IX, may accumulate in the kleptoplastids. The accumulation of Mg-protoporphyrin IX 13-methyl ester and/or Mg-protoporphyrin IX regulates chloroplast development via chloroplast signaling mediated by nuclear genes22,23,24; such partial Chl a biosynthetic pathway may play some role in the regulation of kleptoplastid development.
During the final step of isoprenoid biosynthesis, IspH produces isopentenyl diphosphate (IPP), two isoforms of which were detected in this study. One of these isoforms originated from peridinin dinoflagellates, whereas the other originated from fucoxanthin dinoflagellates (Fig. 2a). Moreover, phytoene synthase, which is involved in carotenoid biosynthesis occurring behind isoprenoid biosynthesis, was identified as having two isoforms that originated from peridinin and fucoxanthin dinoflagellates (Fig. 2b). Interestingly, the highest phytoene synthase level was produced by the gene of fucoxanthin dinoflagellate origin, whereas the highest IspH level was produced by the gene of peridinin dinoflagellate origin (Fig. 2a,b). Thus, the genes of different origins are likely not evolutionarily converged. Dinophysis spp. and their prey contain alloxanthin as a major carotenoid21. Therefore, D. fortii may have deviated from using peridinin to other carotenoids since the usage of different origin of genes is controlled by the regulation of their transcription.
Among the genes involved in photosynthesis, six ascorbate peroxidase isoforms were likely derived from cryptophytes and one from peridinin dinoflagellates, which was highly transcribed (Fig. 3a). Thus, the protein encoded by the gene of peridinin dinoflagellate origin may play predominant functions in Dinophysis spp. In addition, three PetC isoforms originated from cryptophytes (Fig. 3b), suggesting that PetC is acquired via LGT and complements the lack of gene encoding the cytochrome b6/f complex in the T. amphioxeia plastid genome25. Two PetH isoforms were identified as having originated from peridinin and fucoxanthin dinoflagellates (Fig. 3c). In D. fotii, the transcription level of the PetH isoform originating from fucoxanthin dinoflagellates was significantly higher than that of the PetH isoform originated from peridinin dinoflagellates (Fig. 3c). Thus, in D. fortii, the PetH isoform originating from peridinin dinoflagellates may have been replaced by that originating from fucoxanthin dinoflagellates. Furthermore, evolutionary convergence does not appear to have occurred between the two isoforms of this gene in D. fortii. Finally, PsbO was estimated to have originated from haptophytes (Fig. 3d).
Our findings indicated that in Dinophysis, the genes involved in porphyrin, chlorophyll and isoprenoid biosyntheses as well as in photosynthesis are acquired from fucoxanthin dinoflagellates, haptophytes, chlorarachniophytes, cyanobacteria and cryptophytes via LGT (Fig. 4a). Furthermore, the D. fortii genome may harbour other proteins encoded by genes acquired via LGT because approximately half of the analysed proteins were homologues of proteins of the peridinin dinoflagellate Symbiodinium microadriaticum, whereas the remainder were homologues of proteins of other organisms, particularly haptophytes (2.5% of Emiliania huxleyi and 1.6% of Chrysochromulina sp.; Fig. 4b). In contrast, we obtained very little evidence of LGT from cryptophytes (0.7% of Guillardia theta, Fig. 4b). These results suggest a close relationship between ancestral Dinophysis spp. and haptophytes and/or fucoxanthin dinoflagellates during the course of evolution. Conventionally, the phagocytotic digestion of other organisms has been considered the driving force for the acquisition of genes from other organisms (according to the ‘you are what you eat’ ratchet model26). Reportedly, D. fortii possesses digestive food vacuoles in their body27. In addition, our results indicate that the target genes in Dinophysis were derived from various organisms. Therefore, the major LGT events likely occurred within the common ancestors of Dinophysis spp., and their close relationships with symbionts accelerated gene flow, as illustrated in the ‘shopping bag’ model28. Once the ancestral species of Dinophysis began engulfing or living in the proximity of haptophytes and/or fucoxanthin dinoflagellates, the peridinin plastid may have reduced along with the gene flow to the Dinophysis genome from the potential symbionts. Phalacroma mitra belonging to a sister linage of Dinophysis29 predominantly derived kleptoplastids from haptophytes and may have continued to derive these even after the species diverged. Conversely, although plastids of peridinin dinoflagellate origin are generally considered to have been derived from red algae, some studies have postulated these to have been derived from haptophytes2,30. Moreover, the heterotrophic dinoflagellate Pfiesteria piscicida has been reported to harbour genes derived from fucoxanthin dinoflagellates31. Based on this evidence, we suggest that the genes derived from fucoxanthin dinoflagellates and/or haptophytes have been either vertically inherited from the ancestor of dinoflagellates and/or horizontally transferred from haptophytes as in fucoxanthin dinoflagellates. Nonetheless, in the present study, since the major genes acquired via LGT originated from haptophytes and/or fucoxanthin dinoflagellates, the relationship between ancestral Dinophysis and haptophytes and/or fucoxanthin dinoflagellates may have remained steady. Since such kleptoplastids were not permanently retained in Dinophysis, its ancestors may have been required to continue feeding on other organisms to derive plastids. Consequently, LGT may have occurred from various organisms such as cyanobacteria and chlorarachniophytes (Fig. 1b, e). Because the extant Dinophysis spp. feed on other potential prey organisms in addition to M. rubrum32, LGT from other organisms is possible in these species. However, this scenario is only an evolutionary hypothesis (Fig. 5) and remains to be discussed in the light of further evidence and other speculations. During the course of evolution of kleptoplastids in Dinophysis from the time when they began feeding on M. rubrum and utilising the derived plastid, an evolutionary transition towards the retention of plastids obtained from cryptophytes may have begun before the plastids of haptophyte origin were established.
(a) The deduced origins of proteins involved in porphyrin, chlorophyll, isoprenoid and carotenoid biosyntheses as well as in photosynthesis, and (b) the top 10 species in the Basic Local Alignment Search Tool (BLAST) search are shown. Yellow, green, blue, pink and grey boxes in (a) indicate proteins related to the biosynthesis pathways for porphyrins (haem), chlorophylls, isoprenoids and carotenoids, as well as photosynthesis, respectively. Names presented in black and grey indicate identified and unidentified proteins in this study, respectively. Pie charts present identified proteins, and the colours denote proteins originated from peridinin dinoflagellates (orange), fucoxanthin dinoflagellates (yellow), haptophytes (blue), chlorarachniophytes (magenta), cryptophytes (light blue) and cyanobacteria (grey). White pie charts indicate proteins for which the origin is unclear due to a low phylogenetic tree resolution. ‘G’ in the ChlI pie chart indicates that the ChlI gene was coded in the chloroplast genome of T. amphioxeia (accession no. YP_009159192). In (b), homologous species in the BLAST search are arranged by relative abundance in descending order in a clockwise direction.
Schematic illustration of the proposed mechanism of plastid acquisition in Dinophysis. The model proposed by Bodył (2018)2 was modified. (1) A common ancestor of dinoflagellates, perkinsids, apicomplexans, colpodellids and chromerids acquired plastids from Ochrophyta. (2) The ancestor of peridinin dinoflagellates acquired a haptophyte plastid and evolved into both the peridinin and fucoxanthin dinoflagellates that exist today. (3) Dinophysis retained the original haptophyte plastid or evolved a peridinin plastid and then acquired a new plastid from either haptophytes or Kareniaceae, which possess the original haptophyte plastid. Dinopysis subsequently evolved into two lineages: a lineage that primarily possessed the haptophyte plastid, e.g. Phalacroma mitra, and (4) a lineage that switched from using a haptophyte to using a cryptophyte plastid via Mesodinium.
Methods
Establishment of clonal strains
Samples of the ciliate M. rubrum and the cryptophyte T. amphioxeia were isolated from Inokushi Bay in Oita Prefecture, Japan, at the end of February 200733. Cultures were established as prey for D. fortii in modified f/2 medium34,35 with a salinity of 30 practical salinity units (psu) and maintained at 18 °C under a photon irradiance of 100 µmol·m−2·s−1 provided by cool-white fluorescent lamps under a 12:12 h light:dark cycle. T. amphioxeia subculture was maintained by re-inoculating 0.1–0.3 mL of culture (ca. 1.2 × 105 cells mL−1) into fresh modified f/2 medium once per week, and M. rubrum subculture was maintained by re-inoculating 40 mL of culture (2,000–3,000 cells mL−1) into 110 mL of fresh modified f/2 medium and 60 µL of the T. amphioxeia culture (ca. 1.2 × 105 cells mL−1) in 250-mL polycarbonate Erlenmeyer flasks once per week.
D. fortii culture was established from a seawater sample collected from Saloma Lake, Japan (143°148′E, 44°15′N) in October 2015. Cells were picked up by micropipetting and rinsed several times in filtered seawater (0.22 µm). The established D. fortii culture was maintained by re-inoculating 15 mL of culture (500–1,000 cells mL−1) into 150 mL of M. rubrum culture (containing 500–1,000 cells mL−1; 1:4 dilution of fresh culture medium) in 250-mL polycarbonate Erlenmeyer flasks once every 3 weeks under the same conditions as outlined above.
Culture conditions for RNA sequencing
Once established, the clonal strains of T. amphioxeia, M. rubrum and D. fortii were maintained in 150 mL of culture solution at 18 °C under photon irradiances of 100 or 20 µmol·m−2·s−1 provided by cool-white fluorescent lamps under a 12:12 h light:dark cycle.
At the start of the experiment, M. rubrum cultures were transferred D. fortii cultures (in duplicate) at a predator:prey ratio of 1:10, thus allowing D. fortii to acquire and retain plastids from M. rubrum. After 5 days, D. fortii cultures were filtered through a 20-µm nylon mesh to remove any remaining M. rubrum cells, and the filtered culture media were filtered again through 8-µm polycarbonate filters (GE Healthcare, Tokyo, Japan). D. fortii cells were then re-inoculated into culture media devoid of prey and incubated for 1 week. Thereafter, D. fortii cells were once again trapped by filtering the media through a 20-µm nylon mesh and collected by centrifugation at 5,000 × g for 2 min. The cells were immediately immersed in RNALater (Thermo Fisher Scientific, Waltham, MA, USA), left overnight at 4 °C and stored at −80 °C until further use.
T. amphioxeia and M. rubrum sequences from the D. fortii RNA sequences, T. amphioxeia and M. rubrum cells maintained under the highest photon irradiance, followed by 30 min in the dark, were removed from the cultures using 1-µm polycarbonate filters. RNALater was applied to each filter for 5 min to preserve the total RNA. After removing RNALater, the filters were stored at −80 °C until further use.
RNA extraction and cDNA library construction
Total RNA was extracted from T. amphioxeia, M. rubrum and D. fortii preserved in RNALater and stored at –80 °C using the TRIzol Plus RNA Purification Kit (Thermo Fisher Scientific); any contaminating DNA was digested using PureLink DNase (Thermo Fisher Scientific), according to the manufacturer’s instructions. The concentration and purity total RNA were determined using a Qubit RNA HS Assay Kit (Thermo Fisher Scientific) and a Bioanalyzer 2100 with RNA 6000 Nano Kit (Agilent Technologies, Inc., Santa Clara, CA, USA), according to the manufacturer’s instructions.
Duplicate libraries for D. fortii under the two photon irradiance conditions and one library each for T. amphioxeia and M. rubrum under the highest photon irradiance and dark conditions were constructed from the total RNA (1 µg) in accordance with the TruSeq RNA Sample Prep ver. 2 (LS) protocol (Illumina, Inc.). Complementary DNA (cDNA) was synthesised using SuperScript III reverse transcriptase (Thermo Fisher Scientific). The cDNA libraries were sequenced into 150-bp paired-end reads using NextSeq 500 (Illumina, Inc.) at the Research Center for Bioinformatics and Biosciences of the National Research Institute of Fisheries Science, Yokohama, Japan.
Sequence analysis
Sequences from individual samples were generated using the bcl2fastq pipeline ver. 2.17 (Illumina, Inc.). Any adapter sequences, low-quality ends (<QV30) and unpaired reads were removed from the sequences using Trimmomatic36. The sequence length and the quality of the remaining reads were confirmed using FastQC37, then the remaining paired-end reads were assembled using Trinity18 using the ‘-min_kmer_cov = 2′ command option and under default settings for all other options. ORFs of >300 bp were extracted from the assembled sequences and translated into amino acid sequences using TransDecoder38. ORFs of 95% homologous amino acid sequences were clustered using the CD-HIT programme39 using the ‘–c 0.95′ command option and under default settings for all other options to remove redundant amino acid sequences. The remaining amino acid sequences were searched against those of T. amphioxeia and M. rubrum using the Protein Basic Local Alignment Search Tool (BLASTP) programme, with a threshold of sequence homology of >98% identity to remove the amino acid sequences of the prey species.
Proteins derived from D. fortii were annotated based on their homology to sequences in the nr database of NCBI and the UniRef90 database40, using the MMseqs2 programme41, with a threshold e-value of <1e–3. GO numbers, which are shared with the accession numbers used in UniRef9040,42, were assigned from the best hits of the MMseqs2 results against UniRef90. EC numbers were obtained from GO numbers using the Blast2GO software43 to identify proteins related to porphyrin and chlorophyll metabolism, terpenoid backbone biosynthesis and photosynthesis.
Comparison of transcription levels among isoforms
Transcription levels of each gene were determined based on the number of mapped reads. The reads were mapped to each gene using Bowtie244 and counted using RSEM45. Transcription levels were normalised among the libraries using the trimmed mean of M-values method46 with the edgeR package47 in R software ver. 3.3.148. The normalised fragments per kilobase per million mapped fragments (FPKM) of different isoforms were statistically compared using ANOVA and Student’s t-test with R software ver. 3.3.148.
Phylogenetic analysis
Amino acid sequences of several organisms, including Viridiplantae, Rhodophyta, Stramenopiles, Haptophyta, Cryptophyta, Chromerida, Chlorarachniophyta and Dinoflagellates were obtained from public databases (Supplementary Tables 3 and 4). Protein sequences several organisms were retrieved based on their homology to the target proteins of D. fortii using the BLASTP programme. Multiple sequence alignments were performed using MAFFT ver. 7.21249, and gaps were automatically trimmed by trimAl50 using the ‘–automated1′ command option and under default settings for all other options. The best-fit evolutionary model for each alignment was identified by ModelFinder51 using the Akaike information criterion (Supplementary Table 4) and subjected to the maximum-likelihood (ML) and Bayesian phylogenetic analyses. ML trees were inferred using RAxML ver. 8.2.452 with 100 bootstrap replicates, while the posterior probabilities of nodes in ML trees were calculated with MrBayes ver. 3.1.253 using a Metropolis-coupled Markov chain Monte Carlo procedure starting from a random tree and sampled every 100 generations for a total of 1 million generations. One heated and three cold chains were simultaneously started, and the best fitting substitution model for each protein set was used for analyses. The initial 25% of the sampled trees were discarded as ‘burn in’ prior to the construction of the consensus phylogeny.
References
Hackett, J. D., Anderson, D. M., Erdner, D. L. & Bhattacharya, D. Dinoflagellates: A remarkable evolutionary experiment. Am. J. Bot. 91, 1523–1534 (2004).
Bodył, A. Did some red alga-derived plastids evolve via kleptoplastidy? A hypothesis. Biol. Rev. 93, 201–222 (2018).
Tengs, T. et al. Phylogenetic analyses indicate that the 19’Hexanoyloxy-fucoxanthin-containing dinoflagellates have tertiary plastids of haptophyte origin. Mol. Biol. Evol. 17, 718–729 (2000).
Inagaki, Y., Dacks, J. B., Ford Doolittle, W., Watanabe, K. I. & Ohama, T. Evolutionary relationship between dinoflagellates bearing obligate diatom endosymbionts: Insight into tertiary endosymbiosis. Int. J. Syst. Evol. Microbiol. 50, 2075–2081 (2000).
Matsumoto, T. et al. Green-colored plastids in the dinoflagellate genus Lepidodinium are of core chlorophyte origin. Protist 162, 268–276 (2011).
Hackett, J. D., Maranda, L., Su Yoon, H. & Bhattacharya, D. Phylogenetic evidence for the cryptophyte origin of the plastid of Dinophysis (Dinophysiales, Dinophyceae). J. Phycol 39, 440–448 (2003).
Martin, W. & Herrmann, R. G. Gene transfer from organelles to the nucleus: how much, what happens, and Why? Plant Physiol. 118, 9–17 (1998).
Bhattacharya, D., Archibald, J. M., Weber, A. P. M. & Reyes-Prieto, A. How do endosymbionts become organelles? Understanding early events in plastid evolution. BioEssays 29, 1239–1246 (2007).
Takishita, K., Koike, K., Maruyama, T. & Ogata, T. Molecular evidence for plastid robbery (Kleptoplastidy) in Dinophysis, a dinoflagellate causing diarrhetic shellfish poisoning. Protist 153, 293–302 (2002).
Takano, Y., Yamaguchi, H., Inouye, I., Moestrup, O. & Horiguchi, T. Phylogeny of five species of Nusuttodinium gen. nov. (Dinophyceae), a genus of unarmoured kleptoplastidic dinoflagellates. Protist 165, 759–778 (2014).
Gagat, P., Bodył, A., Mackiewicz, P. & Stiller, J. W. Tertiary plastid endosymbioses in dinoflagellates. in Endosymbiosis (ed. Löffelhardt, W.) 233–290 (Springer, 2014).
Taylor, F. J. R., Blackbourn, D. J. & Janice, B. Ultrastucturre of the chloroplasts and associated stuctures within the marine cilitate Mesodinium rubrum (Lohmann). Nature 224, 819–821 (1969).
Myung, G. P. et al. First successful culture of the marine dinoflagellate Dinophysis acuminata. Aquat. Microb. Ecol. 45, 101–106 (2006).
Park, M. G., Park, J. S., Kim, M. & Yih, W. Plastid dynamics during survival of Dinophysis caudata without its ciliate prey. J. Phycol. 44, 1154–1163 (2008).
Hansen, P. J., Nielsen, L. T., Johnson, M., Berge, T. & Flynn, K. J. Acquired phototrophy in Mesodinium and Dinophysis - A review of cellular organization, prey selectivity, nutrient uptake and bioenergetics. Harmful Algae 28, 126–139 (2013).
Wisecaver, J. H. & Hackett, J. D. Transcriptome analysis reveals nuclear-encoded proteins for the maintenance of temporary plastids in the dinoflagellate Dinophysis acuminata. BMC Genomics 11 (2010).
Janouškovec, J. et al. Major transitions in dinoflagellate evolution unveiled by phylotranscriptomics. Proc. Natl. Acad. Sci. 114, E171–E180 (2017).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011).
Oborník, M. & Green, B. R. Mosaic origin of the heme biosynthesis pathway in photosynthetic eukaryotes. Mol. Biol. Evol. 22, 2343–2353 (2005).
Rodgers, K. R. Heme-based sensors in biological systems. Curr. Opin. Chem. Biol. 3, 158–167 (1999).
Rial, P., Garrido, J. L., Jaén, D. & Rodríguez, F. Pigment composition in three Dinophysis species (Dinophyceae) and the associated cultures of Mesodinium rubrum and Teleaulax amphioxeia. J. Plankton Res. 35, 433–437 (2013).
Strand, Å., Asami, T., Alonso, J., Ecker, J. R. & Chory, J. Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrin IX. Nature 421, 79–83 (2003).
Kropat, J., Oster, U., Rudiger, W. & Beck, C. F. Chlorophyll precursors are signals of chloroplast origin involved in light induction of nuclear heat-shock genes. Proc. Natl. Acad. Sci. 94, 14168–14172 (1997).
Kropat, J., Oster, U., Rüdiger, W. & Beck, C. F. Chloroplast signalling in the light induction of nuclear HSP70 genes requires the accumulation of chlorophyll precursors and their accessibility to cytoplasm/nucleus. Plant J. 24, 523–531 (2000).
Kim, J. I. et al. The plastid genome of the cryptomonad Teleaulax amphioxeia. PLoS One 10, 1–17 (2015).
Doolittle, W. F. You are what you eat: a gene transfer ratchet could account for bacterial genes in eukaryotic nuclear genomes. Trends Genet. 14, 307–11 (1998).
Koike, K., Koike, K., Takagi, M., Ogata, T. & Ishimaru, T. Evidence of phagotrophy in Dinophysis fortii (Dinophysiales, Dinophyceae), a dinoflagellate that causes diarrhetic shellfish poisoning (DSP). Phycol. Res. 48, 121–124 (2000).
Larkum, A. W. D., Lockhart, P. J. & Howe, C. J. Shopping for plastids. Trends Plant Sci. 12, 189–195 (2007).
Gómez, F., López-García, P. & Moreira, D. Molecular phylogeny of dinophysoid dinoflagellates: The systematic pos≥ition of Oxyphysis oxytoxoides and the Dinophysis hastata group (dinophysales, dinophyceae). J. Phycol. 47, 393–406 (2011).
Yoon, H. S., Hackett, J. D. & Bhattacharya, D. A single origin of the peridinin- and fucoxanthin-containing plastids in dinoflagellates through tertiary endosymbiosis. Proc. Natl. Acad. Sci. 99, 11724 LP–11729 (2002).
Kim, G. H. et al. Still acting green: continued expression of photosynthetic genes in the heterotrophic dinoflagellate Pfiesteria piscicida (Peridiniales, Alveolata). PLoS One 8, 1–11 (2013).
Kim, M., Kim, S., Yih, W. & Gil, M. The marine dinoflagellate genus Dinophysis can retain plastids of multiple algal origins at the same time. Harmful Algae 13, 105–111 (2012).
Nishitani, G., Nagai, S., Sakiyama, S. & Kamiyama, T. Successful cultivation of the toxic dinoflagellate Dinophysis caudata (Dinophyceae). Plankton Benthos Res. 3, 78–85 (2008).
Guillard, R. R. L. Culture of phytoplankton for feeding marine invertebrates. In Culture of marine invertebrate animals (ed. Smith W.L., Chanley M.H; Springer 1975).
Nagai, S., Matsuyama, Y., Oh, S. J. & Itakura, S. Effect of nutrients and temperature on encystment of the toxic dinoflagellate Alexandrium tamarense (Dinophyceae) isolated from Hiroshima Bay, Japan. Plankt. Biol. Ecol. 2, 103–109 (2004).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible read trimming tool for Illumina NGS data. Bioinformatics 30, 2114–2120 (2014).
Andrews, S. FastQC: a quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc. (Accessed: 22nd February 2018).
Haas, B. J. et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512 (2013).
Li, W. & Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006).
Suzek, B. E., Wang, Y., Huang, H., McGarvey, P. B. & Wu, C. H. UniRef clusters: A comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 31, 926–932 (2015).
Steinegger, M. & Söding, J. MMseqs. 2 enables sensitive protein sequence searching for the analysis of massive data sets. Nature Biotech. 35, 1026–1028 (2017).
Camon, E. The Gene Ontology Annotation (GOA) Database: sharing knowledge in Uniprot with Gene Ontology. Nucleic Acids Res. 32, D262–D266 (2004).
Conesa, A. et al. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676 (2005).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011).
Robinson, M. D. & Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25 (2010).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. Available at: https://www.r-project.org/. (Accessed: 22nd February 2018) (2016).
Katoh, K., Misawa, K., Kuma, K. & Miyata, T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066 (2002).
Capella-Gutiérrez, S., Silla-Martínez, J. M. & Gabaldón, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009).
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A. & Jermiin, L. S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587 (2017).
Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014).
Huelsenbeck, J. P. & Ronquist, F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755 (2001).
Acknowledgements
This study was partly supported by a grant from Cross-ministerial Strategic Innovation Promotion Program, Cabinet Office, Government of Japan. We thank Aiko Watanabe for assistance with the NGS experiment. We also thank Ritsuko Kubota and Ayako Kondo for assistance with D. fortii culture.
Author information
Authors and Affiliations
Contributions
Y. H. performed the experiments, analysed the data and wrote the manuscript. A. Y. and K. F. analysed the data and revised the manuscript. S. N. cultured D. fortii and revised the manuscript.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Hongo, Y., Yabuki, A., Fujikura, K. et al. Genes functioned in kleptoplastids of Dinophysis are derived from haptophytes rather than from cryptophytes. Sci Rep 9, 9009 (2019). https://doi.org/10.1038/s41598-019-45326-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-45326-5
This article is cited by
-
Taming the perils of photosynthesis by eukaryotes: constraints on endosymbiotic evolution in aquatic ecosystems
Communications Biology (2023)
-
The genome of the diatom Chaetoceros tenuissimus carries an ancient integrated fragment of an extant virus
Scientific Reports (2021)
-
Transcriptome characterization of BPG axis and expression profiles of ovarian steroidogenesis-related genes in the Japanese sardine
BMC Genomics (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.