Abstract
Specialist herbivores have often evolved highly sophisticated mechanisms to counteract defenses mediated by major plant secondary-metabolites. Plant species of the herbivore host range often display high chemical diversity and it is not well understood how specialist herbivores respond to this chemical diversity. Pieris larvae overcome toxic products from glucosinolate hydrolysis, the major chemical defense of their Brassicaceae hosts, by expressing nitrile-specifier proteins (NSP) in their gut. Furthermore, Pieris butterflies possess so-called major allergen (MA) proteins, which are multi-domain variants of a single domain major allergen (SDMA) protein expressed in the guts of Lepidopteran larvae. Here we show that Pieris larvae fine-tune NSP and MA gene expression depending on the glucosinolate profiles of their Brassicaceae hosts. Although the role of MA is not yet fully understood, the expression levels of NSP and MA in larvae that fed on plants whose glucosinolate composition varied was dramatically changed, whereas levels of SDMA expression remained unchanged. In addition, we found a similar regulation pattern among these genes in larvae feeding on Arabidopsis mutants with different glucosinolate profiles. Our results demonstrate that Pieris larvae appear to use different host plant adaptive genes to overcome a wide range of glucosinolate profiles in their host plants.
Similar content being viewed by others
Introduction
Understanding the mechanisms that define the host plant ranges of herbivores is key to understanding the coevolution of plants and herbivores. Specialist herbivores have restricted host ranges but usually possess adaptive strategies to overcome the chemical challenges (i.e. defensive secondary metabolites) of their host plants1. Recently, in a number of generalist and specialist herbivores, the molecular adaptive mechanisms to specific groups of secondary metabolites have been partially identified, such as cytochrome P450 in Papilio butterflies against furanocoumarins or UDP-glycosyltransferases from Helicoverpa armigera or Helicoverpa zea to capsaicin1,2,3,4. Although several previous studies have investigated the molecular adaptive mechanisms with which generalist herbivores respond to different host plants5,6,7,8, how specialist herbivores overcome the diversity of the secondary metabolites is not well understood. The same holds true for specialist herbivores that have been exposed to chemical variation in their host plants, since secondary metabolites are often diversified chemically within plant families and even between closely related species9,10.
Glucosinolates (GLSs), a group of secondary metabolites found in Brassicales, are stored in plant cells separated from specific enzymes called myrosinases11. In response to tissue damage in plants, GLSs come into contact with myrosinase enzymes and are hydrolyzed, forming several breakdown products12. Among these, the isothiocyanates (ITCs) are dominant and are toxic to numerous herbivores11. GLSs are divided into three major classes depending on their biosynthetic origins (aliphatic GLSs, benzylic GLSs and indolic GLSs) and are further sorted according to a variable side chain, with more than 140 different GLSs identified so far9,13. Notably, most of these variations seem to be present in the family Brassicaceae, with each species having a specific GLS profile9,14.
Although the GLS myrosinase system in Brassicales is known as an efficient defense against many herbivores15,16, some insects can use Brassicales plants as hosts1,4. Pierid butterflies use Brassicales plants by disarming GLS defense with a protein expressed in the larval gut called nitrile-specifier protein (NSP)17. NSP can redirect the GLS-myrosinase reaction to form less toxic nitriles rather than toxic ITCs. Since Pierid butterfly larvae are generally oligophagous and use a distinct range of Brassicales host plants18, each species will likely be exposed to a range of GLSs in their host plants. Pieris butterflies encounter a large variety of GLS profiles in their host plants because they rely on various genera in the Brassicaceae family, which seems to have the highest GLS diversity in Brassicales14,19.
NSP belongs to the small so-called NSP-like gene family, which also includes genes that encode major allergens (MA) and single domain major allergens (SDMA)20. NSP and MA have a similar genetic structure, as they both possess three replicated domains that originated from SDMA20. SDMA is generally found across Lepidoptera. Although its specific function is still unknown, SDMA is known to be expressed only in larval guts, suggesting this protein plays a general digestion-related role in Lepidoptera21. NSP and MA are specific for Pierid butterfly larvae feeding on Brassicales20, suggesting that both proteins may have GLS-disarming functions. The role of MA proteins in modifying the outcome of the GLS-myrosinase defense system, by which MA causes nitriles rather than ITCs to form, was confirmed in a Brassicaceae-feeding Pierid species, Anthocharis cardamines, which also seems to lack an NSP gene19. However, in general, the role of MA proteins is still unclear in most Pieris species, especially those which encode both NSP and MA19. Given that in at least one Pieris species, not only NSP but also MA have been shown to have GLS-disarming functions, the presence of both genes in some Pieridae could be a molecular adaptive mechanism for overcoming diverse GLSs in the species’ host plants.
Here, we focus on members of the NSP-like gene family and their regulation patterns in Brassicaceae-feeding Pieris larvae exposed to a range of GLS profiles, in order to find out how Pieris butterflies respond and adapt to diverse GLSs in their host plants. We conducted feeding experiments using four Japanese Pieris species (Pieris napi, P. melete, P. rapae and P. brassicae) and two Brassicaceae plants (Arabidopsis kamchatica and Cardamine occulta, both of which have diverse GLS profiles)9,13,22,23. The four closely related Pieris species are known to have different host ranges. P. napi and P. melete mainly use wild Brassicaceae plants such as Arabis or Cardamine in Japan24,25, while P. rapae and P. brassicae are known pests of Brassica crops (Table 1)26,27,28. NSP sequences have been identified only in P. rapae and P. brassicae in this genus19,20,29. We first identified NSP, MA and SDMA sequences in all of the tested Pieris species, combining high throughput RNA sequencing (RNA-seq) of larval samples from our plant-feeding experiments and de novo transcriptome assemblies. We subsequently measured and compared the expression levels of all of the identified NSP, MA and SDMA genes in larvae that fed on two host plants using the RNA-seq data. To confirm the expression levels of the NSP-like gene family observed in our RNA-seq data with a larger number of replicates, we also conducted real-time quantitative PCR (RT-qPCR) on P. melete, considering it to be a representative species and targeting all members of the NSP-like gene family (Fig. 1).
Experimental design of this study. We focused on four Pieris butterflies, and conducted feeding experiments with two Brassicaceae plants in order to see gene expression patterns of NSP-like gene family members to different host plants with distinct GLS profiles by RNA-seq. We confirmed the result of RNA-seq based expression analysis by RT-qPCR using P. melete as a representative with more replicates. We also conducted feeding experiments with Arabidopsis thaliana mutants which differ in their GLS profiles to get gene expression data in more controlled chemical background. For this mutant experiment, we used Pieris napi as a representative.
Since GLS profiles are specific for each Brassicaceae plant species9,13, we expected to find that Pieris larvae feeding on the two Brassicaceae plants regulate NSP-like genes in distinct ways. However, since some plant species also differ in their general chemical profiles – excluding GLSs–in several ways, this chemical difference could also affect the regulation of genes in larvae. To obtain feeding and gene expression data using a more controlled background, we also conducted feeding experiments with Arabidopsis thaliana mutants, which differ in their GLS profiles but share the same chemical background. We prepared wild-type lines (Col-0) and three mutant lines of A. thaliana which have different GLS profiles–namely, MAM1 (which lacks chain elongation genes for the C3/C4 chain length variation of Met-derived GLS), MAM3 (which lacks Met chain elongation gene and has no long-chain aliphatic GLS), and quad-GLS (the quadruple mutant myb28myb29cyp79B2cyp79B3 which has no detectable GLS)30,31,32–and used P. napi as a representative species for this feeding assay. Combining these two approaches, we were able to observe how Pieris butterflies regulate NSP-like gene family members in response to a broad range of GLS defenses in their host plants (Fig. 1).
Results
RNA-seq identified NSP-like gene family sequences of the four Pieris butterflies
We obtained 32–40 million Illumina100 bp paired-end reads from each of the four Pieris larval RNA samples. De novo transcriptome assemblies using Trinity resulted in 64,279; 62,054; 59,327; and 53,004 contigs, and in N50 values of 2,048 bp; 2,132 bp; 2,060 bp; and 2,594 bp for P. napi, P. melete, P. rapae and P. brassicae, respectively. We identified NSP, MA and SDMA sequences from all four Pieris butterfly species with reference sequences (Fig. 2). The newly acquired NSP sequences of P. napi and P. melete have 86% and 84% amino acid sequence identity, respectively, with NSP from P. rapae. MA proteins also showed high identity (89% each), and SDMA proteins showed slightly higher sequence identity (92% each) to P. rapae.
Sequence similarities of NSP-like gene family members in Pieris spp. acquired from RNA-seq analysis. The tree is based on amino acid sequences and was generated using the Maximum Likelihood Method. Values shown at each node are Bootstrap values. Reference sequences from GenBank are provided with accession numbers. According to the RNA-seq data analysis, all four Pieris species encode for NSP, MA and SDMA proteins.
NSP and MA are differentially expressed in response to wild Brassicaceae plants
GLS profiles of Arabidopsis kamchatica and Cardamine occulta, host plants of Pieris butterflies in Japan, were significantly different according to the UPLC-TQMS analysis. In total, we detected 25 types of GLSs in the two tested plant species and sorted these into the three major GLS chemical classes to observe their profile differences (Table S1). A. kamchatica had high amounts of aliphatic and indolic GLSs, whereas C. occulta contained benzylic GLSs, which were much lower in A. kamchatica (Fig. 3a, Table S1). Although none of the four larvae species feeding on the two plant species showed any sign of stopping feeding or of poisoning, we observed that P. rapae and P. brassicae appeared to grow better on C. occulta than on A. kamchatica. However, this difference was not statistically significant (P = 0.12 and 0.064, Fig. 3b).
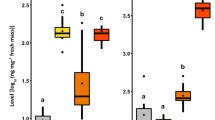
(a) GLS profiles of Arabidopsis kamchatica and Cardamine occulta measured by UPLC-TQMS. Arabidopsis kamchatica had high aliphatic GLS concentration and indolic GLS, whereas C. occulta had high concentration of benzylic GLS. (b) Larval growth of the four Pieris butterflies fed on the two wild Brassicaceae plant species used in the feeding experiment. All four Pieris species fed on both plant species, although not significant growth level differences were observed among the four species (pairwise t test with FDR adjustment, P > 0.05). (c) Relative gene expression levels of NSP-like gene family members in the four Pieris butterflies fed on Arabidopsis kamchatica and Cardamine occulta from digital analyses based on RNA-seq expression levels. NSP and MA showed differential expression levels in larvae fed on the two different host plant species, while SDMA did not. NSP was expressed more highly in larvae fed on C. occulta, whereas MA was expressed more highly in A. kamchatica-feeding larvae. (d) Relative gene expression levels (2−ΔCt) of NSP-like gene family members in Pieris melete larvae compared to larvae feeding on Cardamine occulta and Arabidopsis kamchatica analyzed by RT-qPCR. “*” show statistical significance based on the Mann–Whitney U test (P ≤ 0.05). Significant differences: NSP (P = 0.05), MA (P = 0.05), SDMA (P = 0.90).
We acquired expression levels of the NSP-like gene family from all four species of Pieris larvae feeding on the two Brassicaceae plants by analyzing digital expression based on a single RNA-seq data for each treatment (Fig. 3c). Compared to the observed expression levels of NSP in larvae fed with A. kamchatica, the levels increased when larvae fed on C. occulta, while an inverse gene expression pattern was observed for MA. MA was highly expressed in larvae that fed on A. kamchatica, and less expressed in larvae that fed on C. occulta. This inverse trend was observed in all four Pieris species, whereas expression levels of SDMA (which is not involved in disarming GLS) were similar between the larvae fed with the two Brassicaceae plants in all four Pieris species (Fig. 3c). The result of our P. melete larval RT-qPCR experiments also mirrored the same gene regulation pattern of NSP-like gene family members, supporting our RNA-seq results. Expression levels of NSP and MA differed significantly depending on the plant species on which the larvae fed (NSP and MA: P = 0.05, Mann–Whitney U test), whereas levels of SDMA did not (SDMA: P = 0.90, Mann–Whitney U test, Fig. 3d).
NSP and MA are differentially regulated in P. napi feeding on A. thaliana mutants with different GLS profiles
We identified 9 types of GLSs from the four tested A. thaliana lines (Col-0, MAM1, MAM3 and quad-GLS) by LC-UV, confirming previously described GLS profiles for all the mutant lines (Fig. 4a)30,31,33,34. Col-0 had higher levels of short-chain aliphatic GLSs, and we confirmed that the quad-GLS mutant had no detectable GLSs. In the MAM1 mutant, we detected a higher amount of 3-(Methylsulfinyl)propyl GLS (3MSOP) and 8-(Methylsulfinyl)octyl (8MSOO) but less 4-(Methylsulfinyl)butyl GLS (4MSOB) compared to in the wild type (Col-0)30. MAM3 lacked long-chain aliphatic GLSs (8MSOO or 7-(Methylsulfinyl)heptyl (7MSOH) GLS), as described previously31. The feeding assay performed with P. napi and the four A. thaliana mutant lines showed that P. napi larvae that fed on MAM1 grew more slowly than did those feeding on MAM3 (P = 0.018, FDR-adjusted pairwise t test) (Fig. 4b).
(a) GLS profiles of Arabidopsis thaliana mutants measured by LC-UV (n = 5). MAM1 showed lower 4MSOB concentration but accumulated higher 3MSOP and 8MSOO than Col-0. MAM3 has a lower long-chain aliphatic GLS concentration, and quad-GLS has no or quite low levels of GLSs. (b) Growth rates of P. napi that fed on the four Arabidopsis thaliana mutant lines. “*” shows significance based on the FDR-adjusted pairwise t test (P ≤ 0.05). Significant differences: MAM1 vs. MAM3 (P = 0.018). (c) Relative expression levels of NSP-like gene family members of P. napi against the four Arabidopsis thaliana mutants. Expression level was normalized based on Col-0 for each gene. Different letters on each box show significance (pairwise t test with FDR adjustment, P ≤ 0.05).
Regarding the expression patterns of NSP-like gene family members in P. napi, gene regulation differed significantly for NSP and MA but not SDMA in larvae as a response to the mutant lines they fed on (Fig. 4c). In the quad-GLS mutant, which does not contain GLS, NSP and MA were down-regulated in larvae; these were not down-regulated in larvae that fed on wild-type (Col-0) lines. In MAM1, which had higher levels of 3MSOP but lower levels of 4MSOB, we observed NSP in larvae to be significantly down-regulated compared to larvae that fed on Col-0 (wild type); in contrast, MA did not show this trend and had higher expression levels than in the larvae that fed on wild-type lines. When larvae fed on MAM3 lacking long-chain aliphatic GLS, NSP was expressed at levels similar to those found in larvae that fed on Col-0 but MA expression was highest and comparable to MAM1 mutant plant-feeding larvae.
Discussion
In this study, we conducted feeding experiments combined with gene expression analysis using four Pieris butterflies raised on two Brassicaceae plants and four A. thaliana mutant lines. We aimed to reveal if and how NSP-like gene family members of Pieris butterflies respond to a broad range of GLS profiles. We found that the gene expression levels of NSP and MA in Pieris larvae responded to the presence of GLSs and to GLS profile differences in the plants they fed on, but levels of SDMA remained the same. In addition, gene regulation differed between NSP and MA in response to GLS profiles in host plants; surprisingly, the two members of the NSP-like gene family actually showed inverse expression patterns to the plant species. The results suggest that both NSP and MA are involved in disarming plants’ defense systems by targeting different GLSs, and both of these proteins are the result of an adaptive mechanism Pieris employs to overcome a wide range of GLS profiles in their host plants.
RNA-seq based gene expression analysis showed that NSP was more highly expressed when larvae fed on C. occulta, which had more benzylic GLSs compared to A. kamchatica; in contrast, MA was highly expressed in larvae that fed on A. kamchatica, which had more aliphatic and indolic GLSs (Fig. 3a,c). Unlike NSP and MA, SDMA was expressed at a similar level, regardless of which plant the larvae fed on (Fig. 3c). These expression patterns of NSP-like gene family members were observed in all four Pieris species (Fig. 3c), and this pattern was also confirmed with more replicates by RT-qPCR analysis in P. melete (Fig. 3d). Both P. napi and P. melete are known to be able to use both of these Brassicaceae species as hosts in the field24,25; therefore, these expression patterns cannot be dismissed as a response to host or non-host plant differences. Furthermore, the larval growth rate of the four species showed no correlation with the expression patterns of NSP and MA, suggesting that the observed pattern was not the result of larval performance differences in the two plants (Fig. 3b). The involvement of NSP in disarming GLSs has been shown earlier; however, our results suggest that both NSP and MA are involved in functions related to disarming GLS, since these two genes responded to two host plants with dissimilar GLS profiles. Unlike Pierid-specific NSP or MA, SDMA, which can be found across Lepidoptera, did not seem to respond to GLS profile differences, suggesting SDMA is not involved in GLS-specific detoxification or in processes redirecting the breakdown products (Fig. 3c,d). Interestingly, not only did NSP and MA respond to different host plants, but they also showed inverse expression patterns in larvae feeding on the two plants (Fig. 3c,d). NSP seems to respond to benzylic GLSs, whereas MA appears to respond to aliphatic and indolic GLS. However, since we used different Brassicaceae species in this feeding experiment, it can be assumed that non-GLS based chemical differences between the two plant species exist. Thus, these differences might also affect the regulation of larval NSP and MA gene expression.
We used A. thaliana GLS mutants (wildtype Col-0; MAM1 and MAM3 with different aliphatic GLS compositions compared to Col-0, and quad-GLS with no detectable GLS) in a feeding experiment with P. napi larvae. We found that NSP and MA but not SDMA was down-regulated in larvae that fed on quad-GLS mutants; as these mutants lack GLSs, the expression of NSP and MA must be triggered by the presence of GLSs, supporting our hypothesis that in Pieris larvae, both genes have GLS-disarming roles. Our results are supported by a recent study that also reported a down-regulation of NSP in P. brassicae when larvae fed on an A. thaliana mutant lacking GLSs6. These results suggest that there could be costs associated with high level expression of NSP and MA proteins in the larval gut. Therefore, the observed context-specific down-regulation of either NSP or MA, respectively, could be observed in larvae that feed on plants with overall lower GLS amounts, such as certain Brassicaceae crop plants. Furthermore, mRNA levels of NSP and MA also responded differently to GLS profiles. We observed that NSP was down-regulated in larvae that fed on MAM1 plants compared to those that fed on Col-0 plants (Fig. 4c). The MAM1 mutant has a different aliphatic GLS profile (especially 4MSOB) than Col-0; therefore, the level of NSP might vary in response to a type of aliphatic GLS. MA was only down-regulated in larvae that fed on quad-GLS mutants with no GLSs (Fig. 4c). Since MA was not down-regulated in larvae fed on MAM1 and MAM3 plants (which have different aliphatic GLS profiles), MA might not respond to aliphatic GLSs. Therefore, the observed down-regulation of MA in larvae fed on quad-GLS plants might be caused by a lack of indolic GLS.
When comparing the NSP and MA gene expression profile between the two feeding experiments (with Brassicaceae plant species and with A. thaliana mutants), we found discordant expression patterns only in NSP. While the level of NSP did not respond to the presence of aliphatic GLS in the two plant species assays (Fig. 3a,c), it did respond to differences in the aliphatic GLS profiles in A. thaliana mutants (MAM1 and MAM3). Although the expression response of NSP to aliphatic GLSs seems contradictory, we lack the entire benzylic GLS, which is abundant in C. occulta, in our A. thaliana mutant assay, and so cannot state definitively what such a contradiction means. However, it may be that benzylic GLSs are a major target of NSP and so able to trigger its expression, resulting in the different patterns of NSP expression observed in the two feeding experiments. Therefore, further analyses are necessary, especially those that focus on larval responses to benzylic GLSs.
In both feeding assays, the expression profile of MA was similar. The expression of MA was elevated in larvae that fed on A. kamchatica, which had more indolic GLS than did C. occulta (Fig. 3c,d), as well as in larvae that fed on mutants which have indolic GLSs. If, as mentioned above, the observed down-regulation of MA in quad-GLS A. thaliana mutants was caused by a lack of indolic GLS, MA regulation could be a result of the same GLS trigger. Although the responses of NSP and MA should be tested in specific and controlled experiments, our results suggest that NSP and MA might share GLS-disarming functions that are related but not identical in the many species of Pieris butterflies.
The regulation of detoxification-related gene expression in herbivores fed on different host plants have been compared in many studies5,6,7,8. In most of these, the authors focused mainly on differential gene expression in generalist herbivores as a result of dietary metabolites or on host plant family differences to understand the molecular mechanisms which enable a wider host plant range than in specialist herbivores. However, larval responses to gene regulation in different types of host plants in specialist herbivores have not been well tested6. Here we show that even specialist herbivores may fine-tune adaptive gene expression in response to variations in a class of host plants’ major chemical defenses, the GLSs.
Our study demonstrates that NSP and MA, which are members of an important gene family involved in host plant adaptation in Pieridae, are induced by the presence of GLSs and differentially expressed in larvae that were fed on plants that have different GLS profiles. In addition, we also found that the regulation patterns of NSP and MA were mostly conserved in the four Pieris butterfly species used in this study. Although the function of MA is still unclear, our results strongly support the idea that not only NSP but also MA is involved in the molecular adaptation mechanisms relied on by Pieris butterflies to overcome the GLS defense system in their host plants. This hypothesis is also supported by the fact that A. cardamines, which seems to lack NSP and has only MA, can use Brassicaceae plants as hosts19,35. Furthermore, our results also indicate that NSP and MA have different functions. Although the functional difference between the two genes still needs to be confirmed biochemically, our results suggest that a dynamic gene family has enabled Pieris butterflies to overcome the diversity of GLSs and radiate widely, becoming one of the most successful herbivore groups that feed on Brassicales plants. Further understanding the relationships between gene evolution and the function of the NSP-like gene family and the host plant spectrum of the Pieridae can therefore help to shed light on the molecular mechanisms that mediate the coevolutionary arms race between plants and herbivores.
Materials and Methods
Feeding experiments using two wild Brassicaceae plant species with GLS analysis
We conducted feeding experiments using four closely related Pieris butterfly species (P. napi, P. melete, P. rapae, and P. brassicae) and two Brassicaceae plants from different genera (Arabidopsis kamchatica and Cardamine occulta). We collected female egg-laying butterflies from three out of the four Pieris species from the wild population in Hokkaido (P. napi, P. rapae) and Chiba (P. melete), Japan. For P. brassicae, we collected final instar larvae in Hokkaido (Japan), reared them to adults and mated them by hand-pairing to get fertilized females. We placed the fertilized butterflies in chambers with their host plants (Cardamine leucantha for P. napi and melete, Brassica oleracea var. capitata for P. rapae and brassicae) under high-intensity light conditions. Acquired eggs were incubated at 25 °C and neonates were used for feeding experiments immediately after hatching. We collected seeds from the two species of Brassicaceae plants from the wild population. These seeds were watered, and germinated seeds were transplanted to vermiculite soil. We watered plants once a week with optimally diluted Hyponex solution (N:P:K = 6:10:5; Hyponex, Osaka, Japan). We reared the plants for 2 months under these conditions: 25 °C, with 60% relative humidity and L16:D8. Next, for the feeding experiments, 3 neonates were applied to one plant using a soft-haired brush. We replicated this set twice for each Pieris species and harvested 6 individuals in total from each plant species after 120 hours of feeding. After the harvested larvae were individually weighed (within 0.1 mg), they were flash-frozen in liquid nitrogen immediately and stored at −80 °C until RNA extraction. We conducted FDR-adjusted pairwise t tests to identify statistically significant differences in larval growth among the treatments for each species.
We used these plants not only for the feeding experiments but also for GLS profile analyses. We harvested leaves from three undamaged individual plants and froze them with liquid nitrogen. Leaves were freeze-dried and ground with metal beads. An aliquot of each powdered sample was pooled for each species and analyzed three times by tandem quadrupole mass spectrometry (TQMS) coupled with ultra-performance liquid chromatography (UPLC)36,37. We extracted peaks that showed >30 signal/noise ratios as detected peaks and identified GLSs following Sawada et al. (2009) and Sawada et al. (2017)36,37. The relative concentrations of each GLS among samples were calculated by comparing the peak area with the internal standard (10-camphorsulfonic acid). Detected GLSs were sorted into major GLS chemical classes: short-chain aliphatic (-C5), long-chain aliphatic (C6–8), benzylic and indolic GLS.
RNA extraction, RNA-seq, de novo assembly, NSP-like gene family sequence identification and gene expression level analysis
For each butterfly species, we selected one representative larva from the two treatments (two plant species). We chose 8 larvae for RNA sequencing (larvae of 4 Pieris species fed on 2 different plant species each) in total. We extracted RNA with RNeasy Mini Kit (QIAGEN). Extracted RNA samples were quality checked with an Agilent 2100 Bioanalyzer, and all samples were confirmed to have high-quality total RNA. The library for RNA-seq was prepared by Sure Select Strand-Specific RNA Library Preparation Kit for Illumina Multiplexed Sequencing. We sequenced the samples individually on a HiSeq. 1500 (100 bp paired-end read technology). Acquired reads were trimmed by trimmomatic software with the following options (LEADING:10 TRAILING:10 SLIDINGWINDOW:4:20 MINLEN:40–normalize_reads)38. For de novo assembly, we pooled all of the trimmed reads from the same species. We conducted de novo assembly with Trinity ver. 2.0.6 for each species39. For identifying NSP-like gene family sequences, we used tblastn with setting the assembled contigs (backbone) as databases and NSP-like gene family protein sequences from P. rapae as queries (GenBank accession number AAR84202, ABY88945, ABY88946), and the e-value threshold was set as 1.0e-440. We extracted hit contigs for each query from each species, aligned and trimmed with MEGA6 to reference sequences of NSP-like gene family members from Pieris species stored in GenBank41. We made a ML molecular phylogeny of acquired sequences with reference sequences to confirm our annotation (in amino acid level). To measure the relative expression level of each extracted gene, we excluded redundant isoforms of NSP-like gene family members observed in assembled contig backbones and replaced them with trimmed representative sequences. Relative expression levels of each gene were estimated by mapping trimmed reads on an assembled backbone by RSEM42. Fragments per kilobase of exon per million reads mapped (FPKMs) were used as a relative expression level for each gene.
Gene expression analysis by qPCR
We also conducted RT-qPCR to confirm the gene expression levels of NSP-like gene family members. We chose P. melete as a representative species, and 3 larvae from each treatment were used for RT-qPCR. We designed primers for RT-qPCR analysis with the following Primer3Plus settings: product size = 70–180 bp, Tm = 59–61 °C, GC% = 40–60%, Max Poly-base = 3 for members of NSP-like gene family43. We also designed primers for EF1α, which is frequently used as a housekeeping reference gene in insects for qPCR6. Designed primers are listed in Table S2. We extracted RNA as described above, and after confirming the quality of RNA by Agilent 2100 Bioanalyzer, we digested gDNA from each extracted RNA sample using TURBO DNA-free Kit (QIAGEN). We synthesized cDNA with Prime Script RT reagent Kit with gDNA Eraser (Perfect Real Time) (TAKARA) after RNA purification by RNA Clean & Concentrator kit (ZYMO research). We ran RT-qPCR reactions with a CFX Connect Real-Time PCR Detection System (BIO-RAD) using SYBR Premix Ex Taq (Tli RNase H Plus) with two technical replicates for each sample. We verified specific amplification by performing a melting curve analysis from 65 °C to 95 °C. We calculated relative gene expression levels by the ΔCT method normalized by EF1α44. We conducted one tailed Mann–Whitney U test to see expression level differences between the treatments follow the trend we found in RNA-seq based expression analysis with software Rstudio ver. 1.0.13645. Raw qPCR data are available in Table S3.
Feeding experiments using Arabidopsis thaliana mutants with different GLS backgrounds
We prepared one wild-type (Col-0) and three mutant lines of Arabidopsis thaliana which have different GLS profiles (MAM1, MAM3, quad-GLS). We grew these four lines under short day conditions (25 °C, 8L16D, 60% humidity), and used them for feeding experiments 5 weeks after germination. In this experiment, we used P. napi as a representative species. We collected P. napi larvae in Fukushima, Japan, and reared them to adults. Adults were paired by hand, and acquired neonates were used for the feeding assay. We followed the same protocol as we used for the feeding experiments with the two wild Brassicaceae plants described above. We applied 5 larvae to each mutant individual and replicated this set 4 times (n = 20). We harvested larvae after 120 h feeding and weighed them. We conducted FDR-adjusted pairwise t tests to identify statistically significant differences in larval growth among the treatments. 5 larvae from each treatment were randomly chosen and dissected for further expression level analysis. Mid-gut samples were flash-frozen and stored at −80 °C until RNA extraction. RNA was extracted with innuPREP RNA Mini Kit (Analytik Jena, Germany). We conducted RT-qPCR as described above to measure the expression levels of the NSP-like gene family. We conducted FDR-adjusted pairwise t tests to identify statistically significant differences in gene expression levels among the treatments. Raw qPCR data are available in Table S4.
GLS analysis of A. thaliana mutant lines
We harvested entire rosettes of 5 individuals from each A. thaliana mutant line and froze them with liquid nitrogen. The samples were freeze-dried and ground by metal beads in a shaker. 10 mg of grounded leaf powder was used for chemical analysis. We added 80% of methanol with 50 µM of 4-hydroxybenzyl GLS (Sinalbin), which is absent in A. thaliana, to each mix as an internal standard. After 5 minutes of incubation with 230 rpm of shaking, we spun down the samples with 130,000 rpm for 10 minutes. We added the supernatant to filters conditioned with DEAE sephadex A-25. We washed the filter columns once with 500 µl of 80% MeOH and twice with 1 ml of water. After a final washing step with 1 ml of MES buffer pH5.2, we added 30 µl sulfate to convert GLS into desulfo GLS and incubated each sample overnight at room temperature. We eluted each column with 0.5 ml water and analyzed each using HPLC-UV with a reverse-phase C-18 column (Nucleodur Sphinx RP, 250 mm × 4.6 mm, 5 μm, Machrey-Nagel, Düren, Germany). Desulfo GLSs were identified based on the retention time and UV spectra with known standard libraries46.
Data Availability
The RNA-seq short read data have been deposited in the EBI short read archive (SRA) with the following sample accession numbers: ERX2829492-ERX2829499. The complete study can also be accessed directly using the following URL: http://www.ebi.ac.uk/ena/data/view/PRJEB29048.
References
Futuyma, D. J. & Agrawal, A. A. Macroevolution and the biological diversity of plants and herbivores. Proc. Natl. Acad. Sci. USA 106, 18054–18061 (2009).
Wen, Z., Rupasinghe, S., Niu, G., Berenbaum, M. R. & Schuler, M. A. CYP6B1 and CYP6B3 of the black swallowtail (Papilio polyxenes): Adaptive evolution through subfunctionalization. Mol. Biol. Evol. 23, 2434–2443 (2006).
Ahn, S. J., Vogel, H. & Heckel, D. G. Comparative analysis of the UDP-glycosyltransferase multigene family in insects. Insect Biochem. Mol. Biol. 42, 133–47 (2012).
Heidel-Fischer, H. M. & Vogel, H. Molecular mechanisms of insect adaptation to plant secondary compounds. Curr. Opin. Insect Sci. 8, 8–14 (2015).
Heidel-Fischer, H. M. et al. Phylogenetic relatedness and host plant growth form influence gene expression of the polyphagous comma butterfly (Polygonia c-album). BMC Genomics 10, 506 (2009).
Schweizer, F., Heidel-Fischer, H., Vogel, H. & Reymond, P. Arabidopsis glucosinolates trigger a contrasting transcriptomic response in a generalist and a specialist herbivore. Insect Biochem. Mol. Biol. 85, 21–31 (2017).
Celorio-Mancera, M. de la P. et al. Mechanisms of macroevolution: polyphagous plasticity in butterfly larvae revealed by RNA-seq. Mol. Ecol. 4884–4895, https://doi.org/10.1111/mec.12440 (2013).
Celorio-Mancera, M. de la P. et al. Evolutionary history of host use, rather than plant phylogeny, determines gene expression in a generalist butterfly. BMC Evol. Biol. 16, 59 (2016).
Fahey, J. W., Zalcmann, A. T. & Talalay, P. The chemical diversity and distribution of glucosinolates and isothiocyanates among plants. Phytochemistry 56, 5–51 (2001).
Agrawal, A. A. & Fishbein, M. Plant defense syndromes. Ecology 87, 132–149 (2006).
Wittstock, U. & Halkier, B. A. Glucosinolate research in the Arabidopsis era. Trends Plant Sci. 7, 263–270 (2002).
Halkier, B. A. & Gershenzon, J. Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 57, 303–333 (2006).
Olsen, C. E. et al. Glucosinolate diversity within a phylogenetic framework of the tribe Cardamineae (Brassicaceae) unraveled with HPLC-MS/MS and NMR-based analytical distinction of 70 desulfoglucosinolates. Phytochemistry 132, 33–56 (2016).
Hofberger, J. A., Lyons, E., Edger, P. P., Chris Pires, J. & Eric Schranz, M. Whole genome and tandem duplicate retention facilitated glucosinolate pathway diversification in the mustard family. Genome Biol. Evol. 5, 2155–2173 (2013).
Agrawal, A. A. & Kurashige, N. S. A role for isothiocyanates in plant resistance against the specialist herbivore Pieris rapae. J. Chem. Ecol. 29, 1403–1415 (2003).
Wittstock, U. & Burow, M. Glucosinolate breakdown in Arabidopsis: mechanism, regulation and biological significance. Arabidopsis Book 8, e0134 (2010).
Wittstock, U. et al. Successful herbivore attack due to metabolic diversion of a plant chemical defense. Proc. Natl. Acad. Sci. USA 101, 4859–4864 (2004).
Chew, F. S. Foodplant preferences of Pieris caterpillars (Lepidoptera). Oecologia 347–353 (1980).
Edger, P. P. et al. The butterfly plant arms-race escalated by gene and genome duplications. Proc. Natl. Acad. Sci. USA 112, 8362–8366 (2015).
Fischer, H. M., Wheat, C. W., Heckel, D. G. & Vogel, H. Evolutionary origins of a novel host plant detoxification gene in butterflies. Mol. Biol. Evol. 25, 809–820 (2008).
Randall, T. A., Perera, L., London, R. E. & Mueller, G. A. Genomic, RNAseq, and molecular modeling evidence suggests that the major allergen domain in insects evolved from a homodimeric origin. Genome Biol. Evol. 5, 2344–2358 (2013).
Montaut, S. & Bleeker, R. S. Review on Cardamine diphylla (Michx.) A. wood (Brassicaceae): Ethnobotany and glucosinolate chemistry. J. Ethnopharmacol. 149, 401–408 (2013).
Agerbirk, N., Olsen, C. E., Chew, F. S. & Ørgaard, M. Variable glucosinolate profiles of Cardamine pratensis (Brassicaceae) with equal chromosome numbers. J. Agric. Food Chem. 58, 4693–4700 (2010).
Ohsaki, N. & Sato, Y. Food plant choice of Pieris butterflies as a trade-off between parasitoid avoidance and quality of plants. Ecology 75, 59–68 (1994).
Kitahara, H. Oviposition plants and seasonal migratory movements of sympatric Pieris melete and P. napi japonica (Lepidoptera, Pieridae). Lepid. Sci. 67, 32–40 (2016).
Benson, J., Pasquale, A., Van Driesche, R. & Elkinton, J. Assessment of risk posed by introduced braconid wasps to Pieris virginiensis, a native woodland butterfly in New England. Biol. Control 26, 83–93 (2003).
Harvey, J. A., Poelman, E. H. & Gols, R. Development and host utilization in Hyposoter ebeninus (Hymenoptera: Ichneumonidae), a solitary endoparasitoid of Pieris rapae and P. brassicae caterpillars (Lepidoptera: Pieridae). Biol. Control 53, 312–318 (2010).
Ueno, M. A note on the Large White, Pieris brassicae.(I). Yadoriga 169, 25–41 (1997).
Heidel-Fischer, H. M., Vogel, H., Heckel, D. G. & Wheat, C. W. Microevolutionary dynamics of a macroevolutionary key innovation in a Lepidopteran herbivore. BMC Evol. Biol. 10, 60 (2010).
Kroymann, J. et al. A gene controlling variation in Arabidopsis glucosinolate composition is part of the methionine chain elongation pathway. 127, 1077–1088 (2001).
Textor, S., de Kraker, J.-W., Hause, B., Gershenzon, J. & Tokuhisa, J. G. MAM3 catalyzes the formation of all aliphatic glucosinolate chain lengths in. Arabidopsis. Plant Physiol. 144, 60–71 (2007).
Müller, R. et al. Differential effects of indole and aliphatic glucosinolates on lepidopteran herbivores. J. Chem. Ecol. 36, 905–913 (2010).
Smith, J. D., Woldemariam, M. G., Mescher, M. C., Jander, G. & De Moraes, C. M. Glucosinolates from host plants influence growth of the parasitic plant Cuscuta gronovii and its susceptibility to aphid feeding. Plant Physiol. 172, 181–197 (2016).
Jeschke, V. et al. How glucosinolates affect generalist lepidopteran larvae: growth, development and glucosinolate metabolism. Front. Plant Sci. 8, 1–12 (2017).
Wiklund, C. & Friberg, M. The evolutionary ecology of generalization: among-year variation in host plant use and offspring survival in a butterfly. Ecology 90, 3406–17 (2009).
Sawada, Y. et al. Widely targeted metabolomics based on large-scale MS/MS data for elucidating metabolite accumulation patterns in plants. Plant Cell Physiol. 50, 37–47 (2009).
Sawada, Y. et al. A novel method for single-grain-based metabolic profiling of Arabidopsis seed. Metabolomics 13, 75 (2017).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729 (2013).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011).
Untergasser, A. et al. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35, 71–74 (2007).
Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45 (2001).
RStudioTeam. RStudio: Integrated Development for R (2016).
Reichelt, M. et al. Benzoic acid glucosinolate esters and other glucosinolates from Arabidopsis thaliana. Phytochemistry 59, 663–671 (2002).
Acknowledgements
The metabolome analysis was performed with the technical assistance of M. Sato, Y. Yamada and A. Sakata, and the analysis was supported by the Japan Advanced Plant Science Network. A part of the RNA sequencing was supported by H. Takahashi and Future Medicine Education and Research Organization at Chiba University. The authors thank the greenhouse team of the Max Planck Institute for plant cultivation and also thank Emily Wheeler, Boston, for editorial assistance. This work was supported by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science to YO (No. 15J00320) and MM (No. 24310170), and partially by Max-Planck-Gesellschaft.
Author information
Authors and Affiliations
Contributions
Y.O., A.S., N.T., Y.S., M.Y.H. and M.R. carried out the laboratory work. Y.O., M.M., H.H.F. and H.V. conceived, designed and coordinated the study. Y.O., H.H.F. and H.V. wrote the manuscript. All authors, drafted parts of the manuscript, gave approval for publication and agree to be accountable for the content.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Okamura, Y., Sato, A., Tsuzuki, N. et al. Differential regulation of host plant adaptive genes in Pieris butterflies exposed to a range of glucosinolate profiles in their host plants. Sci Rep 9, 7256 (2019). https://doi.org/10.1038/s41598-019-43703-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-43703-8
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.