Abstract
Identification of functional pathways mediating molecular responses may lead to better understanding of disease processes and suggest new therapeutic approaches. We introduce a method to detect such mediating functions using topological properties of protein-protein interaction networks. We define the concept of pathway centrality, a measure of communication between disease genes and differentially expressed genes. Using pathway centrality, we identify mediating pathways in three pulmonary diseases (asthma; bronchopulmonary dysplasia (BPD); and chronic obstructive pulmonary disease (COPD)). We systematically evaluate the significance of all identified central pathways using genetic interactions. Mediating pathways shared by all three pulmonary disorders favor innate immune and inflammation-related processes, including toll-like receptor (TLR) signaling, PDGF- and angiotensin-regulated airway remodeling, the JAK-STAT signaling pathway, and interferon gamma. Disease-specific mediators, such as neurodevelopmental processes in BPD or adhesion molecules in COPD, are also highlighted. Some of our findings implicate pathways already in development as drug targets, while others may suggest new therapeutic approaches.
Similar content being viewed by others
Introduction
Identification of biological mechanisms underlying disease is crucial to the development of new therapeutic strategies. A common approach to understanding disease processes is through functional analysis of either genes that are differentially expressed in disease, or of putative disease-causing genes identified through GWAS or targeted studies. However, it has long been noted that causal disease genes are not necessarily differentially expressed, and that differential expression does not easily lead to the discovery of disease genes1. In many cases, differential expression predominantly reflects the tissue-specific downstream effects of a disease-causing process that integrates complex genetic and environmental responses. This makes differentially expressed genes useful as diagnostic markers, but often poor as therapeutic targets2. Conversely, functional analysis of causal disease genes often doesn’t fully explain how these downstream responses occur3. To bridge this gap, there is a need to identify the functional pathways mediating the transcriptional response in disease.
We do so by considering the roles that proteins corresponding to disease genes or differentially expressed genes play in protein-protein interaction networks. Such networks are often represented by graphs, where the nodes represent individual proteins and edges represent physical interactions between pairs of proteins. We postulate that the functional pathways mediating disease response will disproportionately reflect communication between these two sets of proteins, and that if we look for such “central” pathways we will find mediators. This topological property is well captured by the concept of betweenness centrality, where fractional betweenness characterizes how many of the shortest paths between all pairs of nodes passes through a given node4,5. We start by introducing a variant of node betweenness called disease fractional betweenness, which counts only the shortest paths between disease genes and differentially expressed genes for a given node. To identify putative mediating pathways in disease response, we then introduce a generalized notion of group centrality6 called pathway centrality, which aggregates disease fractional betweenness scores across all genes in a given pathway.
We apply our novel pathway-centrality method to three pulmonary diseases that primarily affect patients at different stages of life: bronchopulmonary dysplasia (BPD), a neonatal complication of preterm birth; asthma, which is relevant across the lifespan but is often diagnosed in childhood; and chronic obstructive pulmonary disease (COPD), which encompasses a number of progressive lung disorders that predominantly affect the elderly. For each disease, we examine the pathway centrality of all KEGG pathways and all Biological Process gene sets from the Gene Ontology. Approximating significance via permutation, we identify sets with significantly high pathway centrality as functional mediating pathways. We use a separate collection of genetic interaction data to demonstrate systematically that the identified pathways fill the desired topological roles in the network. Our approach finds candidate mediators not discovered through traditional hypergeometric enrichment of the disease gene or differentially expressed gene sets. We also discuss published evidence consistent with our disease-specific findings.
While group centrality has previously been applied via an optimization framework to discover new groups of important nodes in gene networks7, it has not yet been used to identify the functional gene sets playing a pivotal role in disease. This approach is the first to identify functional pathways mediating signals between disease genes and differentially expressed genes by measuring disease-specific communication passing through pathway genes in protein-protein interaction networks. We emphasize that this aim is different from that of finding disease genes themselves. The pathways we identify appear to mediate cellular response to disease states, and yet their component genes may be neither mutated nor significantly differentially expressed in disease.
The prior work most relevant to this effort is a collection of related results linking expression quantitative trait loci (eQTLs) to differentially expressed genes via protein-protein, protein-DNA, and phosphorylation networks. These studies were initially intended to find the causal gene in a disease-linked locus by tracing back the path of information flow from selected target genes that are differentially expressed8,9,10,11. Such efforts are related to ours in the sense that they examine information flow between genes linked to disease and differentially expressed genes. However, these analyses focus on only selected differentially expressed genes, and they do not directly point to mediating biological functions. Our focus is on the disease-related pathways, and our aim is to identify underlying biological functions that mediate cellular response in disease, rather than to identify genes with disease-causing mutations or variants.
We find that pathways involved in innate immunity, and several related signaling pathways including PDGF, JAK/STAT, and toll-like receptor signaling, are common mediators of all three pulmonary disorders. Disease-specific mediators include lipid homeostasis pathways in COPD, integrin mediated cell adhesion in asthma, and insulin-like growth factor receptor signaling in BPD. While a number of our findings have already been proposed as disease mediating pathways, diagnostic tools, or sources of therapeutic targets in previous publications, we discovered several novel mediators that may suggest new therapeutic approaches for these diseases.
Results
A pathway centrality approach to finding mediating pathways in disease
To discover functional processes mediating disease response, we start by identifying a set of disease genes whose mutations or variants have been shown to cause the indicated disease. Additionally, we identify differentially expressed genes from previously-published transcriptomic profiles of disease-relevant tissues. Our protein-protein interaction network is derived from the HIPPIE database12. Details describing network construction, sources of disease genes, and expression data appear in the Methods section.
We define pathway centrality to measure the amount of information a set of pathway genes handles by counting the shortest paths linking disease genes and differentially expressed genes. Specifically, let V be the set of all vertices in a protein-protein interaction network. While the classical definition of fractional betweenness (FB) for a node v ∈ V is the fraction of shortest paths between all pairs of nodes in the network passing through v, our pathway centrality score is based on a modified FB score which we call disease fractional betweenness (DFB). For node v and disease d, disease fractional betweenness only reflects the shortest paths between disease genes and differentially expressed genes that pass through v.
Formally, if D(d) is the set of genes in V associated with disease d, E(d) is the set of differentially expressed genes in V for disease d, Bs,t is the number of shortest paths between s and t, and Bs,t(v) is the number of shortest paths between s and t that pass through v, then disease fractional betweenness is defined as:
We then define pathway centrality (PC) as the average disease fractional betweenness score across all genes in a pathway. Specifically, for a pathway k containing the gene set P(k), pathway centrality PC(k) is defined as:
Once we have computed the pathway centrality score for a pathway k, we need a method to assess significance by characterizing how surprising it is to see a score at least as large as PC(k). The significance of the observed PC(k) score is assessed using a null distribution derived by selecting 10,000 random gene sets of size |P(k)|. The observed fraction of random sets with higher pathway centrality scores than PC(k) is reported as pcent(k), a rough measure of significance. (When the gene set or pathway is clear from context, we omit the argument and just write pcent).
However, developing a valid null distribution requires a non-trivial strategy for random gene set sampling, as pathway genes are known a priori to be relatively central in protein interaction networks. Thus, it is likely that pathway genes have higher fractional betweenness than those that are not involved in well-annotated functional processes. Another fundamental issue is that different centrality measurements are highly correlated13; in particular, fractional betweenness centrality is correlated with degree centrality. This can be problematic because the degree of a gene strongly correlates with how “popular” or well-annotated the gene is. Thus, our pathway centrality analysis could favor pathways containing well-studied genes unless we force the random sampling process to mimic the original degree distribution.
To overcome such biases, we therefore impose restrictions on our random gene set sampling process. First, we require our random samples to be drawn from the collection of genes belonging to at least one pathway. We also want the degree distribution of each random gene set to resemble that of the candidate mediating pathway. However, because there are only a few high-degree genes, we cannot necessarily exactly match the degree of high degree genes in the candidate pathway while choosing from a sufficiently large population of alternatives. Thus, we “bin” the nodes by degree, such that similarly high-degree nodes are placed into a single bin for sampling. This process, described further in the Methods section, ensures that there are a sufficient number of choices for approximately matching the degree of high-degree nodes in mediating pathways.
Finding disease-specific and shared mediators of pulmonary disease
Pathways with significant pathway centrality in individual pulmonary data sets include known disease mediators, along with other pathways whose disease-relevance has not yet been identified. Tables 1 and 2 show a selection of the top Gene Ontology Biological Processes (GO BP) terms and KEGG pathways, respectively, identified in exactly one of the pulmonary disorders; the featured terms were manually selected to represent the functional range of the significant results. There are fewer significant KEGG pathways, but they tend to implicate similar functions to those found using GO BP gene sets. Full results for both pathway collections are available in Supplementary Table S1.
The functional annotations in Table 1 predictably identify immune processes as mediators of each of the three pulmonary disorders, but they highlight different aspects of immunity and signaling that appear to distinguish the individual diseases. Significant immune mediators in asthma include cells of both myeloid and lymphoid origin, emphasizing the role of the innate immune response (e.g., mast cell activation; macrophage activation) in the disease. In contrast, those unique to COPD highlight adaptive immunity. Because both innate and adaptive immune pathways are known to play a role in both diseases14, these results suggest that the innate immune pathways mediating the COPD response (such as Th1) are also implicated in other airway disorders.
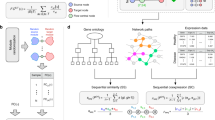
When we look at pathways that play a significant role across all three pulmonary disorders (Table 3), we again find a preponderance of inflammatory and immune processes. Significant immune pathways across all three data sets largely focus on innate immunity, though there are some predominantly adaptive processes (e.g. T cell signaling) and others (IL-1, cytokine signaling) that have roles in both. Several specific signaling pathways and systems are implicated, including JAK/STAT signaling, toll-like receptor signaling, PDGF, interferon gamma, and the renin-angiotensin and complement systems. (Fig. 1(a) shows the JAK/STAT pathway topology in BPD as an example).
(a) Mediating topology of the JAK-STAT signaling pathway in BPD. One of the significant mediating pathways for bronchopulmonary dysplasia (BPD) is the KEGG JAK-STAT signaling pathway. Here, mediating pathway genes are colored dark green and appear between BPD disease genes (red) and differentially expressed genes (blue). Edges directly linking the disease genes to the pathway or the pathway to the differentially expressed genes in BPD are shown. The pathway is visualized using cytoscape49. (b) Idealized topological property of disease-mediating pathways. Let D(d) be a set of disease genes and E(d) be a set of differentially expressed genes for disease d. Genes in P(k)*, a significant mediating pathway of disease d with pcent(k) < 0.05, are expected to play central roles in passing signals from D(d) to E(d).
To distinguish our significant mediating pathways from enriched functions within the disease-related genes, we next assessed whether the identified pathways were significantly enriched (hypergeometric, Benjamini-Hochberg adjusted FDR < 0.05) in either the disease gene set or the differentially-expressed gene set for the indicated disease. Many significant mediating pathways are not detectable by functional enrichment analysis. Pathways in Tables 1 and 2 marked with an asterisk in the right-most column meet a more conservative criterion: no significant enrichment of either the indicated pathways, nor any with a substantially similar function, was detected. For example, in BPD, the GO gene set “Epidermal Growth Factor Receptor Signaling Pathway” is identified as a significant mediator. Yet the disease and differentially expressed genes in BPD did not show significant enrichment of this or any gene sets related to EGFR. The ability of pathway centrality to identify such mediating gene sets suggests that the use of network structure in computing pathway centrality implicates pathways that would not be found by traditional enrichment approaches. Supplementary Table S2 lists the hypergeometric enrichment scores for disease and differentially expressed gene sets for all significant pathways.
Genetic interaction data confirms the mediating topology of identified pathways
One way to verify that the proposed mediating pathways truly include genes mediating responses to disease genes would be to identify an excess of epistatic relationships between them. For example, if a mediating pathway looks like that shown in Fig. 1(b), one might expect a higher likelihood of seeing certain kinds of genetic interactions between a disease gene gD in set D and a mediating gene gP from set P(k)* than between gD and genes that are not in a mediating pathway for that disease. The genetic interactions of most interest are “alleviating” or positive interactions, where the deleterious effect of the double mutant of both gD and gP is less severe than would be predicted by combining the independent effects of individual mutations in gD or gP. Such relationships might arise when gP is part of a pathway mediating the response to gD.
However, because it is difficult to find sufficient numbers of verified human genetic interactions, we additionally collected alleviating (positive) genetic and phenotypic suppression genetic interactions from the model organisms Schizosaccharomyces pombe, Saccharomyces cerevisiae, Drosophila melanogaster, and Caenorhabditis elegans. For each gene set P(k), we then define pmed(k), the probability of finding the observed number of positive genetic interactions between disease genes and genes in P(k) through a similar binning approach to that used for pcent(k).
To assess how surprising it is to see the observed number of such positive genetic interactions between the disease genes and the pathway, we compute a null distribution of the number of alleviating or suppressing interactions between the same set of disease genes and 10,000 random gene sets of the same size as the candidate mediating pathway. Again, we impose restrictions on the source of our random gene sets: they must be drawn from a pool of genes that belong to at least one pathway in the collection and that are downstream genes of any alleviating genetic or phenotypic suppression interactions (Fig. 2(a)). We used a binning strategy analogous to that for pcent to approximately match the in-degree distributions of known downstream genes of disease genes and our random samples.
(a) Systematic confirmation of significant mediating pathways. If identified pathways are truly mediating a disease response, the pathway genes are likely to be downstream of the corresponding disease genes. This topology can be captured by an excess of epistatic relationships between disease genes and a mediating pathway genes. To test whether our significant disease-mediating pathway genes form such topologies with the disease genes more than others, we first collected alleviating genetic interactions from human and a few model organisms. A is the set of all disease genes and B is the set of “downstream” genes that have alleviating genetic interactions with genes in A. We count x, the number of alleviating genetic interactions between disease genes and genes belonging to the specific identified mediating pathway (here x is the number of red edges), and then assess significance by calculating the probability that a random gene set of equal size has at least x alleviating genetic interactions with the disease genes. The null distribution is learned from 10,000 random samples drawn from a pool of genes of any known alleviating genetic interactions (set B). (b) Histogram showing the relationship between pcent and pmed, for GO BP gene sets and BPD. Each bar represents the fraction of pathways with pcent <0.05 in the designated range (from the labeled value minus 0.05, to the labeled value) that also have pmed <0.05. The plot shows that many of pathways with significant pcent scores (below 0.05, shown by the dark bar) also have low pmed scores.
We would then like to assess whether a significant disease-mediating pathway k having a low pcent value is more likely to have a low pmed value. If it does, that indicates an excess of positive genetic interactions between disease genes and genes in P(k), suggesting that the disease genes are potentially located “upstream” of the mediating pathway.
To capture this relationship, we compute a histogram of the percentage of pmed scores below 0.05 for each of 20 quantiles of possible pcent values. A sample plot of these frequencies is shown in Fig. 2(b); plots for all data sets and gene set collections are available as Supplementary Figure S1. One-sided, non-parametric Wilcoxon tests comparing the distribution of pmed values in the first quantile (i.e., pcent below 0.05) to the distribution in all the other quantiles (pcent ≥ 0.05) confirm that the most significant mediating pathways are associated with lower pmed scores in all the pulmonary disease cases. These (raw) significance values are shown in Table 4.
As an example, the KEGG MAPK signaling pathway has a pcent score of 0.0038 in BPD. There are ten supporting alleviating genetic interactions between BPD disease genes and genes in the KEGG MAPK signaling pathway: PLAU-HRAS, PLAU-MAP2K1, TIRAP-MP3K7, TIRAP-TRAF6, TLR4-ECSIT, TNF-CHUK, TNF-RAC, FGFR2-RAC1, FGFR2-KRAS, and FGFR2-CDC42; leading to a corresponding pmed score below 0.0001. This pathway is one of the more than a third of pathways with pcent below 0.05 in BPD whose pmed score is also below 0.05, contributing to the dark bar on the left-hand side of Fig. 2(b).
Both the Wilcoxon tests and the plots support the conclusion that there is enrichment of alleviating genetic relationships between disease genes and pathway genes for the pathways whose pcent values are deemed significant, confirming that pathway centrality is indeed generally finding gene sets with the desired network topology.
Discussion
We have introduced a new centrality-based method to identify functional pathways mediating disease responses by dominating communication between disease and differentially expressed genes. Although there are many issues with the available genetic interaction data, the conservation of genetic interactions across species being one of the most salient, systematic evaluation using genetic interactions confirms that our method finds pathways with the desired network topology. Recent work discussing the plethora of GWAS hits with modest effects suggests that functional network analysis is essential for translating these hits into actionable knowledge15. Our work illustrates one such approach.
The journey from identifying disease-relevant pathways to the discovery of novel and effective therapeutics may involve multiple steps, but it has been widely considered to be a promising approach to drug-discovery in the genomic era. One question is whether the pathway is simply a list of potential targets. In many cases the story will be more complex than that; in particular, it is not always plausible that there is a single targetable molecule that will disrupt the relevant processes16. Still, recent work in airway diseases emphasizes that understanding the relevant pathways is essential to discovering more effective treatments modifying their functions17.
Several of the pathways identified by pathway centrality are already under consideration as therapeutic targets for the indicated disease. For example, the KEGG gene set “Cell Adhesion Molecules (CAMS)” tops the unique KEGG list in COPD (Supplementary Table S1). Prior work suggests that adhesion molecules also play a significant role in the pathogenesis of COPD18, and that clinical trials of therapeutics regulating adhesion and integrin are underway for both COPD and asthma19. “Positive regulation of cell adhesion mediated by integrin,” a non-overlapping GO gene set we uniquely identified as a mediator in asthma, has more recently been considered as a targetable process to reduce airway hyper-responsiveness20. Therefore, it is important that although adhesion and leukocyte chemotaxis are important to all three disorders21,22,23, the pathway centrality approach highlights different sets of genes mediating these responses.
Similarly, the JAK/STAT pathway, implicated as a mediator in all three disorders, has been suggested as an asthma target through inhibitors of activating cytokines and receptors24. JAK pathway inhibitors are in development for a number of inflammatory disorders25. Work in animal models has suggested that targeting this pathway can reduce airway hyperresponsiveness but has widespread effects, leading to efforts to develop inhaled therapeutics targeting the JAK pathway for both COPD and asthma26. The role of JAK/STAT signaling in bronchopulmonary dysplasia is less clear, but it has been suggested that it plays a role in airway smooth muscle mitogenesis, implicated in both asthma and BPD27, and postulated that it may be an alternative mediator of the oxidative stress response in both diseases28. (Fig. 1(a) shows a subset of this pathway and the PPI network for BPD.) Thus, our work suggests that if safe and effective compounds targeting this pathway are developed for asthma or COPD, there may be some potential for their relevance in BPD as well.
Toll-like receptor (TLR) signaling, which activates the innate immune response and was implicated in all three diseases, is another familiar part of the story of airway hyperreactivity and fetal lung development29. TLR polymorphisms have been linked to an increased risk of developing BPD30, and TLR agonists are already being tested for therapeutic efficacy in asthma31. However, the role of this system in COPD is not as well studied. Aspects of the innate immune response are often demonstrably suppressed in COPD patients32, consistent with our results in the previous section showing that most COPD-specific immune response pathways regulate the adaptive immune response. TLR polymorphisms also play a role in disease susceptibility and severity33,34. Our work therefore provides evidence for a role for TLR pathways in the diagnosis, stratification, and treatment of COPD.
Unique to BPD is a collection of mediating neurodevelopmental pathways. BPD has long been known to be associated with worse neurodevelopmental outcomes than those observed in infants without BPD delivered at similar gestational ages35. Recently, BPD has been shown to be associated with a measurable decrease in IQ36. Whether that decrease is a consequence of BPD or arises from a common cause is unknown. Our results, showing neurodevelopmental pathways mediating the expression response in the blood of 5 day old infants who go on to develop BPD, suggest that some of the association is likely due to molecular causes, rather than, say, the consequences of neonatal hypoxia.
The identification of common mediating pathways in airway disease throughout the lifespan may shed light on potential implications of neonatal or childhood respiratory disorders. While the discovery of common immune pathways here is not surprising, the distinction between which pathways appear to be disease specific and which are common may be informative. Meanwhile, the shared involvement of vasoconstriction and adhesion pathways suggests a different commonality that might be exploited to mitigate later pulmonary issues in children with BPD or asthma.
One potential issue affecting our work is that disease genes and differentially expressed genes are not necessarily distinct gene sets. We therefore treat genes in both sets as differentially expressed genes only. However, we also note that the overlap between these groups is sufficiently small (asthma: 1, BPD: 1, COPD: 9) that it is unlikely to substantially affect our results. Similarly, one could imagine that separating up- and down-regulated differentially-expressed genes might provide further power for this type of analysis. Exploring this hypothesis could be a fruitful avenue for future work, although the lack of directionality in many of the considered gene sets makes implementation of such an approach non-trivial.
Our method also suggests a new way to analyze protein-protein interaction networks in the context of a disease of interest. Disease-specific network analysis usually excludes genes not known to be associated with the disease. Our approach enables disease-specific analysis without altering the topology of protein-protein interaction networks, and accounts for roles of neighboring genes in disease-related communication. It also can be applied to any type of molecular network. We see an opportunity to improve the chance for finding novel disease-mediating pathways by combining networks of protein-protein interactions and other types of molecular data, such as transcription factor-target interactions.
Overall, we have demonstrated that our pathway-centrality method finds functional mediators of disease using complementary interaction data. We have seen that the identified pathways include some therapeutic targets already in development, suggesting that others may be similarly promising but novel. Our findings confirm established connections of pulmonary disorders with inflammatory and immune processes, signaling processes, and airway remodeling. We expect that this approach may be applied more generally to discover relevant and informative pathways for any disease or phenotype of interest.
Methods
Protein-protein and genetic interaction networks
We use two biological networks in our experiments. To measure pathway centrality, physical protein-protein interactions were collected from the Human Integrated Protein-Protein Interaction rEference (HIPPIE)12 database. HIPPIE contains experimentally verified protein interactions with confidence scores. We downloaded the interaction data (version 2.1) on September 8, 2017 and selected only those interactions described as “high confidence” (≥0.73), as these interactions are supported by more reliable evidence. We worked with the largest connected component extracted from the network, which contains 62,679 interactions between 12,064 proteins. Note that we use the protein to gene mapping provided by the HIPPIE database to map protein-protein interactions. Thus, although our canonical entities in the network are represented by Entrez gene identifiers, we refer to these interactions as protein-protein interactions throughout the manuscript.
To compute pmed scores based on genetic interaction data, we looked for genetic interaction data featuring alleviating (positive) genetic and phenotypic suppression interactions. Because relatively few of these genetic interactions are known for humans, we additionally collected such interactions from Schizosaccharomyces pombe, Saccharomyces cerevisiae, Drosophila melanogaster, and Caenorhabditis elegans. These interactions came from BioGRID37 (version 3.4.160), the Saccharomyces Genome Database (SGD project, http://www.yeastgenome.org), and Flybase38, all downloaded on May 23, 2018. To find human homologous interaction pairs, we use a mapping downloaded from the HomoloGene database39 on July 19, 2016 (the current version was uploaded on April 14, 2014). This approach yielded 9,395 pairs of putative positive human genetic interactions.
Disease-related genes and functional gene sets
For Asthma and COPD, 111 and 192 disease genes were collected from recent reviews of asthma40 and COPD genes41. Lacking a similar-scale summary of BPD genetics, we collected 81 genes associated with BPD from Online Mendelian Inheritance in Man (OMIM)42 and Genopedia43, as described in44. These datasets were downloaded on April 4, 2018.
Gene expression microarray profiles for Asthma and BPD were obtained from the GEO database (accession numbers GSE4302 and GSE32472, respectively). The first measured differential expression in airway epithelial cells between healthy controls and asthma patients45, while the second examined expression in peripheral blood cells from infants born preterm with or without BPD46. From the preterm study, we used only samples taken on postnatal day 5 (the earliest time point). We selected as differentially expressed genes between disease and control groups those with an adjusted Benjamini-Hochberg t-test p-value below 0.01, yielding 82 and 422 expression-related genes in asthma and BPD, respectively. For COPD, we downloaded RNA-seq EdgeR results comparing expression in lung cells from COPD patients and controls (GSE57148)47. Relying on the analysis methods from the original study, since RNA-seq and Affymetrix analysis pipelines differ, we identified 266 significantly differentially expressed genes with an EdgeR q-value below 10−10.
Note that there may be disease genes that are also differentially expressed in that disease. To avoid confusion about how to use these in computing pathway centrality, we removed genes from the disease gene sets that also appeared in the corresponding set of differentially expressed genes. Disease genes and differentially expressed genes are further excluded from our experiments if they do not have any known interactions with other genes in our protein-protein interaction network data. Removing those genes without known interactions results in 86 (asthma), 70 (BPD) and 146 (COPD) disease genes, and 42 (asthma), 216 (BPD) and 198 (COPD) differentially expressed genes.
Supplementary Figure S2 shows the overlaps between these three disease gene sets and between the three differentially-expressed gene sets. There is very little overlap seen in the differentially-expressed gene sets, and at least half the disease genes for each disease are unique. Thus, common mediating pathways across all three networks are unlikely to have arisen from shared shortest paths between identical sets of genes.
Both the Gene Ontology and the KEGG gene set collections were downloaded from the Molecular Signature DataBase (MSigDB)48 on April 3, 2018 (http://software.broadinstitute.org/gsea/msigdb). This GO gene set collection includes 4,436 Biological Process (BP) terms and 15,578 genes, and the KEGG collection includes 186 pathways and 5,266 genes.
Assessing significance by matching degree distribution of random samples through binning
To compute pcent, we sort nodes by degree and place nodes of increasing degree into one bin, with all nodes of the same degree placed in the same bin, until the size of the bin is above a threshold. The algorithm also checks the last bin, and merges it with the previous bin if the size of the last bin is less than half of the size threshold. We used a bin size of b = 20 for the results presented here, as this appeared to give reasonable sized bins and to avoid combining nodes with too large a degree range. Supplementary Figure S3 shows the bin sizes and degree ranges for b = 20.
To determine the algorithm’s sensitivity to this bin size, we compared pcent values calculated using bin size b = 20 to pcent values calculated using size thresholds of 10 and 40 (i.e., b/2 and 2b). Table 5 shows that pcent values calculated using different bin size thresholds are highly correlated. Our conclusion is that the pcent values are fairly robust to two-fold variation in the bin size, suggesting that our somewhat arbitrary choice of size 20 has at most a modest impact on the results.
The process to compute pmed is identical to that for computing pcent, except for the bin sizes. Here, we chose 5 for the bin size threshold, as it generated a reasonable range of bin sizes. As above, the resulting pmed values were analogously determined to be robust to the choice of this size threshold; Pearson correlations between pmed values calculated using bin sizes 3, 5, and 10 range from 0.9960 to 0.9998, all with Python Scipy p-values for the correlation <0.0001.
Data Availability
All programs we designed and implemented for this study are available for download at https://github.com/TuftsBCB/pathway-centrality. Data generated during our experiments are in supplementary information files, and also accessible at http://bcb.cs.tufts.edu/jpark/pathway-centrality/. This repository includes an excel file containing our full experimental results, two pdf files containing two Venn diagrams and twelve R plots, and three cytoscape session files.
References
Hudson, N. J., Dalrymple, B. P. & Reverter, A. Beyond differential expression: the quest for causal mutations and effector molecules. BMC Genomics 13, 356, https://www.ncbi.nlm.nih.gov/pubmed/22849396, https://doi.org/10.1186/1471-2164-13-356 (2012).
Fox, A. D., Hescott, B. J., Blumer, A. C. & Slonim, D. K. Connectedness of ppi network neighborhoods identifies regulatory hub proteins. Bioinformatics 27, 1135–42, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve db=PubMed dopt=Citation list_uids=21367871, https://doi.org/10.1093/bioinformat-ics/btr099 (2011).
Delude, C. M. Deep phenotyping: The details of disease. Nature 527, S14–5, https://www.ncbi.nlm.nih.gov/pubmed/26536218, https://doi.org/10.1038/527S14a (2015).
Yu, H., Kim, P. M., Sprecher, E., Trifonov, V. & Gerstein, M. The importance of bottlenecks in protein networks: correlation with gene essentiality and expression dynamics. PLoS Comput Biol 3, e59, https://www.ncbi.nlm.nih.gov/pubmed/17447836, https://doi.org/10.1371/journal.pcbi.0030059 (2007).
Freeman, L. C. A set of measures of centrality based on betweenness. Sociometry 40, 35–41, https://www.jstor.org/stable/3033543 (1977).
Everett, M. G. & Borgatti, S. P. The centrality of groups and classes. Journal of Mathematical Sociology 23, 181–201 (1999).
Erdos, D. Centrality measures and analyzing dot-product graphs (Doctoral dissertation). Ph.D. thesis, Boston University, https://open.bu.edu/handle/2144/17127 (2015).
Tu, Z., Wang, L., Arbeitman, M. N., Chen, T. & Sun, F. An integrative approach for causal gene identification and gene regulatory pathway inference. Bioinformatics 22, e489–e496, https://doi.org/10.1093/bioinformatics/btl234 (2006).
Suthram, S., Beyer, A., Karp, R. M., Eldar, Y. & Ideker, T. eqed: an efficient method for interpreting eqtl associations using protein networks. Molecular Systems Biology 4, https://doi.org/10.1038/msb.2008.4 (2008).
Kim, Y.-A., Wuchty, S. & Przytycka, T. M. Identifying causal genes and dysregulated pathways in complex diseases. PLOS Computational Biology 7, e1001095, https://doi.org/10.1371/journal.pcbi.1001095 (2011).
Yeger-Lotem, E. et al. Bridging high-throughput genetic and transcriptional data reveals cellular responses to alpha-synuclein toxicity. Nature genetics 41, 316–323 (2009).
Alanis-Lobato, G., Andrade-Navarro, M. A. & Schaefer, M. H. Hippie v2.0: enhancing meaningfulness and reliability of protein-protein interaction networks. Nucleic Acids Res 45, D408–D414, https://www.ncbi.nlm.nih.gov/pubmed/27794551, https://doi.org/10.1093/nar/gkw985 (2017).
Goh, K. I., Oh, E., Kahng, B. & Kim, D. Betweenness centrality correlation in social networks. Phys Rev E Stat Nonlin Soft Matter Phys 67, 017101, https://www.ncbi.nlm.nih.gov/pubmed/12636633, https://doi.org/10.1103/PhysRevE.67.017101 (2003).
Sabroe, I. et al. Targeting the networks that underpin contiguous immunity in asthma and chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 175, 306–311 (2007).
Boyle, E. A., Li, Y. I. & Pritchard, J. K. An expanded view of complex traits: From polygenic to omnigenic. Cell 169, 1177–1186 (2017).
Parulekar, A. D., Kao, C. C., Diamant, Z. & Hanania, N. A. Targeting the interleukin-4 and interleukin-13 pathways in severe asthma: current knowledge and future needs. Curr Opin Pulm Med 24, 50–55 (2018).
Brusselle, G. & Bracke, K. Targeting immune pathways for therapy in asthma and chronic obstructive pulmonary disease. Ann Am Thorac Soc 11(Suppl 5), S322–328 (2014).
Ishii, Y. Role of adhesion molecules in the pathogenesis of copd. Nihon Rinsho 57, 1965–71, https://www.ncbi.nlm.nih.gov/pubmed/10497391 (1999).
Woodside, D. G. & Vanderslice, P. Cell adhesion antagonists: therapeutic potential in asthma and chronic obstructive pulmonary disease. BioDrugs 22, 85–100, https://www.ncbi.nlm.nih.gov/pubmed/18345706 (2008).
Sundaram, A. et al. Targeting integrin a5b1 ameliorates severe airway hyperresponsiveness in experimental asthma. J. Clin. Invest. 127, 365–374 (2017).
Faura Tellez, G. et al. Protocadherin-1 localization and cell-adhesion function in airway epithelial cells in asthma. PLoS One 11, e0163967, https://www.ncbi.nlm.nih.gov/pubmed/27701444, https://doi.org/10.1371/journal.pone.0163967 (2016).
Ramsay, P. L., O’Brian Smith, E., Hegemier, S. & Welty, S. E. Early clinical markers for the development of bronchopulmonary dysplasia: soluble e-selectin and icam-1. Pediatrics 102, 927–32, https://www.ncbi.nlm.nih.gov/pubmed/9755267 (1998).
Barnes, P. J., Drazen, J. M., Rennard, S. I. & Thomson, N. C. Asthma and copd basic mechanisms and clinical management second edition preface to the 2nd edition. Asthma and Copd: Basic Mechanisms and Clinical Management, 2nd Edition pp. 178ff, <Go to ISI>://WOS:000311345000001, https://doi.org/10.1016/B978-0-12-374001-4.00079-1 (2009).
Vale, K. Targeting the jak-stat pathway in the treatment of ‘th2-high’ severe asthma. Future Med Chem 8, 405–19, https://www.ncbi.nlm.nih.gov/pubmed/26934038, https://doi.org/10.4155/fmc.16.4 (2016).
O’Shea, J. J. et al. The jak-stat pathway: impact on human disease and therapeutic intervention. Annu Rev Med 66, 311–28, https://www.ncbi.nlm.nih.gov/pubmed/25587654, https://doi.org/10.1146/annurev-med-051113-024537 (2015).
Barnes, P. J. Kinases as novel therapeutic targets in asthma and chronic obstructive pulmonary disease. Pharmacol Rev 68, 788–815, https://www.ncbi.nlm.nih.gov/pubmed/27363440, https://doi.org/10.1124/pr.116.012518 (2016).
Simon, A. R., Takahashi, S., Severgnini, M., Fanburg, B. L. & Cochran, B. H. Role of the jak-stat pathway in pdgf-stimulated proliferation of human airway smooth muscle cells. Am J Physiol Lung Cell Mol Physiol 282, L1296–304, https://www.ncbi.nlm.nih.gov/pubmed/12003786, https://doi.org/10.1152/ajplung.00315.2001 (2002).
Zhou, L. & Hershenson, M. B. Mitogenic signaling pathways in airway smooth muscle. Respir Physiol Neurobiol 137, 295–308, https://www.ncbi.nlm.nih.gov/pubmed/14516733 (2003).
Petrikin, J. E., Gaedigk, R., Leeder, J. S. & Truog, W. E. Selective toll–like receptor expression in human fetal lung. Pediatr Res 68, 335–8, https://www.ncbi.nlm.nih.gov/pubmed/20581745, https://doi.org/10.1203/00006450-201011001-00656, https://doi.org/10.1203/PDR.0b013e3181ed1134 (2010).
Malash, A. H., Ali, A. A., Samy, R. M. & Shamma, R. A. Association of tlr polymorphisms with bronchopulmonary dysplasia. Gene 592, 23–8, https://www.ncbi.nlm.nih.gov/pubmed/27457283, https://doi.org/10.1016/j.gene.2016.07.049 (2016).
Biggadike, K. et al. Discovery of 6-amino-2-[(1S)-1-methylbutyl]oxy-9-[5-(1-piperidinyl)pentyl]-7,9-dihydro-8h-pu rin-8-one (gsk2245035), a highly potent and selective intranasal toll-like receptor 7 agonist for the treatment of asthma. J Med Chem 59, 1711–26, https://www.ncbi.nlm.nih.gov/pubmed/26861551, https://doi.org/10.1021/acs.jmedchem.5b01647 (2016).
Shaykhiev, R. & Crystal, R. G. Innate immunity and chronic obstructive pulmonary disease: a mini-review. Gerontology 59, 481–9, https://www.ncbi.nlm.nih.gov/pubmed/24008598, https://doi.org/10.1159/000354173 (2013).
Apostolou, A. et al. Association between tlr2/tlr4 gene polymorphisms and copd phenotype in a greek cohort. Herz, https://www.ncbi.nlm.nih.gov/pubmed/27909766, https://doi.org/10.1007/s00059-016-4510-9 (2016).
Yu, H., Lin, M., Wang, X., Wang, S. & Wang, Z. Toll-like receptor 4 polymorphism is associated with increased susceptibility to chronic obstructive pulmonary disease in han chinese patients with chronic periodontitis. J Oral Sci 58, 555–560, https://www.ncbi.nlm.nih.gov/pubmed/28025440, https://doi.org/10.2334/josnusd.16-0187 (2016).
Anderson, P. J. & Doyle, L. W. Neurodevelopmental outcome of bronchopulmonary dysplasia. Semin. Perinatol. 30, 227–232 (2006).
Twilhaar, E. S. et al. Cognitive Outcomes of Children Born Extremely or Very Preterm Since the 1990s and Associated Risk Factors: A Meta-analysis and Meta-regression. JAMA Pediatr 172, 361–367 (2018).
Stark, C. et al. Biogrid: a general repository for interaction datasets. Nucleic Acids Res 34, D535–9, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve db=PubMed dopt=Citation list_uids=16381927 (2006).
Attrill, H. et al. Flybase: establishing a gene group resource for drosophila melanogaster. Nucleic Acids Res 44, D786–92, https://www.ncbi.nlm.nih.gov/pubmed/26467478, https://doi.org/10.1093/nar/gkv1046 (2016).
Coordinators, N. R. Database resources of the national center for biotechnology information. Nucleic Acids Res 44, D7–19, https://www.ncbi.nlm.nih.gov/pubmed/26615191, https://doi.org/10.1093/nar/gkv1290 (2016).
Bijanzadeh, M., Mahesh, P. A. & Ramachandra, N. B. An understanding of the genetic basis of asthma. Indian Journal of Medical Research 134, 149–161 (2011).
Bossé, Y. Updates on the copd gene list. International Journal of COPD 7, 607–631 (2012).
Amberger, J., Bocchini, C. A., Scott, A. F. & Hamosh, A. Mckusick’s online mendelian inheritance in man (omim). Nucleic Acids Res 37, D793–6, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citationlist_uids=18842627, DOI gkn665 [pii], https://doi.org/10.1093/nar/gkn665 (2009).
Yu, W., Clyne, M., Khoury, M. J. & Gwinn, M. Phenopedia and genopedia: disease-centered and gene-centered views of the evolving knowledge of human genetic associations. Bioinformatics 26, 145–6, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citationlist_uids=19864262, DOI btp618 [pii], https://doi.org/10.1093/bioinformatics/btp618 (2010).
Park, J. et al. Finding novel molecular connections between developmental processes and disease. PLOS Computational Biology 10, e1003578, https://doi.org/10.1371/journal.pcbi.1003578 (2014).
Woodruff, P. G. et al. Genome-wide profiling identifies epithelial cell genes associated with asthma and with treatment response to corticosteroids. Proc Natl Acad Sci USA 104, 15858–63, https://www.ncbi.nlm.nih.gov/pubmed/17898169, https://doi.org/10.1073/pnas.0707413104 (2007).
Pietrzyk, J. J. et al. Gene expression profiling in preterm infants: new aspects of bronchopulmonary dysplasia development. PLoS One 8, e78585, http://www.ncbi.nlm.nih.gov/pubmed/24194948, https://doi.org/10.1371/journal.pone.0078585 (2013).
Kim, W. J. et al. Comprehensive analysis of transcriptome sequencing data in the lung tissues of copd subjects. Int J Genomics 2015, 206937, https://www.ncbi.nlm.nih.gov/pubmed/25834810, https://doi.org/10.1155/2015/206937 (2015).
Liberzon, A. et al. Molecular signatures database (msigdb) 3.0. Bioinformatics 27, 1739–40, https://www.ncbi.nlm.nih.gov/pubmed/21546393, https://doi.org/10.1093/bioinformatics/btr260 (2011).
Shannon, P. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research 13, 2498–504 (2003).
Acknowledgements
We gratefully acknowledge the support of the Eunice Kennedy Shriver National Institute of Child Health & Human Development under award R01HD076140. We thank Diana Bianchi and Jill Maron for helpful discussions, and Noah Daniels for his valuable comments on an earlier draft of the manuscript. We also thank members of the Tufts Bioinformatics and Computational Biology group and the anonymous reviewers for feedback and comments on earlier versions of this work.
Author information
Authors and Affiliations
Contributions
J.P. and D.K.S. conceived the pathway centrality algorithm and wrote the manuscript. J.P., B.J.H., and D.K.S. designed the statistical method to assess significance of our findings and edited the manuscript. J.P. implemented the algorithm and ran the experiments. J.P. and D.K.S. analyzed the data and provided biological interpretation of the results. All authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Park, J., Hescott, B.J. & Slonim, D.K. Pathway centrality in protein interaction networks identifies putative functional mediating pathways in pulmonary disease. Sci Rep 9, 5863 (2019). https://doi.org/10.1038/s41598-019-42299-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-019-42299-3
This article is cited by
-
Network pharmacology of Withania somnifera against stress associated neurodegenerative diseases
Advances in Traditional Medicine (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.